ABSTRACT
The hierarchical three-dimensional folding of the mammalian genome constitutes an important regulatory layer of gene expression and cell fate control during processes such as development and tumorigenesis. Accumulating evidence supports the existence of complex topological assemblies in which multiple genes and regulatory elements are frequently interacting with each other in the 3D nucleus. Here, we will discuss the nature, organizational principles, and potential function of such assemblies, including the recently reported enhancer “hubs,” “cliques,” and FIREs (frequently interacting regions) as well as multi-contact hubs. We will also review recent studies that investigate the role of transcription factors (TFs) in driving the topological genome reorganization and hub formation in the context of cell fate transitions and cancer. Finally, we will highlight technological advances that enabled these studies, current limitations, and future directions necessary to advance our understating in the field.
Introduction
Gene expression during physiological and pathological cell fate changes is controlled in part by the spatiotemporal regulation of enhancers and their target promoters. Despite progress in the genome-wide mapping of putative enhancers based on chromatin accessibility, transcription factor (or cofactor) binding or select histone marks (e.g. H3K27ac and H3K4me1), uncovering functional enhancers and assigning them to target genes remains extremely challenging. The emergence of 3D chromatin organization as an important regulatory layer of gene expression and cell identity [Citation1–7] highlighted the fact that understanding enhancer specificity and activity requires knowledge of its 3D neighborhood [Citation8–12].
Table 1. Glossary for 3D topology terms used in this review.
All different hierarchical levels of 3D chromatin organization, ranging from megabase-scale compartments to Topologically Associating Domains (TADs) and fine-scale chromatin loops contribute to shape the regulatory enhancer landscape [Citation13–15] (see Table 1). At the level of chromosome compartments, active or poised enhancers and genes are expected to be located within the so-called A compartments, which represent more euchromatic, open, and active genomic regions, while repressed and methylated enhancers will often reside within the heterochromatic and transcriptionally inactive B compartments [Citation16]. Interestingly, during processes such as differentiation and somatic cell reprogramming A-to-B and B-to-A compartment switches can occur, consistent with enhancer silencing or de novo activation, respectively, and a global 3D rewiring [Citation17–19]. Although enhancers can function over large distances, their activity is largely restricted within TADs. TADs are chromatin domains with high degree of self-association that are insulated from neighboring genomic regions by boundary elements [Citation5]. Albeit TAD boundaries are largely conserved across cell types and species and are thought to spatially confine transcriptional regulatory units [Citation20], boundary insulation and intra-TAD enhancer–promoter contacts are dynamically changing during cell fate transitions instructing cell-type-specific transcriptional programs [Citation21]. Here, we will focus on the most recent 3D chromatin studies reporting complex and dynamic enhancer and promoter contacts and discuss the nature, organizational principles, and potential function of such intricate interactions.
Enhancer–promoter networks and 3D hubs
Over the last two decades, hundreds of studies using either DNA fluorescence in situ hybridization (FISH) or chromosome conformation capture (3C)-based approaches have described pairwise enhancer–promoter interactions in various cellular contexts [Citation21–23]. These studies were instrumental assigning enhancers to the right target genes and revealed cases in which this communication may happen over very large distances (over a Megabase) and by skipping one or more intervening genes. Intriguingly, in addition to the typical one-to-one enhancer–promoter contacts, increasing evidence from multiple studies and experimental systems support the existence of complex and highly dynamic enhancer–promoter networks.
The first observations of multi-loci connections were based on 4C-based approaches around selected viewpoints in multiple different cellular contexts and conditions, including antiviral response [Citation24], reprogramming [Citation3,Citation4,Citation25], olfactory neurons [Citation26,Citation27], erythroid cell differentiation [Citation28] and other developmental processes [Citation29]. These studies, often combined with single-cell 3D-DNA FISH validations, revealed contacts among multiple genes and regulatory elements both in cis and in trans at variable frequencies providing important insights into the dynamic nature of 3D chromatin organization and the factors that may drive formation of multiconnected hubs. In addition, although limited to few select loci, these studies established the first functional links between long-range interactions and transcriptional regulation or co-regulation of implicated genes. The development of Hi-C technology which allows genome-wide mapping of regions in physical proximity further improved our understanding of the organizational principles of 3D chromatin topology and the association with gene expression [Citation30,Citation31]. However, the ability of Hi-C to detect fine-scale long-range enhancer–promoter contacts is limited by the resolution and requires very high sequencing depth. Moreover, findings from several groups hint to the fact that, even at high resolution, Hi-C is mostly able to detect structural loops (such as those mediated by CTCF) but promoter–enhancer contacts are underrepresented [Citation32,Citation33]. Recent technological advances that combine Hi-C with antibody-based or oligo-based enrichment steps, such as ChIA-PET [Citation34], PLAC-seq [Citation35], HiChIP [Citation36] and Capture Hi-C [Citation37] have drastically improved the resolution and the discovery rate of promoter–enhancer interactionss. A number of recent studies that utilize these technologies in different cellular systems and combined with antibodies against transcriptional and architectural factors (e.g. RNA PolII or Smc1) or characteristic “active” histone modifications (H3K4me3 or H3K27ac) have provided unique insights into the frequency, nature, dynamics, and roles of enhancer–promoter contacts and multiconnected hubs [Citation8,Citation38–41].
By performing HiChIP with an antibody against histone H3 lysine 27 acetylation (H3K27ac), we have recently generated 3D maps of active enhancer and promoter contacts in murine somatic and pluripotent stem cells (PSCs) and described a drastic rewiring during somatic cell reprogramming [Citation33]. This assay captured thousands of high-confidence and statistically significant enhancer–enhancer (41%), enhancer–promoter (41%), and promoter–promoter (18%) contacts. Although the majority of cis-regulatory elements were engaged in only one interaction, we found few hundreds of genomic regions communicating with 10 or more (up to 33) distant genes and/or enhancers at high frequencies. Importantly, the degree of interactivity (number of distinct contacts per anchor), which was validated by H3K27ac-independent approaches such as 4C-seq and Hi-C, was strongly associated with the transcriptional levels of connected genes.
Among the highest connected HiChIP anchors in pluripotent stem cells were many of the previously described super-enhancers (SE) [Citation42] as well as known stem cell regulators, including Mycn, Esrrb, and Mir290. In agreement with these findings, an independent study, which applied Hi-C in 21 primary human tissues, identified tissue-specific SEs as hotspots of local chromatin interactions termed FIREs (frequently interacting regions) [Citation43]. FIREs showed high levels of transcriptional activity in the respective cell types and included many cell-identity genes. The increased 3D interactivity of SEs has been further supported by the detection of SEs within recently described Hi-C stripes [Citation44] (described later in this review) and the reported inter-connectivity amongSEs in cis and trans [Citation45]. Together, these results suggest that a high degree of connectivity might be an inherent characteristic of regulatory elements that control high transcriptional levels of cell-type-associated genes, such as SEs or stretch enhancers [Citation46,Citation47].
