ABSTRACT
Background/Aims: CircABCB10 function as an endogenous miRNA sponge plays an important role in various tumors. This experimental design was based on circABCB10 to explore the pathogenesis of non-small cell lung cancer (NSCLC). Methods: CircRNA microarray was used to examine circRNA expression profiles in lung cancer from 3 NSCLC patients and paired healthy lung tissues. The expression of circABCB10 and miR-584-5p was detected by q-PCR. CCK-8, colony formation, and transwell assays to study the circABCB10 effects on tumor cell growth and cell migration invasiveness. To validate downstream target genes of circABCB10 and miR-584-5p detected by luciferase reporter assays. RT-qPCR and Western blotting were used to study E2F5 expression. The tumor growth was detected by nude mice in vivo. Results: We analyzed the human circRNA expression profile in NSCLC tissues. CircABCB10 was identified as a circRNA that increased in NSCLC tissues. CircABCB10 was noticeably raised in NSCLC, and high circABCB10 expression was related to low survival in NSCLC patients. Silencing of circABCB10 suppressed non-small cell lung cancer cell migration, cell proliferation, and invasion.CircABCB10 can act as a sponge of miR-584-5p to up-regulate E2F5 expression level. E2F5 knockdown or overexpress of miR-584-5p gene reversed the circABCB10 who has carcinogenic effects. There was a negative correlation expression between the circABCB10 and miR-584-5p gene, and There was a positive relationship between the expression of circABCB10 and E2F5 in NSCLC tumors. Conclusion: CircABCB10 promoted the progression of NSCLC by modulating the miR-584-5p/E2F5 axis.
Abbreviation: NSCLC: non-small cell lung cancer; circ RNA: circular RNA; miRNA: micro RNA
Background
As a leading cause of death worldwide, cancer is increasing with population aging, as well as apparent cancer risk factors, such as smoking, obesity, lack of exercise, and changes in fertility patterns [Citation1,Citation2]. In underdeveloped countries, the incidence of lung cancer exceeds the incidence of breast cancer, colon cancer and prostate cancer as the number one killer of male cancer patients [Citation3]. Non-small cell lung cancer (NSCLC) accounts for 80-90% of lung cancer [Citation4,Citation5]. Most patients diagnosed with lung cancer when the symptoms is in an advanced stage. Generally, lung cancer deaths are 90% caused by distant metastasis [Citation6]. Studies have shown that if the diagnosis is made before metastasis, the 5-year survival rate is as high as 50%-70%. Once metastasis occurs, the five years survival rate will be 5%. Thus, early prevention of lung cancer is vital [Citation4].
In recent years, molecular diagnosis and targeted therapy of NSCLC have provided a new pathway to understand diagnosis and treatment of lung cancer [Citation7,Citation8]. The correlation between individualized gene expression of NSCLC and personalized tumor staging and patient prognosis has gradually become a study direction of lung cancer. Circular RNA (circ RNA) is a type of non-coding RNA that is abundant in mammals, which can participate in gene regulation in vivo [Citation9]. In the research progress of tumors, a large number of studies have found that circular RNA acts as a miRNA “cavernous body” to regulate downstream target genes [Citation10]. It has been found that multiple circRNAs function as a “cavernous body” of RNA to adsorb miRNA because of its miRNA binding site, thereby regulating miRNA inhibition by the mechanism of competitive endogenous RNAs (ceRNAs) [Citation11,Citation12]. CircABCB10 is a novel circRNA that overexpress in tumor tissues and displays oncological effect, which is disclosed in breast cancer [Citation13], clear cell renal cell carcinoma [Citation14], and ovarian cancer [Citation15]. Taken together, circABCB10 plays a significant role in tumor process.
