ABSTRACT
Long noncoding RNAs (lncRNAs) are key players in cancer progression. However, the function of lncRNA NNT-AS1 on glioma is unclear. In the present study, a total of 73 tumor tissues and matched adjacent non-tumor tissues were collected, and glioma cell lines were cultured in vitro. mRNA expression was tested using RT-qPCR. The protein expression level was determined using the western blot assay, cell proliferation was measured using the CCK-8 and BrdU proliferation assay, and the cell cycle, cell migration and invasion were determined using flow cytometry analysis, the wound healing assay and transwell, respectively. The results showed that lncNNT-AS1 is significantly up-regulated during the early stages of glioma. In particular, high levels of NNT-AS1 are observed in glioma cell lines compared to human astrocyte (HA) cells. Furthermore, the inhibition of lnc-NNT-AS1 by siRNA interfere attenuates the cell viability, proliferation, migration and invasion of glioma cell lines. Mechanistically, the inhibition of NNT-AS1 directly interacted with miRNA-494-3p, and positively regulated the downstream target PRMT1 in vitro. Further study proved that the overexpression of miRNA-494-3p and the inhibition of PRMT1 could attenuate both glioma cell proliferation and metastases. Collectively, our results indicated that the miR-494-3p-PRMT1 axis is involved the tumor-suppressive effects of NNT-AS1 inhibition, which sheds new light on lncRNA-directed diagnostics and the therapeutics of glioma.
KEYWORDS:
Introduction
Glioma is the leading cause of brain cancer related-deaths in the world [Citation1]. Gliomas with extended life expectancy showed differences in cell types, location, and severity, such as astrocytoma, brain stem glioma, ependymoma, oligodendroglioma, mixed glioma and optic nerve glioma. Although great achievements have been made in the diagnosis and targeted therapy of glioma, the prognosis of patients is still poor [Citation2–Citation5]. Therefore, it is crucial to explore the relative molecular mechanism involved in the cell proliferation and metastasis of gliomas.
Long non-coding RNAs (lncRNAs) are unable to encode proteins, but are longer than 200 nucleotides. Studies have reported that lncRNAs are associated with tumorigenesis, including cell proliferation, and migration. For instance, the lncRNA NNT-AS1 (transcribed in the opposite direction to nicotinamide nucleotide transhydrogenase) was found to exert an oncogenic role in gastric cancer, breast cancer, ovarian cancer, small cell lung cancer and glioma [Citation6–Citation9]. Recently, miRNAs have been widely reported to interact with lncRNAs, forming the ceRNA pathway [Citation5]. Zheng et al. found that the lncRNA AGAP2-AS1 could promote cell proliferation by sponging miR-15a/b-5p and activating Wnt/β-catenin [Citation10]. This suggests that lncRNA interacts with miRNA to regulate cell proliferation and invasion in glioblastoma. However, although NNT-AS1 was predicted to be strongly expressed in glioma tissue, its role in glioma cell proliferation still remains unknown.
In the present study, we showed that NNT-AS1 expression was up-regulated in human glioma tissues and different glioma cells lines. Correlation analysis revealed that the expression of NNT-AS1 was correlated with early tumor stage. NNT-AS1 knock-down could significantly attenuate glioma cell proliferation and invasion ability. Furthermore, bioinformatics analysis and mechanism experiments were used to detect miRNAs which interact with NNT-AS1 in glioma. In the mechanism study, our data indicated that NNT-AS1 could act as a sponge of miR-494-3p/arginine N-methyltransferase 1 (PRMT1) protein during glioma progression.
Materials and methods
Sample collection
A total of 73 tumor tissues and matched adjacent non-tumor tissues were collected from patients who were diagnosed with glioma at Shunde Hospital, Southern Medical University (The First People’s Hospital of Shunde Foshan). Consecutive enrollment samples were all collected in 2018. The diagnosis was confirmed by pathology, and according to the WHO grade. Patients were divided into two groups: I+II (n = 41) and III+IV (n = 32) [Citation11]. The last group was the most aggressive and the fastest-growing glioma type. Patients did not receive radiation or chemotherapy before surgery. Tissues were immediately frozen in liquid nitrogen and kept at −80°C before use. Tissues were used for qRT-PCR analysis. This study was approved by the Ethics Committees of Shunde Hospital. All patients gave their written informed consent to participate in this study, and the data did not contain any information which could identify the patients.