An important consideration when evaluating results from these classical pairwise 3C-based studies is that they represent the “average” 3D chromatin conformation of cell populations and cannot distinguish between co-occurring interactions in the same cells and mutually exclusive, or competitive, interactions that occur in different cells. Recently developed technologies, such as multi-contact 4C and Tri-C, enabled the detection of simultaneous multi-loci interactions within a single allele [Citation48–52]. These studies identified high-order complexes formed by multi-way chromatin interactions between enhancers and promoters at selected mouse gene loci, including the globin locus [Citation48,Citation49] and Pcdh locus [Citation50] as well as antigen receptor genes [Citation52], confirming the existence of “regulatory hubs” within single alleles in which enhancer elements can simultaneously contact and potentially regulate multiple genes. Overall, these results confirm the presence of multi-way contacts in single cells but are restricted to a very small number of tested loci. Single-cell Hi-C could potentially overcome this issue by providing a genome-wide view of multi-way contacts in single cells [Citation53–55]; however, this method is limited by the sparsity of data that is inherent to current single-cell assays and cannot achieve high enough resolution to be informative at the level of individual regulatory elements.
Other approaches that do not rely on pairwise proximity ligation have recently emerged to study chromatin conformation [Citation56], including genome architecture mapping [Citation57] (GAM), split-pool recognition of interactions by tag extension [Citation58] (SPRITE), transposase-mediated analysis of chromatin looping [Citation59] (Trac-looping), optical reconstruction of chromatin architecture (ORCA) [Citation60] and chromatin-interaction analysis via droplet-based and barcode-linked sequencing [Citation61] (ChIA-Drop). Using these techniques, the authors were able to identify multi-loci chromatin interactions involving three or more genomic regions, albeit at low frequencies. For example, ChIA-Drop in Drosophila cells revealed that the vast majority (80%) of RNA polymerase II-mediated complexes included only one promoter, supporting that most promoters are not spatially interconnected in individual cells and alleles. Nonetheless, the same study captured 2,700 complexes that simultaneously connect 3 or more promoters, indicating that rare multi-way interactions do occur in single molecules . This is in agreement with recent high-resolution FISH-based approaches [Citation62]. These results suggest caution in the interpretation of high 3D interactivity from population-averaged 3C-based data and support that highly connected hubs likely represent different pairwise (instead of multi-way) contacts that occur within the population. This could either reflect population heterogeneity (different stable loops in cell subpopulations) and/or dynamic pairwise contacts across the population (see also ).
Functional implications of hubs
As discussed above, high 3D chromatin interactivity strongly correlates with increased regulatory and transcriptional activity. To understand the degree to which multiple contacts have functional impact on the expression of involved genes, we will consider two types of multi-way interactions (): (i) promoter-centric hubs, in which one promoter can contact multiple enhancers and (ii) enhancer-centric hubs, in which a single enhancer can interact with multiple genes.
Figure 1. Different types of 3D hubs and their biological relevance. Schematic diagram of different types of dynamic multi-way interactions. (a) Promoter-centric hubs, where a single promoter can interact with multiple enhancers. (b) Enhancer-centric hubs, where one enhancer is in contact with multiple promoters. (c) Multi-hubs represent networks of interconnected enhancer and promoter hubs. The potential biological relevance of each type of 3D hub is indicated below each diagram.
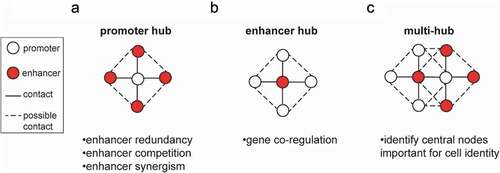
Promoter-centric hubs have been extensively described in various contexts and have been suggested to confer phenotypic robustness through enhancer redundancy ()). For example, deleting individual enhancers in loci required for mammalian limb formation did not affect limb development and morphology, however, deleting pairs of enhancers resulted in discernible phenotypes [Citation63]. Another example is the alpha-globin gene, where no single enhancer element is critical for the activation of alpha-globin transcription [Citation64]. In this case too, it seems that one enhancer can compensate for the loss of a different enhancer, in a redundant fashion.
Our integrative analysis of RNA-seq data and H3K27ac HiChIP data in MEFs (mouse embryonic fibroblast) and PSCs [Citation33] indicated that genes with higher number of enhancer contacts have higher levels of cell type-specific expression, suggesting that different enhancers may function synergistically to ensure tight and robust target gene activation. Future systematic interrogation of individual enhancers within promoter hubs in established PSCs or in the context of reprogramming is needed to directly test this hypothesis and further dissect potential functional hierarchies for the initial activation and maintenance of the transcriptional status of the target genes.
While enhancers are typically assigned to the single most proximal gene, 3C-based assays have revealed cases where enhancers contact multiple different genes over distance [Citation36,Citation38,Citation47] ()), expanding the repertoire of candidate target genes per enhancer and raising the possibility of gene co-regulation by the same enhancer. Our H3K27ac HiChIP analysis in PSCs [Citation33] identified a few hundred 3D enhancer hubs, where enhancers or SEs formed significant interactions with two or more genes (up to 10 in cis). Interestingly, when tested in the dynamic context of reprogramming, gene pairs within such hubs showed significantly higher percentage of transcriptional co-activation, compared to gene pairs of similar linear distance or residing in the same TADs, suggesting that hubs are responsible for co-regulation of spatially connected genes. Functional validation experiments further supported this notion, since CRISPR/(d)Cas9 targeting of select enhancer hubs, e.g. the Tbx3 and Zic2/Zic5 hubs, induced the concerted downregulation of multiple hub-associated genes without affecting neighboring non-hub genes.
In agreement with these results, a parallel study in human pancreatic islets applied promoter Capture-C technology and found that 42% of enhancers had high-confidence interactions with more than one gene, forming 3D enhancer hubs [Citation46,Citation47]. Genes within hubs enriched for islet-related functions, including insulin secretion and diabetes, and accordingly showed coordinated transcriptional changes upon perturbation of glucose levels. CRISPR-mediated activation or inactivation experiments further supported that enhancers hubs are responsible for the co-regulation of multiple hub-connected genes, while non-hub transcripts were only moderately affected [Citation47]. Importantly, both studies report cases where only some of the hub-connected genes respond to the enhancer perturbations, while others remain unaffected. We envision two possible scenarios that might explain this outcome. The first would suggest the existence of compensatory mechanisms due to the connection of non-responsive genes to additional enhancers that confer redundancy, as described in the “promoter-centric” hubs. An alternative possibility is that only some of the detected long-range contacts play direct regulatory roles, stressing the need to establish robust criteria for the prediction and identification of such interactions and their distinction from structural or bystander contacts.
Predicting the regulatory effects of chromatin loops has been an area of intense investigation and several parameters have been proposed to dictate the regulatory effects of enhancer–promoter contacts [Citation65,Citation66]. The relative strengths and frequencies of different enhancer–promoter contacts, the linear distance between them and the presence of intervening insulators /boundaries are some of them. The local chromatin structure (accessibility, histone marks, and transcription factor binding) of enhancers and candidate target promoters might also play important role in defining the specificity of these interactions. Utilizing combinations of some these parameters, various predictive models have been proposed with variable degrees of success when independently validated for specific loci [Citation32,Citation67–69]. Moreover, integration of experimentally validated enhancers as a training set could help in the construction of models with high predictive value.