For the past few years, miRNAs synergistically interacts with other transcription factors to form a complex gene expression regulatory network system [Citation16]. Researches have shown that the expression levels and types of miRNAs in human tumor cells and tumor tissues are significantly different from those in normal cells and tissues, suggesting that miRNAs may be involved in the occurrence and growth of tumors and serve as a tumor suppressor or tumor promoter [Citation17,Citation18]. Studies have found that miR-584-5p is closely related to tumorigenesis [Citation19–22]. For instance, miR-584-5p functions as an oncogenic miRNA by targeting KCNE2, and KCNE2 in turn mediates the effects of miR-584-5p on hepatocellular carcinoma HCC cell behaviors [Citation21]. In gastric cancer, miR‐584‐5p overexpression promotes apoptosis and inhibits proliferation but promotes apoptosis via targeting WW domain‐containing E3 ubiquitin protein ligase 1 [Citation20]. In addition, miR‐584‐5p suppressed cell growth, invasion, metastasis and angiogenesis by recruiting enhancer of zeste homolog 2 to facilitate the methylation of matrix metalloproteinase 14 in human neuroblastoma [Citation22]. However, there are little reports on the biological effects of miR-584-5p expression on NSCLC. In recent years, several studies have found that circRNA may play a role through adsorption miRNA [Citation23–26], For instance, in Pancreatic Ductal Adenocarcinoma (PDAC), circular RNA hsa_circRNA_0007334 promotes MMP7 and COL1A1 expression by functioning as a miRNA Sponge that competitively adsorb hsa-miR-144-3p and hsa-miR-577, so that promotes the development of PDAC [Citation24].
Based on the previous studies, it was hypothesized that circRNA might regulate the progression of NSCLC through miR-584-5p expression. The primary purpose of this research was to investigate the mechanism of action of circRNA-regulating NSCLC and provide a theoretical basis for finding new drug targets.
Materials and method
Research object
40 NSCLC samples and paired para-cancerous tissue specimens were collected from the clinical sample bank of Peking Union Medical College Hospital. The patient had not previously received chemotherapy or radiation therapy. The study was approved by the Peking Union Medical College Hospital research ethics committee, and all tissue samples get patients’ approved signed with written consent. All patients had unreceived radiotherapy or chemotherapy. The clinicopathological characteristics of patients (gender, age, pathological stage, pathological lymph node metastasis) are presented in .
Table 1. Clinicopathological characteristics of patients (n = 40)
Expression profiling of circRNAs
A circRNA chip (Arraystar Human circRNAs chip; Array Star, Rockville, MD, USA) containing 2566 probes specific for human circRNA splice sites was used. Seven pairs of NSCLC samples (tumor tissue and paired healthy tissue) were analyzed on a circRNA chip. The Gene Expression Omnibus accession number was GSE97332.
Cell culture and transfection
Human NSCLC cell lines (SPC-A1, H1975, HCC827, H1650, PC9, and A549) and 16HBE cells (human bronchial epithelial cells) were ordered from the Shanghai Center for Life Sciences Cell Center. The human NSCLC cell line A549 was cultured in DMEM medium with 10% fetal bovine serum, while other cells mentioned above were cultured in RPMI 1640 basic medium with 10% fetal bovine serum.
si-circABCB10, si-E2F5, miR-584-5p agomir, miR-584-5p mimetic and control vector (GenePharma, China) were individually transfected into target cells with Lipofectamine 2000 reagen. The sequence of si-circABCB10 was as follows: 5ʹ-TGGTGAAATAAATATGCGCAA-3ʹ. The cDNA sequence of human circABCB10 was cloned into the vector of pcD-ciR to construct a circABCB10 overexpression plasmid.
Cell proliferation assay
The transfected target cells were split into 96-well plates, and cell growth was measured every 24 hours. The absorbance values were finally determined at 450 nm with a Biotek microplate reader (ELx 800, Biotek, USA).
Cell clone formation assay
The transfected cells were seeded into a 6-well plate. After a period of two weeks culture, the cells were treated with methanol fixation, stained with crystal violet(0.1%), and then captured all the colonies and counted.
Transwell migration and invasion assay
The upper basement membrane of the Transwell chamber was pre-coated with 20 μg Matrigel and cultured overnight in a 24-well plate. The cell suspension was transferred into the first chamber, and the lower chamber was filled with medium. After 12 hours of culture, it was washed for three times with PBS, fixed with 90% of the formaldehyde, and then stained in the crystal violet solution. The photograph was taken under an inverted microscope. In the cell migration experiment, except the upper part of the well chamber, the rest operation was consistent with the invasion experiment.