Cell culture
The glioma cell lines A172, LN229, and U251 (Shanghai Institute of Life Sciences Cell Resource Center, Shanghai, China) were grown in DMEM media (Hyclone), with 10% fetal calf serum, 100 U/mL penicillin, and 0.1 mg/mL streptomycin, at 37°C in a humidified CO2 incubator (95% air, 5% CO2) [Citation12,Citation13].
The miR-494-3p mimics were purchased from RiboBio (Guangzhou, China) [Citation14]. Cell transfections were performed by lipofectamine 2000 (Invitrogen) [Citation15]. Briefly, cells were plated in 6-well plates to 80% confluence. Small interfering RNA (siRNA) sequences for NNT-AS1(si-NNT-AS1) or its negative control (si-NC), miR-494-3p or miRNA mimic negative control, miR-494-3p inhibitor or inhibitor negative control, PRMT1 expression vector (PRMT1 onexpre) or the empty pcDNA3.1, were added to 50 μl OptiMEM medium, and 4 μL Lipofectamine 2000 in 50 μl Opti-MEM medium, and then the siRNAs were mixed with Lipofectamine 2000 for 20 min at room temperature. The mixture was added to 2 mL cell culture medium and cells were incubated for 6 h before replacing the medium. Total RNA for qRT-PCR and protein samples for western blot analysis were prepared 48 h after transfection. All experimental procedures were performed according to the manufacturer’s instructions.
RNA extraction and reverse transcription and quantitative real-time PCR (qRT-PCR)
Total RNA from tissues and cells was isolated using TRIZOL (Invitrogen) in accordance with the manufacturer’s instructions [Citation16]. mRNA and miRNA were initially reverse-transcribed to cDNA using the Revert Aid First Strand cDNA Synthesis Kit (Thermo, USA). Then, the cDNA was amplified using iQ SYBR Green (Bio-Rad, USA), and β-actin was used as a control. The xonditions used in qRT-PCR were as follows: 30 cycles consisting of denaturation at 95°C for 30 s, annealing at 56°C for 30 s, and extension at 72°C for 30 s. Lastly, relative fold expressions were calculated using the 2-ddCt method. In addition, the miR-494-3p expression was obtained by TaqMan miRNA-assay kit (Applied Biosystems, USA). Experiments were all performed 3 times.
siRNA and primer sequences are listed as follows: PRMT1 siRNA: 5ʹ-CCA UCG ACC UGG ACUUCAATT-3ʹ, 5ʹ-UUGAAGUCCAGGUCGAUGGTT-3ʹ; PRMT1 primer: Forward 5ʹ-TTGACTCCTATGCCCACT-3ʹ, Reverse: 5ʹ-CCACATCCAGCACCACC-3ʹ; siNNT-AS1: 5ʹ-GAAAAGAAAAAGAAGCUUATT-3ʹ; NNT-AS1 primer: Forward: 5ʹ-CTGGAATCCC TGCTACTCAGGA-3ʹ, Reverse: 5ʹ-GCCATGTGATATGCCTGCTC-3ʹ; miR-494-3p mimic primer: 5ʹ-UGAAACAUACACGGGAAACCUC-3ʹ and 5ʹ-GGUUUCCCGUGUAUGU UUCAUU-3ʹ and anti-miR-494-3p 5ʹ-GAGGUUUCCCGUGUAUGUUUCA-3ʹ.
Luciferase activity assay
Cells were co-transfected with miR-494-3p inhibitors or control and the pGL3/Luc-PRMT1 3ʹUTR or the mutant 3ʹUTR-PRMT1, together with the controls. 48 h after transfection, the cells were lysed using RIPA buffer, and luciferase intensity was measured using a fluorescence spectrophotometer. The 3ʹ UTR of PRMT1 and the mutant 3ʹUTR of PRMT1 was amplified using the pGL3/Luc-PRMT1, and then cloned downstream of the pGL3/Luc vector.
CCK-8 assay
Cells were selected in the logarithmic growth phase and seeded in 96-well plates at 1 × 103 cells/well for 1d, 2d, 3d, 4d and 5d. Ten μL CCK8 solution (Dojindo, Japan) was added and the plate was incubated at 37°C for 1 h in the dark. The absorbance was read at 450 nm using a microplate reader (Bio-Rad Laboratories, USA) [Citation17].