A recent landmark study performed a systematic interrogation of thousands of putative enhancers around 30 target genes providing unprecedented insights into the principles of enhancer–promoter functionality [Citation70]. By using a novel technology called CRISPRi-FlowFISH in human erythroleukemia cells, the authors performed a large-scale enhancer perturbation screen measuring the effects on the expression of endogenous candidate target genes through FISH [Citation70]. Comparing their experimental results to the outcomes of various predictive models, they show that the recently proposed “activity-by-contact” (ABC) model significantly outperforms both in accuracy and sensitivity previous predictive models based either only on linear distance or on 3D genomic features (Hi-C contact or domain) or on chromatin states using machine learning approaches. The ABC model calculates the relative contribution of a regulatory element on target gene activity by using only two factors: the strength of the enhancer (as measured by H3K27ac ChIP-seq) weighted by the frequency of 3D contact with the target promoter (by using Hi-C, PolII ChIA-PET, or H3K27ac HiChIP information) divided by the total effect of all regulatory elements. Importantly, the perturbation-based maps built in this study uncovered complex connections where individual enhancers regulated up to 5 genes and individual genes were regulated by up to 14 distal elements, further supporting the functional role of highly connected hubs in living cells.
Promoter-centric or enhancer-centric hubs represent a simplistic view of potential promoter–enhancer networks. An unbiased analysis focusing on the highly interacting hubs in both murine somatic and pluripotent cells revealed that the majority of enhancer and promoter hubs are inter-connected to each other forming complex high-order networks, as depicted in ). Based on the H3K27ac HiChIP [Citation33] we were able to estimate that more than 80% of enhancer hubs contained genes that were also parts of promoter-hubs, suggesting an extensive “cross-talk” and interconnectivity (unpublished data).
Together the abovementioned studies demonstrate the complexity of the 3D regulatory networks and highlight the challenges to accurately predict enhancer efficacy and specificity. Addressing these challenges will enable better understanding of the function of non-coding regulatory elements in health and disease and will ultimately enable identifying central nodes in the regulatory networks that control cell-type-specific or pathogenic programs.
Mechanisms of loop and hub formation
One of the most well-described mechanisms that has been proposed to shape the 3D genome is the loop extrusion model (see Table 1). Loop extrusion nicely explains the formation of TADs [Citation71–73] and relies on progressive extrusion of chromatin through the cohesin ring until it reaches convergent CTCF-bound sites. Loop extrusion is also the underlying mechanism of the so-called “architectural stripes” ()), which are formed by a cluster of unidirectional CTCF binding sites that blocks cohesin-mediated loop extrusion from progressing in that direction (Citation30, Citation44, Citation73, Citation74). In the other direction, the DNA is being extruded and each segment forms a contact with the anchor, resulting in a stripe. This model can explain how one regulatory region (at the stripe anchor) could contact several other genomic loci, forming a hub. Architectural stripes are associated with both poised and active chromatin landscapes and have been shown to facilitate cognate promoter–enhancer interactions [Citation44,Citation74]. Intriguingly, upon targeted degradation of either CTCF or Cohesin subunits, long-range chromatin loops, TADs and stripes are drastically eliminated, while compartments and cis and trans contacts around super-enhancers remain unaffected or become even stronger [Citation45,Citation75–78], suggesting additional or competing forces of 3D chromatin organization. Despite the global effects on chromatin architecture, the effects on gene expression are less severe and selective, suggesting the presence of additional mechanisms that preserve transcriptional regulation and gene-enhancer communication. In agreement, recent work from several groups supports the existence of CTCF/cohesin-dependent and independent chromatin contacts [Citation79–82] and speculated on their different function and organizational principles.
Figure 2. Mechanisms of hub formation. Two main forces have been reported to contribute to hub formation involving promoter–enhancer contacts. (a) Architectural stripes are formed by cohesin-mediated loop extrusion, when a cluster of unidirectional CTCF binding sites on one boundary blocks extrusion in that direction, while in the other direction, the DNA is being extruded and each segment forms a contact with the anchor, resulting in a stripe. (b) phase separation is based on weak, low affinity, multivalent interactions mediated by large intrinsically disordered regions (IDRs) within proteins, that lead to the formation of subnuclear biomolecular condensates. These membrane-less structures are enriched, among others, in transcription factors, epigenetic writers and readers, nascent RNAs and activating histone marks that facilitate hub chromatin contacts.
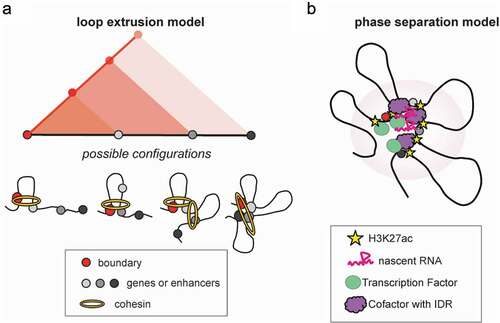
Other mechanisms have been proposed to explain the formation of chromatin loops such as the handcuff mechanism [Citation83] and the depletion/attraction mechanism [Citation84]. However, it is becoming commonly accepted that the chromatin 3D architecture is shaped through an interplay between loop extrusion and segregation/self-organization [Citation13–15,Citation23]. The latter describes the process of “self-attraction” among loci with similar transcriptional and chromatin states [Citation16], such as epigenetic modifications (e.g. histone marks) and protein composition (e.g. transcription factors and cofactors) and exclusion of regions with different chromatin characteristics. This process nicely explains the formation of chromosome compartments and subnuclear membrane-less structures, including nucleoli and speckle bodies.
One of the proposed mechanisms for self-organization is through phase separation [Citation85–88] ()), a process based on weak, low affinity, multivalent interactions mediated by large intrinsically disordered regions (IDRs) within proteins, that lead to the formation of subnuclear biomolecular condensates (see Table 1). It is plausible that phase separation could drive the formation of chromatin contacts as well as 3D hubs independently of cohesin-mediated extrusion by bringing multiple distal regions in physical proximity. In support to this notion, a large number of transcription factors [Citation85,Citation88,Citation89], epigenetic modulators [Citation87,Citation90], (BRD4 PRC2), transcriptional cofactors [Citation91,Citation92] (Mediator, PolII) [Citation91], long non-coding RNAs [Citation93] and histone modifications [Citation94] (H3K9me3, etc.) have been reported to promote nuclear condensation and form large nuclear clusters, which are particularly sensitive to concentration changes. Many of these factors are found preferentially enriched and at high density on multiconnected hubs [33Citation33], FIREs [Citation43] and super-enhancers [Citation42], further supporting the hypothesis that 3D hubs are formed through biomolecular condensation, promoted by the high density of bound TFs and recruited cofactors. The high levels of histone acetylation and transcriptional activity that characterize 3D enhancer hubs and interacting genes may further facilitate multivalent interactions and self-organization.