Dual-luciferase reporter gene assay
The wild type or mutant sequence of the E2F5 and circABCB10 3ʹ-UTR (3ʹ-untranslated region) was inserted into the vector of pmirGLO. Cells were co-transfected with miR-584-5p inhibitors or mimetics, and these reporter plasmids, respectively. After two days of co-transfection and co-expression, luciferase activity was detected by a dual luciferase assay system (Promega).
Animal research
Twelve 6-week-old female BALB/C nude mice were randomly divided into two groups. Each mouse was subcutaneously injected with A549 cells to establish a mouse xenotransplantation model. On 9th day, tumor-injected mice were injected with cholesterol-conjugated si-circABCB10, miR-584-5p agomir, and negative control (GenePharma). Two weeks later, tumor size was measured twice a week. All nude mice were sacrificed after two weeks, and tumors were dissected for qPCR or western blot analysis experiments. The study was conducted in strict accordance with the National Institutes of Health Laboratory Animal Care and Use Guidelines.
Western blot
The transfected cells were collected, total protein was extracted, and the protein concentration was quantified with a kit of BCA Protein Kit. Anti-GAPDH antibody (1:1000 dilution, Abcam, UK) and the anti-E2F5 antibody (1:1000, Proteintech, USA) were added and mixed and incubated at 4°C overnight. After that, it was incubated with 1:5,000 labeled anti-rabbit secondary antibody (Abcam, UK) for 1 h. Western blot analysis Specific experimental methods were performed concerning the literature [Citation27].
Statistical method
The monitoring data were analyzed by SPSS19.0 statistical software. The results of data analysis were shown as mean ± standard deviation (mean ±SD). Multigroup data analysis was based on one-way ANOVA. LSD test was used for subsequent analysis. P < 0.05 meant the difference was significant.
Results
Dysregulated circRNAs in HCC tissues
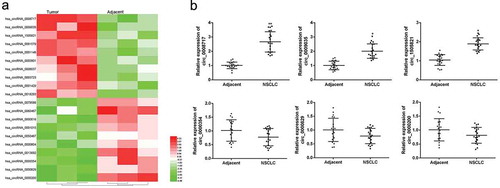
First, tissue samples from 3 pairs of matched patients were selected for circRNA microarray detection. By analyzing the intensity of expression within the HCC tumor and non-tumor groups, the 20 most abnormal circular RNAs in HCC tumor tissues were shown in ). Next, the expression of three major altered circRNAs in HCC and the expression in non-tumor tissue samples was analyzed, as shown in ). Compared with the matched control group, circRNA hsa_circ_0008717 (circBase, www.circbase.org/) was significantly increased in HCC tumor tissue (p < 0.05). Hsa_circ_0008717 was located at chr1: 229665945–229678118, and the gene symbol ABCB10 was 724 in length and was identified as circ-ABCB10. Therefore, we were interested in investigating the biological effects of circ ABCB10 in HCC.
Figure 1. Disordered circRNA in HCC tumor tissue. (a) Heat map analysis results. (b) Expression of six higher levels of circRNA in 20 HCC tumor tissues and adjacent non-tumor tissues. * p < 0.05
CircABCB10 was significantly overexpressed and predicted poor prognosis in NSCLC
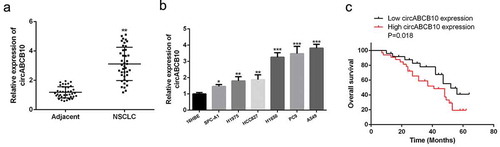
As shown in ), CircABCB10 expression level was elevated in NSCLC tissues with a comparison to that in paired adjacent normal tissues (), p < 0.01). The expression yield of circABCB10 was significantly enhanced in NSCLC cell lines (SPC-A1, H1975, HCC827, H1650, PC9 and A549) with a comparison to that in human bronchial epithelial cells (16HBE) ()). Patients were organized into high expression group (n = 20) and low expression group (n = 20) using the median level (circABCB10) as the cutoff value. As shown in ), with a comparison to the circABCB10 low expression group, the overall survival rate of the circABCB10 high expression group significantly lower (P < 0.05). These data indicated that circABCB10 had a potential carcinogenic effect in NSCLC.