BrdU proliferation assay
BrdU (10 μg/mL) was added 2 h before section fixation. Sections were first treated with 2 N HCl at 37°C for 30 min to denature the DNA and then neutralized in 0.1 M borate buffer (pH = 8.5) for 10 min. Sections were then incubated for 1 h at room temperature in 5% (w/v) BSA, and 0.3% Triton X-100. After blocking, the sections were treated with a primary anti-mouse BrdU monoclonal antibody (1:400, overnight, 4°C, Sigma Aldrich Canada). After that, sections were then incubated with an FITC-labeled secondary antibody. Finally, the cell nucleus was counterstained with PI (Millipore, USA) for 10 min. The number of proliferative cells (BrdU-positive) was counted in three random fields/slides [Citation18].
Western blotting`
Proteins were extracted using RIPA buffer for reaction on ice for 30 min (Invitrogen, USA). Supernatant was collected and protein concentration was determined using a BCA assay kit (Takara, Japan). Ten μg/well of protein was then separated by 10% SDS-PAGE and transferred to PVDF membranes (Roche, USA). After blocking with 5% milk, the membranes were subsequently incubated with primary antibodies for PRMT1 (Abcam, USA), β-actin (Proteintech, USA), and Vimentin (Abcam, USA) at 4°C overnight. The blots were then treated by Horseradish peroxidase-conjugated secondary antibodies at room temperature for 2 h. Finally, the blots were visualized by using the ECL detection reagent (Millipore, USA) and an automated chemiluminescence image analysis system (Biogene, UK). Densitometric analysis was performed using the Image J software.
Flow cytometry analysis for cell cycle and apoptosis
Annexin V and propidium iodide (PI) double staining was used for cellular apoptosis. For cell cycle detection, the glioma cells were treated with siNNT-AS1 for 6 h, and then the medium was changed to DMEM medium (Hyclone), containing 10% fetal calf serum. Three days after siRNA treatment, cell proliferation were detected using flow cytometry analysis. Cells were collected, rinsed with PBS and fixed with 75% pre-cold ethanol at −20°C for 24 h. Cells were then washed with PBS twice and stained with propidium iodide (PI) by the cycle test plus DNA reagent kit (BD, USA) at room temperature for 1 min. The proportion of cells in S phase was determined by flow cytometry (Becton Dickinson, USA) using the Cell Quest Modfit software [Citation19].
Wound healing assay
Cell migration was determined by using the wound healing assay. In brief, U251 cells were cultured in 24-well plates. Six hours after siRNA transfection, DM4 (M01781-NPG, inhibits cell cycle) was used to inhibit cell division. Then, a wound was scratched with a 200 μL pipette tip. The detached cells were removed before incubating in DMEM with 10% serum. The image of the scratch was photographed after wounding 0 and 72 h later. Lastly, the healing rate was calculated as a percentage of healing area relative to the initial wound area [Citation20].
Invasion assay by transwell
Cells were seeded in the upper chamber of the 0.6 μM polycarbonate membrane-coated Matrigel (Corning, USA) in serum-free DMEM medium and incubated overnight at 37°C. Medium containing DMEM, and DMEM with 10% FBS, were then added to the lower chamber. Cells were incubated under normal conditions for 24 h. Non-invaded cells on the upper surface of the polycarbonate membrane were removed. Cells at the low-side of the membrane were fixed, stained, washed, and then counted under an optical microscope [Citation21].
Dual luciferase activity assay
All cells were seeded in 24-well plates. For the NNT-AS1 miR-494-3p interaction, cells were co-transfected with wild type (wt), and miR-494-3p mimics, or miR-negative control (miR-NC). Luciferase assays were performed using the Dual Luciferase Reporter Assay System (Promega, USA) 24 h after transfection [Citation22].
Statistical analysis
For the correlation test, the relevance between miR-494-3p and NNT-AS1 was detected using the Pearson correlation coefficient method. Experiments were repeated three times. The difference between two groups was compared by the two-tailed unpaired student’s t test. Moreover, the significance test of the mean difference between 3 or more groups was analyzed by one-way ANOVA, followed by the Bonferroni test. Differences were considered to be statistically significant when P < 0.05.