The relative contribution of each hub-associated component (transcriptional factors, protein cofactors, histone marks, and nascent transcripts) in hub organization and regulation has not been systematically investigated. However, recent studies that perturbed the activity, binding, or concentration of individual factors either by genetic interference or chemical inhibition have started shedding light into their roles in enhancer–promoter communication and gene regulation. For example, it has been shown that inhibition of BRD4 [Citation87] binding or general perturbation of protein condensation by 1,6-hexanediol [Citation91] treatment result in dissolution of large Med1, BRD4, and PolII condensates in mouse PSCs, preferential loss of SE activity and downregulation of many associated genes. Interestingly, some recent studies demonstrated that although these treatments have drastic effects on enhancer activity and gene expression, they do not affect 3D chromatin organization and enhancer–promoter contacts [Citation95], arguing against the role of BRD4 and/or protein condensation in long-range interactions. Similarly, inhibition of transcription per se seems to have very little impact on promoter–enhancer contacts [Citation95,Citation96]. These data together argue that the transcriptional activity of a gene and the physical interaction with enhancer(s) are separable events.
Transcription, transcription factors, and hubs
The role of transcription and transcription-associated factors in 3D chromatin organization becomes increasingly appreciated over the years [Citation97–99]. Back in 1993 Jackson et al. [Citation100] had identified the so-called “transcription factories,” a concept that has been defined and re-defined over the years and which, in the broader sense, indicates nuclear foci of concentrated RNA polymerase II and basic transcriptional machinery at which nascent transcription occurs (see Table 1). Active genes and their regulatory elements have been shown to coalesce at transcription factories, and the dynamic relocation of genes toward and away from transcription factories correlates with their transcriptional “on” and “off” cycles. Transcription factories may thus represent the first report of a multiprotein nuclear body containing at least two different transcription units at any given time (P-P, E-E, P-E), resulting in the formation of chromatin 3D hubs.
The potential architectural function of TFs was initially based on integrative analyses of 4C-seq or Hi-C datasets with ChIP-seq results [Citation19,Citation101]. More recent functional studies in various cellular contexts provided important experimental support either by directly depleting selected TFs or modulating TF binding sites and quantifying the negative effects on enhancer–promoter looping and gene expression [Citation33,Citation38,Citation96,Citation102–104]. For example, we have recently showed [Citation33] that Cas9-mediated disruption of individual KLF4 binding sites from PSC-specific enhancers resulted in a reduced expression of multiple genes within the enhancer hubs, partly by reducing contact frequency ()). In addition, inducible CRISPR-Cas9-mediated KO of KLF4 in stem cells, together with KLF2 and KLF5 KO (two closely related family members with overlapping binding targets) caused an overall decrease of hub and super-enhancer connectivity, supporting that KLF factors play a critical role in the maintenance of the 3D enhancer network in PSCs ()). Other examples of TFs mediating enhancer–promoter contacts include Nanog [Citation105] and Sox2 [Citation103] in pluripotent stem cells, GATA1 [Citation106] and Klf1 [Citation22] in erythrocytes, Pax3 [Citation104] and MyoD [Citation96] during myogenesis, Pax5 [Citation102] in B cell differentiation, Notch [Citation38] in the context of cancer and YY1 in various cell types [Citation39,Citation107].
Figure 3. Roles of KLF4 in the organization of 3D hubs. (a) KLF4 plays a role in the maintenance of 3D enhancer hubs. Depletion of KLF4 protein or disruption of its binding site within enhancers was shown to have a major effect on the maintenance of promoter–enhancer chromatin contacts within the hub and expression of the linked genes [Citation1]. (b) KLF4 HiChIP in pluripotent stem cells showed that KLF4 is involved both in active and repressive/poised loops and hubs around pluripotency or developmental genes, respectively. For each category the potential KLF4 cofactors and example genes are shown. O/S/N: Oct4, Sox2, and Nanog proteins, PRC: polycomb repressive complex.
![Figure 3. Roles of KLF4 in the organization of 3D hubs. (a) KLF4 plays a role in the maintenance of 3D enhancer hubs. Depletion of KLF4 protein or disruption of its binding site within enhancers was shown to have a major effect on the maintenance of promoter–enhancer chromatin contacts within the hub and expression of the linked genes [Citation1]. (b) KLF4 HiChIP in pluripotent stem cells showed that KLF4 is involved both in active and repressive/poised loops and hubs around pluripotency or developmental genes, respectively. For each category the potential KLF4 cofactors and example genes are shown. O/S/N: Oct4, Sox2, and Nanog proteins, PRC: polycomb repressive complex.](/cms/asset/17bb431a-37e0-476c-b06d-5566d93b53d7/kccy_a_1805238_f0003_oc.jpg)
Mechanistically, TFs can mediate promoter–enhancer contacts in a variety of ways, which have been extensively described in several recent reviews [Citation21,Citation97,Citation98]. In addition to the phase separation model mentioned above, these mechanisms include direct protein homo-dimerization (e.g. YY1 [Citation39]), recruitment of co-factor proteins that in turn form oligomers (e.g. Ldb1 [Citation104,Citation108,Citation109]), interaction with loop extruders (such as cohesin [Citation25,Citation105,Citation110,Citation111] and CTCF [Citation74–Citation76,Citation107,Citation112]) and recruitment of chromatin modifiers, which can either methylate DNA or modify histone tails [Citation113,Citation114], therefore leading to recruitment of other loop-mediating factors. To gain a better mechanistic understanding on how TFs shape the 3D genome organization, further experiments would be required in order to (i) identify all interacting proteins and nucleo-protein complexes in a locus-specific manner (e.g. by applying proximity labeling technologies around highly-interacting hubs), (ii) systematically perturb TF-mediated interactions either by mutating specific protein domains or block specific interfaces.
Using a combination of 3D genomics, functional and proteomics assays we recently demonstrated the role of KLF4 in mediating long-range chromatin interactions during reprogramming and in established PSCs and its potential architectural co-factors [Citation33]. By performing KLF4 HiChIP, we identified several thousands of KLF4-enriched chromatin contacts in ESC, which could be clustered into two categories ()): (A) loops that overlap with enhancer–promoter contacts as detected by H3K27ac HiChIP and are enriched in active marks and transcribed genes and (B) loops that contained mostly poised developmental genes and are devoid of active histone marks. Bioinformatics and proteomics analyses revealed that the first class of KLF4-bound contacts were also co-occupied by Mediator, BRD4, cohesin and other pluripotency TFs (e.g. Nanog, Oct4), whereas the second class were bound by components of the Polycomb Repressive Complexes 1 and 2. These results suggest that TFs might collaborate with different cofactors to establish activating or repressive chromatin contacts. Given that many of these cofactors have been shown to form condensates and multi-connected hubs [Citation87,Citation110,Citation115], it is plausible that TFs might nucleate the formation of distinct types of topological assemblies (activating or repressive hubs) by a process of self-organization.