Figure 2. Expression levels of circABCB10 in NSCLC. (a) CircABCB10 expression levels in NSCLC tissues and matched adjacent normal tissues (n = 40). (b) CircABCB10 expression levels in NSCLC cell lines and normal liver cell lines. (c) Survival curve of NSCLC patients with low (n = 20) or high (n = 20) CircABCB10 expression.** P < 0.01, *** P < 0.001
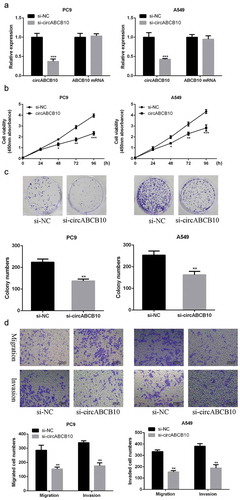
Silencing of circABCB10 inhibited NSCLC growth in vitro
As indicated in ), the expression level of circABCB10 was obviously decreased after siRNA transfection, indicating successful transfection (P < 0.001). As noted in ), the cell growth rate of the circABCB10-silencing group was significantly decreased in A549 and PC9 cells with a comparison to that of the control group in a time-dependent manner (48 h, P < 0.05; 72 h, P < 0.01; 96 h, P < 0.001). As shown in ), compared with the control group, the number of colony formation in the circABCB10-silencing group was obviously decreased (P < 0.01). As demonstrated by ), with a comparison to the control group, the number of cell invasion and migration was significantly reduced in the circABCB10-silencing group (P < 0.01). The above results indicated that circABCB10 exerted a carcinogenic effect and promoted the metastasis and growth of NSCLC cells.
Figure 3. CircABCB10 promoted the malignant behavior of NSCLC cells. (a) Th expression levels of CircABCB10. (b) Effect of circABCB10 expression on cell viability. (c) Effect of circABCB10 on cell proliferation. (d) Effect of circABCB10 on cell migration and invasion. *** p < 0.001, ** p < 0.01, *p < 0.05
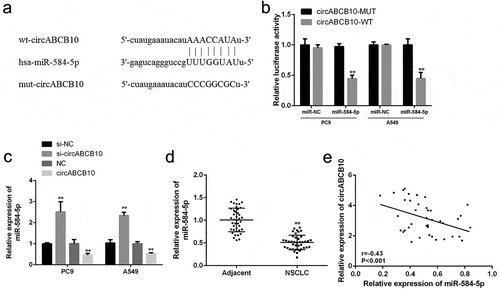
CircABCB10 served as a sponge of miR-584-5p
In addition, via bioinformatics, miR-584-5p was identified as a potential target for CircABCB10 ()). In order to validate the anticipated results, luciferase reporter assays were performed using the WT-CircABCB10 or mutant (mut)-CircABCB10 luciferase reporter plasmid. As shown in ), ectopic expression of miR-584-5p significantly suppressed the luciferase bioactivity of WT-CircABCB10 but had no noticeable effect on the luciferase bioactivity of mut-CircABCB10. Furthermore, overexpression of circABCB10 was significant compared with that of the normal group ()). As indicated in ), miR-584-5p was significantly down-regulated in NSCLC tissues compared with that in normal tissues (P < 0.01). In addition, a direct negative correlation between circABCB10 expression and miR-584-5p levels in NSCLC tissues was found ()). Taken together, these results suggest that circabcb10 may play a role through miR-584-5p.