Results
NNT-AS1 was found to be up-regulated in glioma tissues and glioma cell lines
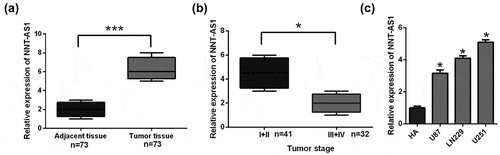
To detect the expression of NNT-AS1 in glioma, the NNT-AS1 expression levels were determined in 73 pairs of glioma tissue samples and adjacent non-tumor tissues. NNT-AS1 expression was higher (P < 0.01) in 70% (51/73) of glioma tissues compared with normal tissues ()). NNT-AS1 expression levels in glioma are significantly related to grade I and II levels of glioma ()). Moreover, high levels of NNT-AS1 were detected in U87, LN229, and U251 ()) compared with normal HA cells. Collectively, our results proved that NNT-AS1 is overexpressed in glioma tissues and glioma cell lines. However, whether the inhibition of NNT-AS1 could affect glioma cell biologic activity needs further investigation.
Inhibition of NNT-AS1 suppressed glioma cell proliferation, migration, invasion and apoptosis
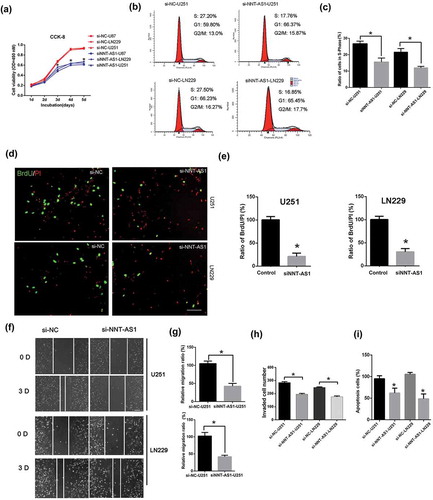
The siRNA of NNT-AS1 was used to knockdown the expression of NNT-AS1. Cell viability was significantly decreased in the NNT-AS1 siRNA-treated groups compared to the NNT-AS1-siRNA negative control groups ()). The cell cycle and cell proliferation assay demonstrated that NNT-AS1 knockdown decreased the ratio of cells in S-Phase in U251 and LN229 cells ()). In addition, cells transfected with NNT-AS1 siRNA showed a lower migratory ability than the NNT-AS1 siRNA negative control group in U251 and LN229 cells ()). Furthermore, the invasion of U251 and LN229 cells was significantly inhibited by NNT-AS1 siRNA ()). Lastly, cell apoptosis was also detected, and the NNT-AS1 siRNA significantly inhibited cell apoptosis in U251 and LN229 cell lines ()).
NNT-AS1 directly interacts with miRNA-494-3p
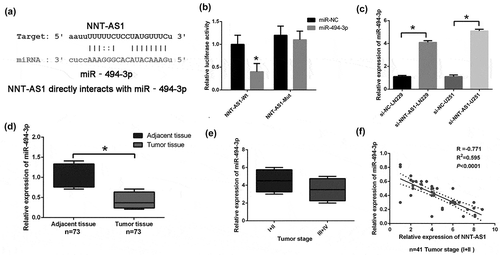
It was largely reported that lncRNAs could regulate post-transcription, interfere with miRNA and function as miRNA sponges to reduce the binding of miRNA to target genes; thus, the CLIP-seq data and miRanda program were used to predict potentially related miRNAs. The results showed that HAS-miR-494-3p interacts with NNT-AS1 ()), and the Pan-Cancer analysis of HAS-miR-494-3p and NNT-AS1 in 525 samples proved that the miR-494-3p was strongly co-expressed with NNT-AS1 (http://starbase.sysu.edu.cn). The results of dual luciferase activity assay showed that the luciferase activity was reduced by miR-494-3p mimic transfection in the NNT-AS1-wt group ()), but there was no significant change in the NNT-AS1-mut group ()). Moreover, the miR-494-3p level was significantly higher in the NNT-AS1 siRNA group compared to the NNT-AS1 negative control groups in LN229 and U251 cell lines ()). Furthermore, the miR-494-3p expression was significantly decreased and correlated with NNT-AS1 expression in glioma tissues ()). The relative expression of NNT-AS1 with miR-494-3p was also analyzed. The results indicated that miR-494-3p and NNT-AS1 expression in 41 glioma patients was positively correlated ()).