Dynamics of hub establishment and maintenance
Most of the studies that aimed to address the role of 3D chromatin organization in gene regulation and the involved architectural factors have mostly focused on transcriptional and topological effects in steady-state conditions and in established cell lines. Therefore, the mechanisms that control establishment rather than maintenance of long-range chromatin interactions and the dynamic interplay between protein factor binding, chromatin looping, and transcriptional activation remain largely elusive. Few recent studies have started shedding light into these dynamic processes in the context of inducible gene expression [Citation116], directed differentiation [Citation96] or reprogramming [Citation3,Citation19,Citation33,Citation117,Citation118] as well as cell cycle [Citation119–123]. For example, it was recently shown that in contrast with the moderate and localized effects of cohesin depletion on gene expression, inducible activation of genes and enhancers in macrophages as response to microbial signals was directly dependent on cohesin [Citation116]. Similarly, reduced cohesin levels or mutations can compromise proper activation of lineage-specific genes and perturb the balance between self-renewal and differentiation [Citation116,Citation124], supporting an instructive role of cohesin in the establishment of lineage-specific programs.
To gain insights into these dynamic processes and dissect the order of molecular events that dictate 3D enhancer rewiring and transcriptional reprogramming of somatic cells toward pluripotency, we recently combined KLF4 HiChIP datasets with ChIP-seq and RNA-seq at different time points during iPSC reprogramming [Citation33]. We found that KLF4 binding was strongly associated with de novo establishment of enhancer–promoter contacts, and often preceded chromatin looping and 3D hub organization. On the other hand, looping formation and enhancer/gene activation were closely linked and usually coincided (). These data would suggest that KLF4 binding is important, but not always sufficient to induce long-range chromatin interactions and/or transcriptional activation. The temporal delay between TF binding and subsequent loop/hub formation suggests the existence of additional barriers that need to be removed (e.g. local chromatin remodeling, switch of chromatin state or compartment) or the absence of critical architectural and regulatory cofactors (e.g. proteins or potentially ncRNAs that are not yet expressed at early time-points during reprogramming).
Figure 4. Kinetics of KLF4 binding, hub formation and transcription activation during somatic cell reprogramming to Pluripotent Stem Cells (PSC). KLF4 bound hubs as detected by both KLF4 and H3K27ac HiChIP are split into three categories depending on the relative timing of KLF4 binding and looping [Citation1]. The median expression of genes within these hubs during reprogramming is plotted on the right. Overall, looping appears to coincide with gene activation, while KLF4 binding often precedes looping and expression, suggesting the requirement of additional cofactors.
![Figure 4. Kinetics of KLF4 binding, hub formation and transcription activation during somatic cell reprogramming to Pluripotent Stem Cells (PSC). KLF4 bound hubs as detected by both KLF4 and H3K27ac HiChIP are split into three categories depending on the relative timing of KLF4 binding and looping [Citation1]. The median expression of genes within these hubs during reprogramming is plotted on the right. Overall, looping appears to coincide with gene activation, while KLF4 binding often precedes looping and expression, suggesting the requirement of additional cofactors.](/cms/asset/1166b7b3-6b57-4323-a120-932e178067cb/kccy_a_1805238_f0004_oc.jpg)
The abovementioned examples highlight the need for future studies to carefully dissect the different mechanisms that control the formation or stability of 3D chromatin loops. For example, it will be interesting to test whether inducible degradation of hub-associated factors or perturbation of protein condensation, which appeared to have minimal effects on the maintenance of enhancer–promoter contacts, will have a strong impact in the de novo establishment of such loops during dynamic processes, such as cell cycle or activation/differentiation.
Conclusions and future directions
Genetic or epigenetic alterations that either perturb enhancer activity or enhancer specificity by enabling communication with aberrant target genes have been described in various disorders as the potential molecular basis of pathogenic transcriptional programs [Citation125–131]. These findings highlight that studying the nature and mechanisms of enhancer rewiring in disease has a considerable potential for understanding and treating disorders, such as cancer. Using 3D genomics assays several groups started to assign non-coding genetic variants, associated with disease, and putative enhancers to their interacting target genes in the contexts of cancer [Citation132], neuronal disorders [Citation133,Citation134], autoimmune and cardiovascular diseases [Citation36]. However, the computational and biological interpretation of these results remain very challenging. Predicting the actual regulatory impact of a long-range interaction and the potential synergies or redundancies with other interacting elements are very active topics of research both by applying mathematical modeling and through large-scale functional screens in various cellular contexts. Generating high-quality 4D genomics datasets in the course of dynamic biological processes, such as differentiation or disease progression, and developing statistically rigorous and standardized analyses pipelines will be essential steps toward this direction. Moreover, building 3D regulatory networks and applying advanced machine learning approaches may enable a better prediction of central 3D regulatory nodes that ca be targeted to efficiently modulate cell-type-specific or disease-specific transcriptional programs. Finally, implementation of existing CRISPR-Cas9 technologies or development of novel assays that enable large-scale interrogation of putative enhancers in the context of dynamic cell fate transitions invivo or ex vivo would be extremely informative.
The existence of highly interacting hubs and their association with highly active genes and enhancers that are critical for cell identity highlights the need for additional studies in order to dissect the organizational principles of such hubs and their functional roles in controlling cell fate decisions. Development or improvement of technologies that enable detection of multiway interactions and quantifying the frequency of such events at the single-cell level will be very relevant. In addition, orthogonal validations with imaging or recently developed approaches that do not depend on cross-linking or proximity ligation will help to reevaluate the hub connectivity, dynamics, and heterogeneity overcoming potential technical biases.
Another challenge that needs to be addressed in the following years is to unravel the mechanisms that promote or stabilize long-range enhancer–promoter interactions and hubs. As discussed above, important insights have been gained for the contribution of loop extrusion mechanisms versus self-organization through phase separation. However, the relative synergy or competition between these forces and the contribution of individual factors remain elusive. Systematic identification of all involved protein or RNA factors using technologies such as APEX2 [Citation135], which allow proximity labeling around target proteins or target loci, followed by mass-spectrometry, will be a critical first step to find top candidate genome organizers, that are either “universal” or hub-specific. Additional studies where top candidate factors are systematically perturbed either by inducible depletion/degradation or by blocking their binding and activities will allow to appreciate regulatory hierarchies and uncouple effects on transcriptional activity versus chromatin topology. It is also important to keep in mind that different mechanisms may mediate the establishment or the maintenance of 3D contacts. For example, it is possible that one protein factor is essential for the initial activation of a gene or the establishment of an enhancer–promoter interaction, but is not relevant for steady-state activity due to compensation by additional factors and/or enhancers. Therefore, the choice of treatment (e.g. protein knock-out versus reversible degradation), the duration (few hours versus days) and the study system (e.g. cycling asynchronous cells versus postmitotic or synchronized at specific cell cycle stages) need to be carefully designed and selected for proper interpretation of the results.
Acknowledgements
We thank members of the Apostolou and the Stadtfeld groups for input on our manuscript , Aristotelis Tsirigos and Ari Melnick for brainstorming on hub ideas. We apologize to colleagues whose work was not cited due to space limitations.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Gorkin DU, Leung D, Ren B. The 3D genome in transcriptional regulation and pluripotency. Cell Stem Cell. 2014;14:762–775.