Figure 4. MiR-584-5p was a target gene for circABCB101. (a) Bioinformatics analysis revealed predicted binding sites between circABCB101 and miR-584-5p. (b) Analysis of luciferase activity in PC9 and A549 cells co-transfected with miR-584-5p mimic and pmirgLO-circABCB10-WT or pmirgLO-circABCB10-Mut vector. (c) Expression levels of miR-584-5p. (d) Expression levels of miR-584-5p in adjacent normal tissues and NSCLC tissues (n = 40). (e) Correlation analysis of circabcb10 and mir-584-5p in NSCLC tissues (n = 40) (r = −0.43, P < 0.001). ** P < 0.01, *** P < 0.001
CircABCB10 sponged miR-584-5p to upregulate E2F5 expression
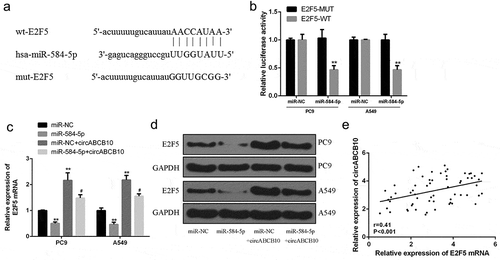
Furthermore, E2F5 was identified as a potential target for miR-584-5p according to bioinformatics ()). Luciferase reporter assays were performed using the WT-miR-584-5p or mutant (Mut)-miR-584-5p luciferase reporter plasmid to validate the predicted results. As shown in ), ectopic expression of E2F5 significantly inhibited the luciferase activity of WT-miR-584-5p but had no significant effect on the luciferase activity of mut-miR-584-5p. In addition, overexpression of miR-584-5p reduced E2F5 mRNA and protein expression levels compared with that in the control group. Overexpression of CircABCB10 increased E2F5 expression levels, while co-transfection with CircABCB10 and miR-584-5p reversed the effect of CircABCB10 and miR-584-5p on E2F5 mRNA and protein expression levels (). Furthermore, circABCB10 expression was positively correlated with E2F5 expression in NSCLC tissues, In addition, the expression of circabcb10 was positively correlated with the expression of E2F5 in NSCLC tissues (P < 0.001) ()). These data indicated that circABCB10 enhanced the expression level of the oncogene E2F5 by acting as a sponge of miR-584-5p.
Figure 5. CircABCB10 up-regulated E2F5 by sponging miR-584-5p. (a) Putative binding sites for miR-584-5p and E2F5 3ʹ-UTR. (b) The luciferase activity with miR-584-5p mimic and pmirgLO-E2F5 3ʹ-UTR-WT or pmirgLO-E2F5 3ʹ-UTR-Mut vector. (c) mRNA expression levels of E2F5. (d) Protein expression levels of E2F5. (e) Pearson correlation analysis of circABCB10 and E2F5 in NSCLC tissues (n = 40).* vs control group, ** P < 0.01; #vs miR-584-5p mimics group, #P < 0.05
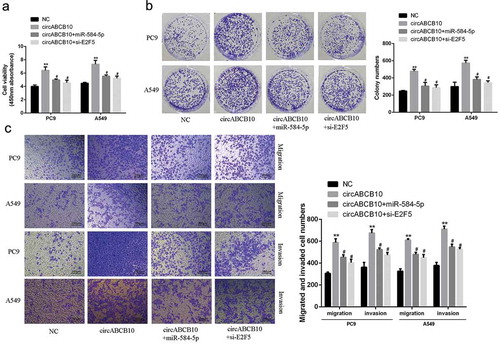
E2F5 silencing or miR-584-5p overexpression effectively reversed circABCB10-induced promotion of NSCLC progression
The results were shown in ). Compared with the control group, overexpression of circABCB10 significantly increased the proliferation rate of NSCLC cells, but miR-584-5p overexpression and E2F5 silencing significantly reversed the proliferation, counteracting the stimulating effect of circABCB10 on proliferation. The results were shown in . Compared with the control group, Overexpression of circABCB10 dramatically increased the number of colony formation, cell invasion and migration of NSCLC cells, but miR-584-5p overexpression and E2F5 silencing significantly reversed the effect of circABCB10 on cell invasion and migration. These results demonstrated that E2F5 silencing or miR-584-5p overexpression reversed the progression of circABCB10 induced NSCLC cells.