PRMT1 is decreased by NNT-AS1 inhibition
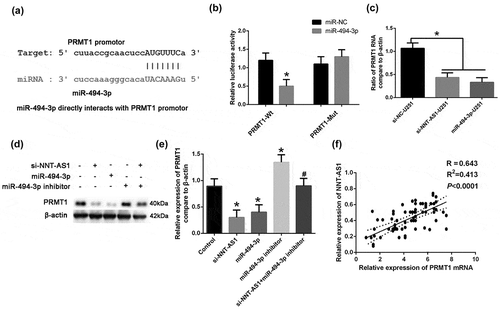
We explored the role of the target gene of miR-494-3p, PRMT1, in glioma. The results of the CLIP-seq data and miRanda program predicted that HAS-miR-494-3p interacts with the PRMT1 gene ()). Using the dual luciferase activity kit, we found that the miR-494-3p could significantly increase the luciferase activity in the PRMT1-Mutant (PRMT1-Mut) group ()). Results from qRT-PCR and western blotting showed that the PRMT1 mRNA and protein expression levels were significantly decreased in the miR-494-3p treated group, and increased in the miR-494-3p inhibitor group ()). Moreover, si-NNT-AS1 remarkably down-regulated the protein expression level of PRMT1; this effect was attenuated by the miR-494-3p inhibitor ()). The relative expression of PRMT1 was correlated with NNT-AS1 expression ()). These results suggest that NNT-AS1 can regulate PRMT1 expression through miR-494-3p.
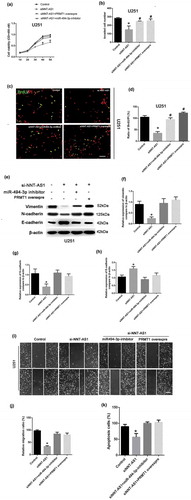
Figure 4. PRMT1 act as an downstream target in NNT-AS1/miR494-3p axis.(a) The interaction sites of HAS-miR-494-3p and the PRMT1 promoter was predicted. (b) By using the dual luciferase activity kit, the interaction of miR-494-3p and the PRMT1 promoter was illustrated. *P < 0.05 versus PRMT1-Wt. (c) U251 cells were treated with NNT-AS1-siRNA (si-NNT-AS1) or miR-494-3p, miR-494-3p inhibitor, and si-NNT-AS1+miR-494-3p inhibitors. The expression of PRMT1 mRNA was detected using qRT-PCR. *P < 0.05 versus control, #P < 0.05 versus si-NNT-AS1. (d-e) U251 cells were treated with NNT-AS1-siRNA (si-NNT-AS1) or miR-494-3p, miR-494-3p inhibitor, and si-NNT-AS1+miR-494-3p inhibitors. The protein expression of PRMT1 was detected using Western blotting. *P < 0.05 versus the control, #P < 0.05 versus the si-NNT-AS1. (f) The correlation of NNT-AS1 and PRMT1 mRNA was calculated in tissues from tumor stages I and II (P < 0.0001). All of the data are presented as the mean ± SD. All of the data are presented as the mean ± SD.
The miR-494-3p-PRMT1 axis is involved the tumor-suppressive effects of NNT-AS1 inhibition
To explore the involvement of the miR-494-3p/PRMT1 axis in the NNT-AS-regulated tumor proliferation and invasion, the CCK8 assay was used to detect the role of miR-494-3p/PRMT1 in U251 cell viability. Our results showed that the reduced cell viability induced by siNNT-AS1 could be rescued by the miR-494-3p inhibitor. Nevertheless, the cell viability in the siNNT-AS1+PRMT1 overexpression group was also increased compared with the siNNT-AS1 group ()). More importantly, the invaded cell number was increased in the NNT-AS1 siRNA+miR-494-3p inhibitor group and the siNNT-AS1+PRMT1 overexpression group compared to the siNNT-AS1 group ()). The BrdU assay showed that the inhibition of cell proliferation by siNNT-AS1 could be rescued by the miR-494-3p inhibitor. Moreover, the overexpression of PRMT1 could increase cell proliferation in NNT-AS1 siRNA-treated groups ()). Western blotting was used to detect EMT-related factors, E-cadherin, N-cadherin, and vimentin in control, siNNT-AS1, siNNT-AS1+PRMT1 overexpressing, and si-NNT-AS1+miR494-3p inhibitor groups in U251 cells. Our results indicated that siNNT-AS1 could attenuate the EMT level in U251 cells, which could be rescued by the miR-494-3p inhibitor or PRMT1 overexpression ()). Results from the cell migration assay confirmed that siNNT-AS1 could attenuate the cell migration level in U251 cells, and that the cell migration level is up-regulated in the siNNT-AS1+miR-494-3p inhibitor or siNNT-AS1+PRMT1 groups ()). Lastly, the results from the cell apoptosis assay illustrated that siNNT-AS1 could attenuate the cell apoptosis level in U251 cells, and that cell apoptosis is up-regulated in the siNNT-AS1+miR-494-3p inhibitor and siNNT-AS1+PRMT1 groups.