- Dekker J, Belmont AS, Guttman M, et al. The 4D nucleome project. Nature. 2017;549:219–226.
- Apostolou E, Ferrari F, Walsh RM, et al. Genome-wide chromatin interactions of the Nanog locus in pluripotency, differentiation, and reprogramming. Cell Stem Cell. 2013;12:699–712.
- Denholtz M, Bonora G, Chronis C, et al. Long-range chromatin contacts in embryonic stem cells reveal a role for pluripotency factors and polycomb proteins in genome organization. Cell Stem Cell. 2013;13:602–616.
- Dixon JR, Selvaraj S, Yue F, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380.
- Beagan JA, Gilgenast TG, Kim J, et al. Local genome topology can exhibit an incompletely rewired 3D-folding state during somatic cell reprogramming. Cell Stem Cell. 2016;18:611–624.
- Di Giammartino DC, Apostolou E. The chromatin signature of pluripotency: establishment and maintenance. Curr Stem Cell Rep. 2016;2:255–262.
- Li G, Ruan X, Auerbach RK, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98.
- Yao L, Berman BP, Farnham PJ. Demystifying the secret mission of enhancers: linking distal regulatory elements to target genes. Crit Rev Biochem Mol Biol. 2015;50:550–573.
- Sun F, Chronis C, Kronenberg M, et al. Promoter-enhancer communication occurs primarily within insulated neighborhoods. Mol Cell. 2019;73:250–63 e5.
- Hnisz D, Day DS, Young RA. Insulated neighborhoods: structural and functional units of mammalian gene control. Cell. 2016;167:1188–1200.
- Dowen JM, Fan ZP, Hnisz D, et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159:374–387.
- Nuebler J, Fudenberg G, Imakaev M, et al. Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proc Natl Acad Sci U S A. 2018;115:E6697–E706.
- Rada-Iglesias A, Grosveld FG, Papantonis A. Forces driving the three-dimensional folding of eukaryotic genomes. Mol Syst Biol. 2018;14:e8214.
- Beagan JA, Phillips-Cremins JE. On the existence and functionality of topologically associating domains. Nat Genet. 2020;52:8–16.
- Lieberman-Aiden E, van Berkum NL, Williams L, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293.
- Takebayashi S, Dileep V, Ryba T, et al. Chromatin-interaction compartment switch at developmentally regulated chromosomal domains reveals an unusual principle of chromatin folding. Proc Natl Acad Sci U S A. 2012;109:12574–12579.
- Dixon JR, Jung I, Selvaraj S, et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–336.
- Stadhouders R, Vidal E, Serra F, et al. Transcription factors orchestrate dynamic interplay between genome topology and gene regulation during cell reprogramming. Nat Genet. 2018;50:238–249.
- Szabo Q, Bantignies F, Cavalli G. Principles of genome folding into topologically associating domains. Sci Adv. 2019;5:eaaw1668.
- Schoenfelder S, Fraser P. Long-range enhancer-promoter contacts in gene expression control. Nat Rev Genet. 2019;20:437–455.
- Drissen R, Palstra RJ, Gillemans N, et al. The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev. 2004;18:2485–2490.
- Robson MI, Ringel AR, Mundlos S. Regulatory landscaping: how enhancer-promoter communication is sculpted in 3D. Mol Cell. 2019;74:1110–1122.
- Apostolou E, Thanos D. Virus infection induces NF-kappaB-dependent interchromosomal associations mediating monoallelic IFN-beta gene expression. Cell. 2008;134:85–96.
- Wei Z, Gao F, Kim S, et al. Klf4 organizes long-range chromosomal interactions with the oct4 locus in reprogramming and pluripotency. Cell Stem Cell. 2013;13:36–47.
- Markenscoff-Papadimitriou E, Allen WE, Colquitt BM, et al. Enhancer interaction networks as a means for singular olfactory receptor expression. Cell. 2014;159:543–557.
- Lomvardas S, Barnea G, Pisapia DJ, et al. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413.
- Schoenfelder S, Sexton T, Chakalova L, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42:53–61.
- Noordermeer D, Leleu M, Splinter E, et al. The dynamic architecture of hox gene clusters. Science. 2011;334:222–225.
- Rao SS, Huntley MH, Durand NC, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680.
- Bonev B, Mendelson Cohen N, Szabo Q, et al. Multiscale 3D genome rewiring during mouse neural development. Cell. 2017;171:557–72 e24.
- Gasperini M, Hill AJ, McFaline-Figueroa JL, et al. A genome-wide framework for mapping gene regulation via cellular genetic screens. Cell. 2019;176:1516.
- Di Giammartino DC, Kloetgen A, Polyzos A, et al. KLF4 is involved in the organization and regulation of pluripotency-associated three-dimensional enhancer networks. Nat Cell Biol. 2019;21:1179–1190.
- Li G, Cai L, Chang H, et al. Chromatin interaction analysis with paired-end tag (ChIA-PET) sequencing technology and application. BMC Genomics. 2014;15(Suppl 12):S11.
- Fang R, Yu M, Li G, et al. Mapping of long-range chromatin interactions by proximity ligation-assisted ChIP-seq. Cell Res. 2016;26:1345–1348.
- Mumbach MR, Satpathy AT, Boyle EA, et al. Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nat Genet. 2017;49:1602–1612.
- Davies JO, Telenius JM, McGowan SJ, et al. Multiplexed analysis of chromosome conformation at vastly improved sensitivity. Nat Methods. 2016;13:74–80.
- Petrovic J, Zhou Y, Fasolino M, et al. Oncogenic notch promotes long-range regulatory interactions within hyperconnected 3D cliques. Mol Cell. 2019;73:1174–90 e12.
- Weintraub AS, Li CH, Zamudio AV, et al. YY1 is a structural regulator of enhancer-promoter loops. Cell. 2017;171:1573–88 e28.
- Ji X, Dadon DB, Powell BE, et al. 3D chromosome regulatory landscape of human pluripotent cells. Cell Stem Cell. 2016;18:262–275.
- Song M, Pebworth M-P, Yang X, et al. 3D epigenomic characterization reveals insights into gene regulation and lineage specification during corticogenesis. bioRxiv. 2020. 2020.02.24.963652.
- Whyte WA, Orlando DA, Hnisz D, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319.
- Schmitt AD, Hu M, Jung I, et al. A compendium of chromatin contact maps reveals spatially active regions in the human genome. Cell Rep. 2016;17(8):2042–2059.
- Vian L, Pekowska A, Rao SSP, et al. The energetics and physiological impact of cohesin extrusion. Cell. 2018;175:292–294.
- Rao SSP, Huang SC, Glenn St Hilaire B, et al. Cohesin loss eliminates all loop domains. Cell. 2017;171:305–20 e24.
- Parker SC, Stitzel ML, Taylor DL, et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci U S A. 2013;110:17921–17926.
- Miguel-Escalada I, Bonas-Guarch S, Cebola I, et al. Human pancreatic islet three-dimensional chromatin architecture provides insights into the genetics of type 2 diabetes. Nat Genet. 2019;51:1137–1148.