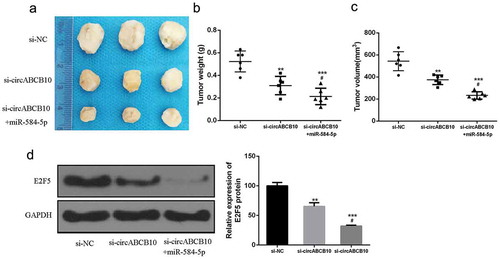
circABCB10 promoted NSCLC progression in vivo by regulating the miR-584-5p/E2F5 axis
The effect of circABCB10 on the progression of NSCLC in vivo was further determined. The results showed that compared with that in the control group, circABCB10 knockdown inhibited tumor weight and tumor volume, while miR-584-5p overexpression also enhanced the inhibition of si-circABCB10 (). In addition, as shown in ), circABCB10 knockdown was able to inhibit E2F5 expression compared with that in the control group, while miR-584-5p overexpression further inhibited E2F5 expression. These data suggested that circABCB10 silencing can inhibit NSCLC progression by modulating the miR-584-5p/E2F5 axis.
Figure 7. Silencing of circABCB10 inhibited NSCLC growth by modulating the miR-584-5p/E2F5 axis. (a) Representative images of subcutaneous tumors from three groups. (b, c) Tumor volume and tumor weight of nude mice. (d) Protein expression level of E2F5. Compared with si-circABCB10, #P < 0.05, ** p < 0.01, compared with p-NC, *** p < 0.001)
Discussion
Non-small cell lung cancer (NSCLC), is the most common type of lung cancer [Citation28]. According to related reports, the five-year survival rate of advanced NSCLC is only 16% [Citation29]. However, despite the great efforts of scholars all over the world, the research on the mechanism of the development of NSCLC is still not perfect. Finding potential regulatory targets and mechanisms of NSCLC is still a significant problem that plagues the world. Through the research and discussion on the possible regulatory targets and mechanism of action of NSCLC, it is expected to provide new ideas and treatment plans for basic research and clinical treatment of NSCLC.
Increasing evidences indicate that circRNAs function as microRNA sponge as well as transcription regulators to regulate gene expressions, which are involved in cancer [Citation30,Citation31]. For example, Circ_0067934 can inhibit the invasion and migration of NSCLC cells [Citation32]. CircABCB10 is another up-and-coming circular RNA that has received increasing attention. It has been found that circABCB10 promoted cancer progression by sponge activity of miR-203 and upregulation of Bmi-1 expression in osteosarcoma [Citation33]. In breast cancer, circABCB10 promotes breast cancer proliferation and progression through sponging miR-1271 [Citation13]. In addition, CircABCB10 promotes nonsmall cell lung cancer cell proliferation and migration by sponging miR‐1252 to increase FOXR2 synthase expression and consequently promote NSCLC progression [Citation34]. These results suggest that circABCB10 as a endogenous miRNA sponge plays a significant role in tumor process.
As a target of circRNA, miRNA is the most widely studied non-coding RNA, which can regulate cell proliferation, differentiation, and apoptosis by degrading target mRNA or inhibiting its translation and participate in individual development, body metabolism, and other processes [Citation35]. It has been found that miR-584-5p plays an anti-tumor role in gastric cancer by inhibiting the transcription of the WW domain [Citation20]. We screened miR-584-5p as a target gene for circABCB10 by the database. Moreover, circABCB10 regulated its expression by targeting the 3ʹUTR of the miR-584-5p gene. Also, circABCB10 knockdown or overexpression can ectopically express the expression level of miR-584-5p. While miR-584-5p was down-regulated in NSCLC tissues, and miR-584-5p between circABCB10 had a negative correlation expression in NSCLC tissues.
The transcription factor E2F5 binds to its upstream and downstream target gene promoters, thereby altering target gene expression [Citation36]. Its most familiar function is to regulate cell cycle progression, DNA synthesis, and cell proliferation and apoptosis [Citation37]. Recent studies have shown that E2F5 expression levels are reduced in some tumors such as breast cancer, uterine tumors, kidney cancer, and gastric cancer [Citation38,Citation39]. In this study found that E2F5 was a potential target for miR-584-5p and that miR-584-5p regulated by targeting the 3ʹUTR of the E2F5 gene. In addition, overexpression of circABCB10 increased E2F5 mRNA and protein expression levels, while circABCB10 and miR-584-5p co-transfection was able to reverse the effects of CircABCB10 and miR-584-5p on E2F5 mRNA and protein expression levels. CircABCB10 expression was positively correlated with E2F5 expression in NSCLC tissues. In addition, overexpression of circABCB10 significantly increased the proliferation rate of NSCLC cells and enhanced the invasion and migration ability of cells. However, miR-584-5p overexpression and E2F5 silencing can substantially reverse the proliferation of circABCB10 and the promotion of cell invasion and migration. These indicated that circABCB10 upregulated the expression level of oncogene E2F5 by acting as a sponge of miR-584-5p. Therefore, circRNA-ABCB10 may serve as a promising predictive biomarker and therapeutic target for NSCLC patients.