Discussion
LncRNAs have been found to be pervasively transcribed in the genome. They are often associated with human disease, especially cancer [Citation23]. The lncRNA NNT-AS1 is transcribed in the opposite direction to nicotinamide nucleotide transhydrogenase. Recent studies have revealed that NNT-AS1 plays a crucial role in cancers, such as lung cancer [Citation9,Citation24], human ovarian cancer [Citation8], gastric cancer [Citation25], cervical cancer [Citation26], and colorectal cancer [Citation27].
Glioma is a kind of aggressive malignancy in the central nervous system; half of all primary brain tumors worldwide are glioma [Citation28]. Although advances have been made in glioma treatment, the prognosis of patients still remains poor [Citation29–Citation31]. Therefore, this study aimed to explore the novel molecular mechanisms underlying the progression of glioma. Due to the dysregulation of lncRNAs, they have been reported in human malignant tumors[Citation32]. We proved that NNT-AS1 was highly expressed in the tumor tissue of glioma patients. In addition, in this study, we revealed that miR-494-3p, which is down-regulated by NNT-AS1, is important in lncRNA NNT-AS1-promoted cell proliferation and metastases. Suppressing NNT-AS1 could attenuate the proliferations of glioma cell lines, and miRNA129p has been shown to be the downstream target of NNT-AS125. Moreover, we demonstrated that NNT-AS1 overexpression is more significant in grade I and II glioma. This suggests that not only is NNT-AS1 a tumor promoter in glioma, but it may contribute to the diagnosis in the early stages of glioma. We found that NNT-AS1 could promote proliferation and cell viability in glioma cell lines. Moreover, NNT-AS1 siRNA could decrease the cells in S-Phase, and presented the slower closing of scratch wounds, with inhibited invasion ability. However, the mechanisms of NNT-AS1 in the promotion of glioma still remains little known.
Emerging evidence has indicated that lncRNAs act as a sponge for miRNAs, which could regulate the functions of miRNAs [Citation33–Citation36]. Previously, it was reported that NNT-AS1 presented as a miRNA sponge in different kinds of cancers. For instance, NNT-AS1 promoted gastric cancer tissue growth and metastasis by miRNA-424 [Citation27]. To determine the regulatory pattern of NNT-AS1 in glioma, we identified miRNA-494-3p as a potential target of NNT-AS1 by DIANA tools and the luciferase reporter assay. In this study, our qRT-PCR results showed that miRNA-494-3p expression was significantly decreased in glioma cell lines and glioma tissues. Moreover, it was correlated with NNT-AS1 expression in glioma tissues. si-NNT-AS1 could significantly increase the miRNA-494-3p expression in glioma cell lines.
miRNA-494-3p has been found to suppress the proliferation, invasion, and migration of prostate cancer [Citation37], suggesting the tumor suppressive effect of miRNA-494-3p. In addition, the downstream target of miRNA-494-3p is rarely reported. Therefore, the miRanda program predicted the interaction sites of miRNA-494-3p and PRMT1. PRMT1 is a major type of protein arginine methyltransferase and is also the most studied PRMT. PRMT1-dependent methylation of Arg3 on H4 tail peptides was studied in vitro. Importantly, PRMT1 null mice die at an early stage, indicating its essential role in embryonic development. However, the role of PRMT1 in glioma has not been studied. Our result further confirmed that the inhibition of NNT-AS1 in glioma cell lines increased miRNA-494-3p expression, but in return attenuated the expression of PRMT1. Interfering with the expression of PRMT1 revealed the same results for the inhibition of NNT-AS1; this can both decrease cell viability and invasion cell number. Western blotting showed that the NNT-AS1 siRNA could significantly decrease PRMT1 expression. However, the expression of PRMT1 could be rescued by the overexpression of miR-494-3p in the NNT-AS1 siRNA-treated group. This suggests that the miRNA-494-3p-PRMT1 axis is involved in the tumor-suppressive effects of NNT-AS12 inhibition.
Collectively, our research indicated that NNT-AS1 was ectopically expressed in glioma tissues and cell lines. It acted as an oncogene in glioma progression. The inhibition of NNT-AS1 suppressed the proliferation and invasion ability in vitro of tumor growth via the miR-494-3p/PRMT1 axis. Therefore, inhibiting NNT-AS1 might be a promising therapy target for the treatment of glioma in the early stages in glioma patients.