- Oudelaar AM, Davies JOJ, Hanssen LLP, et al. Single-allele chromatin interactions identify regulatory hubs in dynamic compartmentalized domains. Nat Genet. 2018;50:1744–1751.
- Oudelaar AM, Harrold CL, Hanssen LLP, et al. A revised model for promoter competition based on multi-way chromatin interactions at the alpha-globin locus. Nat Commun. 2019;10:5412.
- Allahyar A, Vermeulen C, Bouwman BAM, et al. Enhancer hubs and loop collisions identified from single-allele topologies. Nat Genet. 2018;50:1151–1160.
- Jiang T, Raviram R, Snetkova V, et al. Identification of multi-loci hubs from 4C-seq demonstrates the functional importance of simultaneous interactions. Nucleic Acids Res. 2016;44:8714–8725.
- Weiterer SS, Meier-Soelch J, Georgomanolis T, et al. Distinct IL-1alpha-responsive enhancers promote acute and coordinated changes in chromatin topology in a hierarchical manner. EMBO J. 2020;39:e101533.
- Nagano T, Lubling Y, Yaffe E, et al. Single-cell Hi-C for genome-wide detection of chromatin interactions that occur simultaneously in a single cell. Nat Protoc. 2015;10:1986–2003.
- Ramani V, Deng X, Qiu R, et al. Massively multiplex single-cell Hi-C. Nat Methods. 2017;14:263–266.
- Ramani V, Deng X, Qiu R, et al. Sci-Hi-C: a single-cell Hi-C method for mapping 3D genome organization in large number of single cells. Methods. 2020;170:61–68.
- Kempfer R, Pombo A. Methods for mapping 3D chromosome architecture. Nat Rev Genet. ;2020;21(4):207-226..
- Beagrie RA, Scialdone A, Schueler M, et al. Complex multi-enhancer contacts captured by genome architecture mapping. Nature. 2017;543:519–524.
- Quinodoz SA, Ollikainen N, Tabak B, et al. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell. 2018;174:744–57 e24.
- Lai B, Tang Q, Jin W, et al. Trac-looping measures genome structure and chromatin accessibility. Nat Methods. 2018;15:741–747.
- Mateo LJ, Murphy SE, Hafner A, et al. Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature. 2019;568:49–54.
- Zheng M, Tian SZ, Capurso D, et al. Multiplex chromatin interactions with single-molecule precision. Nature. 2019;566:558–562.
- Finn EH, Pegoraro G, Brandao HB, et al. Extensive heterogeneity and intrinsic variation in spatial genome organization. Cell. 2019;176:1502–15 e10.
- Osterwalder M, Barozzi I, Tissieres V, et al. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature. 2018;554:239–243.
- Hay D, Hughes JR, Babbs C, et al. Genetic dissection of the alpha-globin super-enhancer in vivo. Nat Genet. 2016;48:895–903.
- Whalen S, Truty RM, Pollard KS. Enhancer-promoter interactions are encoded by complex genomic signatures on looping chromatin. Nat Genet. 2016;48:488–496.
- Cao Q, Anyansi C, Hu X, et al. Reconstruction of enhancer-target networks in 935 samples of human primary cells, tissues and cell lines. Nat Genet. 2017;49:1428–1436.
- Fulco CP, Munschauer M, Anyoha R, et al. Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science. 2016;354:769–773.
- Sanjana NE, Wright J, Zheng K, et al. High-resolution interrogation of functional elements in the noncoding genome. Science. 2016;353:1545–1549.
- Schmidt F, Kern F, Schulz MH. Integrative prediction of gene expression with chromatin accessibility and conformation data. Epigenetics Chromatin. 2020;13:4.
- Fulco CP, Nasser J, Jones TR, et al. Activity-by-contact model of enhancer-promoter regulation from thousands of CRISPR perturbations. Nat Genet. 2019;51:1664–1669.
- Fudenberg G, Abdennur N, Imakaev M, et al. Emerging evidence of chromosome folding by loop extrusion. Cold Spring Harb Symp Quant Biol. 2017;82:45–55.
- Sanborn AL, Rao SS, Huang SC, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci U S A. 2015;112:E6456–65.
- Fudenberg G, Imakaev M, Lu C, et al. Formation of chromosomal domains by loop extrusion. Cell Rep. 2016;15:2038–2049.
- Barrington C, Georgopoulou D, Pezic D, et al. Enhancer accessibility and CTCF occupancy underlie asymmetric TAD architecture and cell type specific genome topology. Nat Commun. 2019;10:2908.
- Nora EP, Goloborodko A, Valton AL, et al. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell. 2017;169:930–44 e22.
- Wutz G, Varnai C, Nagasaka K, et al. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. Embo J. 2017;36:3573–3599.
- Busslinger GA, Stocsits RR, van der Lelij P, et al. Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature. 2017;544:503–507.
- Haarhuis JHI, van der Weide RH, Blomen VA, et al. The cohesin release factor WAPL restricts chromatin loop extension. Cell. 2017;169:693–707 e14.
- Sima J, Chakraborty A, Dileep V, et al. Identifying cis elements for spatiotemporal control of mammalian DNA replication. Cell. 2019;176:816–30 e18.
- Thiecke MJ, Wutz G, Muhar M, et al. Cohesin-dependent and independent mechanisms support chromosomal contacts between promoters and enhancers. Cell Rep. 2020;21;32(3):107929.
- Bintu B, Mateo LJ, Su JH, et al. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science. 2018:362(6413):eaau1783
- Hsieh TS, Cattoglio C, Slobodyanyuk E, et al. Resolving the 3D landscape of transcription-linked mammalian chromatin folding. Mol Cell. 2020;78(3):539–553.e8.
- Brackley CA, Johnson J, Michieletto D, et al. Extrusion without a motor: a new take on the loop extrusion model of genome organization. Nucleus. 2018;9:95–103.
- Marenduzzo D, Finan K, Cook PR. The depletion attraction: an underappreciated force driving cellular organization. J Cell Biol. 2006;175:681–686.
- Boija A, Klein IA, Sabari BR, et al. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell. 2018;175:1842–55 e16.
- Hnisz D, Shrinivas K, Young RA, et al. A phase separation model for transcriptional control. Cell. 2017;169:13–23.
- Sabari BR, Dall’Agnese A, Boija A, et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018:361(6400):eaar3958
- Chong S, Dugast-Darzacq C, Liu Z, et al. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science. 2018:361(6400):eaar2555
- Hnisz D, Abraham BJ, Lee TI, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947.
- Tatavosian R, Kent S, Brown K, et al. Nuclear condensates of the polycomb protein chromobox 2 (CBX2) assemble through phase separation. J Biol Chem. 2019;294:1451–1463.
- Cho WK, Spille JH, Hecht M, et al. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science. 2018;361:412–415.
- Zamudio AV, Dall’Agnese A, Henninger JE, et al. Mediator condensates localize signaling factors to key cell identity genes. Mol Cell. 2019;76:753–66 e6.