Conclusion
CircABCB10 promoted proliferation, invasion, and metastasis of NSCLC by inhibiting miR-584-5p and increasing E2F5, suggesting that circABCB10 may be a potential oncogene of glioma. It could provide experimental evidence for the clinical prognosis and further targeted intervention of this tumor.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Peking Union Medical College Hospital. The research has been carried out in accordance with the World Medical Association Declaration of Helsinki. All patients and healthy volunteers provided written informed consent prior to their inclusion within the study. The study was conducted in strict accordance with the National Institutes of Health Laboratory Animal Care and Use Guidelines.
Consent for publication
All authors have read and approved the final manuscript.
Authors contribution
Hongsheng Liu supervised the whole study, data analysis, manuscript preparation
Dongjie Ma, Yingzhi Qin, Cheng Huang, Yeye Chen, Zhijun Han, Xiaoyun Zhou data collection and analysis, manuscript preparation
Data availability
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Siegel R, Ms DNM, Dvm AJ. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29.
- Siegel R, Ward E, Brawley O, et al. Cancer statistics, 2011. CA Cancer J Clin. 2011;61(4):212–236.
- Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359(13):1367–1380.
- Rizvi NA, Hellmann MD, Snyder A, et al. Mutational landscape determines sensitivity to PD-1 blockade in non–small cell lung cancer. Science. 2015;348(6230):124–128.
- Shaw AT, Yeap BY, Minokenudson M, et al. Clinical features and outcome of patients with non–small-cell lung cancer who harbor EML4-ALK. J Clin Oncol Off J Am Soc Clin Oncol. 2009;27(26):4247.
- Hung JJ, Jeng WJ, Hsu WH, et al. Predictors of death, local recurrence, and distant metastasis in completely resected pathological stage-I non-small-cell lung cancer. J Thorac Oncol. 2012;7(7):1115–1123.
- Mender I, Laranger R, Luitel K, et al. Telomerase-mediated strategy for overcoming non–small cell lung cancer targeted therapy and chemotherapy resistance. Neoplasia. 2018;20(8):826–837.
- Papadimitrakopoulou V, Lee JJ, Wistuba II, et al. The BATTLE-2 study: a biomarker-integrated targeted therapy study in previously treated patients with advanced non–small-cell lung cancer. J Clin Oncol Off J Am Soc Clin Oncol. 2016;34(30):3638–3647.
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461.
- Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015c;25(8):981–984.
- Bachmayrheyda A, Reiner AT, Auer K, et al. Correlation of circular RNA abundance with proliferation – exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015;5(8057):8057.
- Li P, Chen S, Chen H, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015b;444:132–136.
- Liang HF, Zhang XZ, Liu BG, et al. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res. 2017;7(7):1566–1576.
- Huang Y, Zhang Y, Jia L, et al. Circular RNA ABCB10 promotes tumor progression and correlates with pejorative prognosis in clear cell renal cell carcinoma. Int J Biol Markers. 2019;34(2):176–183.
- Chen Y, Ye X, Xia X, et al. Circular RNA ABCB10 correlates with advanced clinicopathological features and unfavorable survival, and promotes cell proliferation while reduces cell apoptosis in epithelial ovarian cancer. Cancer Biomark. 2019;26(2):151–161.
- Zhong X, Coukos G, Zhang L. miRNAs in human cancer. J Pathol. 2015;223(2):102–115.
- Foekens JA, Sieuwerts AM, Smid M, et al. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc Natl Acad Sci U S A. 2008;105(35):13021–13026.