Author contributions
Xiaobing Xu contributes to research conception and design as well as manuscript drafting, manuscript review, guarantor of integrity of the entire study. Dahai Zheng and Shi Luo are responsible for definition of intellectual content, study concepts, study design and manuscript editing, data analysis, manuscript preparation. Xiang Wang and Lenian Lu analyzed and interpreted the data and made statistical analysis; Famu Lin Daliang Chen, and Jianmin Chen performed the clinical studies.
Ethics declarations
This study was approved by the Ethics Committees of Shunde Hospital. All patients gave their written informed consent to participate in this study.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Chi JH, Panner A, Cachola K, et al. Increased expression of the glioma-associated antigen ARF4L after loss of the tumor suppressor PTEN. Laboratory investigation. J Neurosurg. 2008;108:299–303.
- Gupta R, Webb-Myers R, Flanagan S, et al. Isocitrate dehydrogenase mutations in diffuse gliomas: clinical and aetiological implications. J Clin Pathol. 2011;64:835–844.
- Braunstein S, Raleigh D, Bindra R, et al. Pediatric high-grade glioma: current molecular landscape and therapeutic approaches. J Neurooncol. 2017;134:541–549.
- Chen R, Smith-Cohn M, Cohen AL, et al. Glioma subclassifications and their clinical significance. Neurotherapeutics. 2017;14:284–297.
- Li C, Zhang S, Qiu T, et al. Upregulation of long non-coding RNA NNT-AS1 promotes osteosarcoma progression by inhibiting the tumor suppressive miR-320a. Cancer Biol Ther. 2019;20:413–422.
- Wang X, Ren M, Li Y, et al. Long noncoding RNA NNT-AS1 promotes gastric cancer proliferation and invasion by regulating microRNA-363 expression. J Cell Biochem. 2019;120:5704–5712.
- Li Y, Lv M, Song Z, et al. Long non-coding RNA NNT-AS1 affects progression of breast cancer through miR-142-3p/ZEB1 axis. Biomed Pharmacothe. 2018;103:939–946.
- Huang Y, Shi J, Xu Y. Long non-coding RNA NNT-AS1 contributes to cell proliferation, metastasis and apoptosis in human ovarian cancer. Oncol Lett. 2018;15:9264–9270.
- Shen Q, Jiang Y. LncRNA NNT-AS1 promotes the proliferation, and invasion of lung cancer cells via regulating miR-129-5p expression. Biomed Pharmacothe. 2018;105:176–181.
- Zheng Y, Lu S, Xu Y, et al. Long non-coding RNA AGAP2-AS1 promotes the proliferation of glioma cells by sponging miR-15a/b-5p to upregulate the expression of HDGF and activating Wnt/beta-catenin signaling pathway. Int J Biol Macromol. 2019;128:521–530.
- Mittelbronn M, Simon P, Loffler C, et al. Elevated HLA-E levels in human glioblastomas but not in grade I to III astrocytomas correlate with infiltrating CD8+ cells. J Neuroimmunol. 2007;189:50–58.
- Liu L, Shi Y, Shi J, et al. The long non-coding RNA SNHG1 promotes glioma progression by competitively binding to miR-194 to regulate PHLDA1 expression. Cell Death Dis. 2019;10:463.
- Lan F, Qin Q, Yu H, et al. Effect of glycolysis inhibition by miR-448 on glioma radiosensitivity. J Neurosurg. 2019;1–9. DOI:https://doi.org/10.3171/2018.12.JNS181798
- Lin H, Huang ZP, Liu J, et al. MiR-494-3p promotes PI3K/AKT pathway hyperactivation and human hepatocellular carcinoma progression by targeting PTEN. Sci Rep. 2018;8:10461.
- Hannafon BN, Cai A, Calloway CL, et al. miR-23b and miR-27b are oncogenic microRNAs in breast cancer: evidence from a CRISPR/Cas9 deletion study. BMC Cancer. 2019;19:642.
- Hagemann C, Meyer C, Stojic J, et al. High efficiency transfection of glioma cell lines and primary cells for overexpression and RNAi experiments. J Neurosci Methods. 2006;156:194–202.