- Fox AH, Nakagawa S, Hirose T, et al. Paraspeckles: where long noncoding RNA meets phase separation. Trends Biochem Sci. 2018;43:124–135.
- Wang L, Gao Y, Zheng X, et al. Histone Modifications regulate chromatin compartmentalization by contributing to a phase separation mechanism. Mol Cell. 2019;76:646–59 e6.
- Crump NT, Ballabio E, Godfrey L, et al. BET inhibition disrupts transcription but retains enhancer-promoter contact. bioRxiv. 2019;848325.
- Dall’Agnese A, Caputo L, Nicoletti C, et al. Transcription factor-directed re-wiring of chromatin architecture for somatic cell nuclear reprogramming toward trans-differentiation. Mol Cell. 2019;76:453–72 e8.
- Stadhouders R, Filion GJ, Graf T. Transcription factors and 3D genome conformation in cell-fate decisions. Nature. 2019;569:345–354.
- Kim S, Shendure J. Mechanisms of interplay between transcription factors and the 3D genome. Mol Cell. 2019;76:306–319.
- Elemento O, Rubin MA, Rickman DS. Oncogenic transcription factors as master regulators of chromatin topology: a new role for ERG in prostate cancer. Cell Cycle. 2012;11:3380–3383.
- Iborra FJ, Pombo A, Jackson DA, et al. Active RNA polymerases are localized within discrete transcription “factories’ in human nuclei. J Cell Sci. 1996;109(Pt 6):1427–1436.
- de Wit E, Bouwman BA, Zhu Y, et al. The pluripotent genome in three dimensions is shaped around pluripotency factors. Nature. 2013;501:227–231.
- Johanson TM, Lun ATL, Coughlan HD, et al. Transcription-factor-mediated supervision of global genome architecture maintains B cell identity. Nat Immunol. 2018;19:1257–1264.
- Bertolini JA, Favaro R, Zhu Y, et al. Mapping the global chromatin connectivity network for Sox2 function in neural stem cell maintenance. Cell Stem Cell. 2019;24:462–76 e6.
- Magli A, Baik J, Pota P, et al. Pax3 cooperates with Ldb1 to direct local chromosome architecture during myogenic lineage specification. Nat Commun. 2019;10:2316.
- Nitzsche A, Paszkowski-Rogacz M, Matarese F, et al. RAD21 cooperates with pluripotency transcription factors in the maintenance of embryonic stem cell identity. PLoS One. 2011;6:e19470.
- Vakoc CR, Letting DL, Gheldof N, et al. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol Cell. 2005;17:453–462.
- Beagan JA, Duong MT, Titus KR, et al. YY1 and CTCF orchestrate a 3D chromatin looping switch during early neural lineage commitment. Genome Res. 2017;27:1139–1152.
- Deng W, Lee J, Wang H, et al. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012;149:1233–1244.
- Monahan K, Horta A, Lomvardas S. LHX2- and LDB1-mediated trans interactions regulate olfactory receptor choice. Nature. 2019;565:448–453.
- Kagey MH, Newman JJ, Bilodeau S, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435.
- Huang D, Wei Z, Lu W. Genome organization by Klf4 regulates transcription in pluripotent stem cells. Cell Cycle. 2013;12:3351–3352.
- Hansen AS, Hsieh TS, Cattoglio C, et al. Distinct classes of chromatin loops revealed by deletion of an RNA-binding region in CTCF. Mol Cell. 2019;76:395–411 e13.
- Yin Y, Morgunova E, Jolma A, et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science. 2017;356(6337):eaaj2239.
- Zhu F, Farnung L, Kaasinen E, et al. The interaction landscape between transcription factors and the nucleosome. Nature. 2018;562:76–81.
- Schoenfelder S, Sugar R, Dimond A, et al. Polycomb repressive complex PRC1 spatially constrains the mouse embryonic stem cell genome. Nat Genet. 2015;47:1179–1186.
- Cuartero S, Weiss FD, Dharmalingam G, et al. Control of inducible gene expression links cohesin to hematopoietic progenitor self-renewal and differentiation. Nat Immunol. 2018;19:932–941.
- Ferrari F, Apostolou E, Park PJ, et al. Rearranging the chromatin for pluripotency. Cell Cycle. 2014;13:167–168.
- Hu JF, Hoffman AR. Chromatin looping is needed for iPSC induction. Cell Cycle. 2014;13:1–2.
- Teves SS, An L, Hansen AS, et al. A dynamic mode of mitotic bookmarking by transcription factors. Elife. 2016:5:e22280.
- Festuccia N, Gonzalez I, Owens N, et al. Mitotic bookmarking in development and stem cells. Development. 2017;144:3633–3645.
- Zhang H, Emerson DJ, Gilgenast TG, et al. Chromatin structure dynamics during the mitosis-to-G1 phase transition. Nature. 2019;576:158–162.
- Abramo K, Valton AL, Venev SV, et al. A chromosome folding intermediate at the condensin-to-cohesin transition during telophase. Nat Cell Biol. 2019;21:1393–1402.
- Hsiung CC, Bartman CR, Huang P, et al. A hyperactive transcriptional state marks genome reactivation at the mitosis-G1 transition. Genes Dev. 2016;30:1423–1439.
- Viny AD, Bowman RL, Liu Y, et al. Cohesin members stag1 and stag2 display distinct roles in chromatin accessibility and topological control of hsc self-renewal and differentiation. Cell Stem Cell. 2019;25:682–96 e8.
- Flavahan WA, Drier Y, Liau BB, et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529:110–114.
- Chen X, Zhang M, Gan H, et al. A novel enhancer regulates MGMT expression and promotes temozolomide resistance in glioblastoma. Nat Commun. 2018;9:2949.
- Sur I, Taipale J. The role of enhancers in cancer. Nat Rev Cancer. 2016;16:483–493.
- Morton AR, Dogan-Artun N, Faber ZJ, et al. Functional enhancers shape extrachromosomal oncogene amplifications. Cell. 2019;179:1330–41 e13.
- He Y, Long W, Liu Q. Targeting super-enhancers as a therapeutic strategy for cancer treatment. Front Pharmacol. 2019;10:361.
- Sengupta S, George RE. Super-enhancer-driven transcriptional dependencies in cancer. Trends Cancer. 2017;3:269–281.
- Lupianez DG, Kraft K, Heinrich V, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–1025.
- Li X, Shi L, Wang Y, et al. OncoBase: a platform for decoding regulatory somatic mutations in human cancers. Nucleic Acids Res. 2019;47:D1044–D55.
- Song M, Yang X, Ren X, et al. Mapping cis-regulatory chromatin contacts in neural cells links neuropsychiatric disorder risk variants to target genes. Nat Genet. 2019;51:1252–1262.
- Kikuchi M, Hara N, Hasegawa M, et al. Enhancer variants associated with Alzheimer’s disease affect gene expression via chromatin looping. BMC Med Genomics. 2019;12:128.
- Hung V, Udeshi ND, Lam SS, et al. Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nat Protoc. 2016;11:456–475.
- Dekker J, Rippe K, Dekker M, et al. Capturing chromosome conformation. Science. 2002;295:1306–1311.