- Hergenreider E, Heydt S, Tréguer K, et al. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat Cell Biol. 2012;14(3):249.
- Guo T, Zheng C, Wang Z, et al. miR‑584‑5p regulates migration and invasion in non‑small cell lung cancer cell lines through regulation of MMP‑14. Mol Med Rep. 2019;19(3):1747–1752.
- Li Q, Li Z, Wei S, et al. Overexpression of miR-584-5p inhibits proliferation and induces apoptosis by targeting WW domain-containing E3 ubiquitin protein ligase 1 in gastric cancer. J Exp Clin Cancer Res. 2017;36(1):59.
- Wei H, Wang J, Xu Z, et al. miR-584-5p regulates hepatocellular carcinoma cell migration and invasion through targeting KCNE2. Mol Genet Genomic Med. 2019 Jun;7(6):e702.
- Xiang X, Mei H, Qu H, et al. miRNA-584-5p exerts tumor suppressive functions in human neuroblastoma through repressing transcription of matrix metalloproteinase 14. BBA Mole Basis Dis. 2015;1852(9):1743–1754.
- Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151.
- Wang Y, Zhang J, Li J, et al. CircRNA_014511 affects the radiosensitivity of bone marrow mesenchymal stem cells by binding to miR-29b-2-5p. Bosn J Basic Med Sci. 2019;19(2):155–163.
- Xie H, Ren X, Xin S, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7(18):26680–26691.
- Yang J, Cong X, Ren M, et al. Circular RNA hsa_circRNA_0007334 is predicted to promote MMP7 and COL1A1 expression by functioning as a miRNA sponge in pancreatic ductal adenocarcinoma. J Oncol. 2019;2019:7630894.
- Croppo GP, Visvesvara GS, Leitch GJ, et al. Western blot and immunofluorescence analysis of a human isolate of Encephalitozoon cuniculi established in culture from the urine of a patient with AIDS. J Parasitol. 1997;83(1):66–69.
- Han SW, Kim TY, Hwang PG, et al. Predictive and prognostic impact of epidermal growth factor receptor mutation in non–small-cell lung cancer patients treated with Gefitinib. J clin oncol. 2005;23(11):2493–2501.
- Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362(25):2380.
- Li J, Yang J, Zhou P, et al. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am J Cancer Res. 2015a;5(2):472–480.
- Xu L, Zhang M, Zheng X, et al. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2017;143(1):17–27.
- Wang J, Li H. CircRNA circ_0067934 silencing inhibits the proliferation, migration and invasion of NSCLC cells and correlates with unfavorable prognosis in NSCLC. Eur Rev Med Pharmacol Sci. 2018;22:3053–3060.
- Xiang Z, Natino D, Qin Z, et al. Identification and functional characterization of circRNA-0008717 as an oncogene in osteosarcoma through sponging miR-203. Oncotarget. 2018;9(32):22288–22300.
- Tian X, Zhang L, Jiao Y, et al. CircABCB10 promotes nonsmall cell lung cancer cell proliferation and migration by regulating the miR-1252/FOXR2 axis. J Cell Biochem. 2018;120(3):3765–3772.
- Rong D, Sun H, Li Z, et al. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. 2017;8(42):73271–73281.
- Polanowska J, Le CL, Orsetti B, et al. Human E2F5 gene is oncogenic in primary rodent cells and is amplified in human breast tumors. Genes Chromosomes Cancer. 2000;28(1):126–130.
- Tian H, Hou L, Xiong YM, et al. miR-132 targeting E2F5 suppresses cell proliferation, invasion, migration in ovarian cancer cells. Am J Transl Res. 2016;8(3):1492.
- Lu GF, Sun YL, An SL, et al. MicroRNA-34a targets FMNL2 and E2F5 and suppresses the progression of colorectal cancer. Exp Mol Pathol. 2015;99(1):173–179.
- Yao YL, Wu XY, Wu JH, et al. Effects of MicroRNA-106 on proliferation of gastric cancer cell through regulating p21 and E2F5. Asian Pac J Cancer Prev. 2013;14(5):2839–2843.