- Liu X, Zhu Q, Guo Y, et al. LncRNA LINC00689 promotes the growth, metastasis and glycolysis of glioma cells by targeting miR-338-3p/PKM2 axis. Biomed Pharmacothe. 2019;117:109069.
- Huang D, Wang Y, Xu L, et al. GLI2 promotes cell proliferation and migration through transcriptional activation of ARHGEF16 in human glioma cells. J Exp Clin Cancer Res. 2018;37:247.
- Qiao H, Wang YB, Gao YM, et al. Prucalopride inhibits the glioma cells proliferation and induces autophagy via AKT-mTOR pathway. BMC Neurol. 2018;18:80.
- Zhang C, Zhang J, Hao J, et al. High level of miR-221/222 confers increased cell invasion and poor prognosis in glioma. J Transl Med. 2012;10:119.
- Mair DB, Ames HM, Li R. Mechanisms of invasion and motility of high-grade gliomas in the brain. Mol Biol Cell. 2018;29:2509–2515.
- Li L, Zhang H. MicroRNA-379 inhibits cell proliferation and invasion in glioma via targeting metadherin and regulating PTEN/AKT pathway. Mol Med Rep. 2018;17:4049–4056.
- DiStefano JK. The emerging role of long noncoding RNAs in human disease. Methods Mol Biol. 2018;1706:91–110.
- Cai Y, Dong ZY, Wang JY. LncRNA NNT-AS1 is a major mediator of cisplatin chemoresistance in non-small cell lung cancer through MAPK/Slug pathway. Eur Rev Med Pharmacol Sci. 2018;22:4879–4887.
- Chen B, Zhao Q, Guan L, et al. Long non-coding RNA NNT-AS1 sponges miR-424/E2F1 to promote the tumorigenesis and cell cycle progression of gastric cancer. J Cell Mol Med. 2018;22:4751–4759.
- Hua F, Liu S, Zhu L, et al. Highly expressed long non-coding RNA NNT-AS1 promotes cell proliferation and invasion through Wnt/beta-catenin signaling pathway in cervical cancer. Biomed Pharmacothe. 2017;92:1128–1134.
- Wang Q, Yang L, Hu X, et al. Upregulated NNT-AS1, a long noncoding RNA, contributes to proliferation and migration of colorectal cancer cells in vitro and in vivo. Oncotarget. 2017;8:3441–3453.
- Wang W, Zhao Z, Yang F, et al. An immune-related lncRNA signature for patients with anaplastic gliomas. J Neurooncol. 2018;136:263–271.
- Del Sol-Fernandez S, Portilla-Tundidor Y, Gutierrez L, et al. Flower-like Mn-doped magnetic nanoparticles functionalized with alphavbeta3-integrin-ligand to efficiently induce intracellular heat after alternating magnetic field exposition, triggering glioma cell death. ACS Appl Mater Interfaces. 2019;11:26648–26663.
- Guerra-Rebollo M, Garrido C, Sanchez-Cid L, et al. Targeting of replicating CD133 and OCT4/SOX2 expressing glioma stem cells selects a cell population that reinitiates tumors upon release of therapeutic pressure. Sci Rep. 2019;9:9549.
- Peng G, Yang C, Liu Y, et al. miR-25-3p promotes glioma cell proliferation and migration by targeting FBXW7 and DKK3. Exp Ther Med. 2019;18:769–778.
- Peng Z, Liu C, Wu M. New insights into long noncoding RNAs and their roles in glioma. Mol Cancer. 2018;17:61.
- D’Angelo D, Mussnich P, Sepe R, et al. RPSAP52 lncRNA is overexpressed in pituitary tumors and promotes cell proliferation by acting as miRNA sponge for HMGA proteins. J Mol Med. 2019;97:1019–1032.
- Zhang X, Wang S, Wang H, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18:20.
- Zhang PF, Wei CY, Huang XY, et al. Circular RNA circTRIM33-12 acts as the sponge of MicroRNA-191 to suppress hepatocellular carcinoma progression. Mol Cancer. 2019;18:105.
- Liu H, Zhang Q, Lou Q, et al. Differential analysis of lncRNA, miRNA and mRNA expression profiles and the prognostic value of lncRNA in esophageal cancer. Pathol Oncol Res. 2019. DOI: https://doi.org/10.1007/s12253-019-00655-8.
- Shen PF, Chen XQ, Liao YC, et al. MicroRNA-494-3p targets CXCR4 to suppress the proliferation, invasion, and migration of prostate cancer. Prostate. 2014;74:756–767.