ABSTRACT
The involvement of propofol and circular RNAs (circRNAs) in lung cancer progression has been identified. However, the relationship between propofol and circRNAs as well as the underlying molecular mechanisms on lung cancer development remain unclear. Cell viability, migration and invasion were measured by cell counting kit-8 assay, 5-bromo-2-deoxyuridine (BrdU) and transwell assay. Glycolytic metabolism was calculated by measuring the glucose consumption, lactate production and extracellular acidification. Western blot was used to detect the protein of glucose transporter 1 (GLUT1), glycolysis enzymes, and forkhead box M1 (FOXM1). The expression of circRNA transcriptional adaptor 2A (circTADA2A), microRNA (miR)-455-3p and FOXM1 mRNA was detected by quantitative real-time polymerase chain reaction. The interaction between miR-455-3p and circTADA2A or FOXM1 was analyzed using the dual-luciferase reporter assay. Murine xenograft model was established to perform in vivo experiments. We found propofol treatment alleviated lung cancer cell proliferation, migration, invasion and aerobic glycolysis in vitro as well as inhibited tumor growth in vivo. Propofol decreased the level of circTADA2A and exerted anti-tumor effects by regulating circTADA2A. MiR-455-3p directly interacted with circTADA2A and FOXM1 in lung cancer cells, and circTADA2A could regulate FOXM1 expression by binding to miR-455-3p. Subsequently, rescue assay showed that propofol inhibited cell proliferation, migration, invasion and aerobic glycolysis by regulating circTADA2A/miR-455-3p/FOXM1 axis in lung cancer. Collectively, propofol suppressed cell carcinogenesis and aerobic glycolysis by regulating circTADA2A/miR-455-3p/FOXM1 axis in lung cancer, providing an effective clinical implication for propofol to prevent the development of lung cancer.
Introduction
Ranking as the highest incidence and mortality rates of all cancers, lung cancer is the major threat to public health issues with an about 5-year survival rate of 16.6% around the world [Citation1,Citation2]. Despite great advances have been made in diagnosis and oncological therapeutic strategies, the effective improvements of the long-term outcome in lung cancer patients remain unsatisfactory [Citation3]. Therefore, further investigations on the molecular mechanism underlying lung cancer pathogenesis and progression are necessary for managing the clinical treatments.
Increasing evidence indicates that anesthetic agents can increase long-term survival rates of patients after surgery, especially in preventing cancer recurrence [Citation4,Citation5]. Propofol is the most extensively used intravenous anesthetic in clinical surgery, with the highlights of rapid effect, short action and little side effects [Citation6,Citation7]. In addition to multiple anesthetic effects, recent studies have investigated that propofol also has a variety of non-narcotic effects, such as anticancer activity [Citation8,Citation9]. Propofol inhibits the malignancy of numerous human cancers, including breast cancer [Citation10], gastric cancer [Citation11], and lung cancer [Citation12]. However, the molecular mechanisms underlying propofol-induced anti-tumor activity are complex and still unclear. Currently, it has been identified that most cancer cells reprogram their metabolic type from oxidative phosphorylation to aerobic glycolysis to meet their metabolic needs for sustaining cell proliferation even in the presence of oxygen [Citation13–18]. Compared with normal cells, this metabolic shift favors tumor cell growth advance in tumor environments regardless of oxygen availability, and also contributes to oncogenic signaling to facilitate cancer malignancy [Citation19,Citation20]. Thus, targeting this glucose metabolism may be a potential way to develop clinically therapeutic interventions [Citation20,Citation21].
Circular RNAs (circRNAs) are a class of noncoding RNA (ncRNA) molecules with covalently closed loop structures, which elevate the stability of these molecules to resistant exonuclease degradation compared with their linear RNAs decay [Citation22]. Due to the tissue- and developmental-specific expression, growing evidence has reported that circRNAs act as important modulators in a variety of cellular processes in cancers, including cell growth, proliferation, invasion, migration, drug resistance, and metabolism, to affect tumor progression and development [Citation23–27]. CircRNA transcriptional adaptor 2A (circTADA2A) is a novel identified circRNA molecule. The recent study showed that circTADA2A was elevated in non-small cell lung cancer (NSCLC), and interacted with miR-520 f to contribute to cell proliferation and migration in NSCLC [Citation28], indicating the potential carcinogenic roles in lung cancer progression. However, large-scale identification of circTADA2A expression in propofol-induced lung cancer was not yet reported.
In this study, we focused mainly on investigating the potential role of propofol in the process of carcinogenesis and aerobic glycolysis, explored the relationship between propofol and circRNAs as well as identified the underlying mechanisms by which they affected cell progression in lung cancer.
Materials and methods
Clinical specimens
Tumor samples and matched normal tissue samples were obtained from 30 paired lung cancer patients who underwent surgical resection at The First Affiliated Hospital of Zhengzhou University, and were immediately stored at −80°C until further analysis . All patients were diagnosed by histopathological examination. This study was permitted by the Ethics Committee of The First Affiliated Hospital of Zhengzhou University and written informed consent had been provided from all subjects.
Cell culture, transfection and propofol exposure
Two human lung cancer cell lines (A549 and H1299) and human airway epithelial cell line BEAS-2B were purchased from Shanghai Academy of Life Science (Shanghai, China) and grown in the Roswell Park Memorial Institute 1640 medium (RPMI 1640, Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (Gibco) at 37°C with 5% CO2.
The small interfering RNA (siRNA) sequences targeting circTADA2A covalent closed junction (si-circTADA2A, 5’-CCAGCUGGACUGCUCAAGAAG-3’), siRNA targeting forkhead box M1 (FOXM1) (si-FOXM1, 5’-GGAAATGCTTGTGATTCAACA-3’), siRNA negative control (si-NC, 5’- UUCUCCGAACGUGUCACGUTT-3’; 5’-CAGUACUUUUGUGUAGUACAA-3’), pcDNA-circTADA2A overexpression vector (pcDNA-circTADA2A), and pcDNA empty vector (pcDNA-NC) were synthesized by Invitrogen (Carlsbad, CA, USA). Inhibitor and mimic of miR-455-3p and the negative control (miR-NC and anti-miR-NC) were purchased from RIBOBIO (Guangzhou, China). The plasmids or miRNAs were transfected into 60–80% confluent A549 and H1299 cells using LipofectamineTM 2000 transfection reagent (Invitrogen).
Propofol obtained from Sigma (St. Louis, MO, USA) was dissolved in dimethyl sulfoxide (DMSO; Sigma) following the protocol of manufacturer. Cells were maintained and treated with propofol in a concentration gradient (2, 4, 6, 8, and 10 µg/mL) for 48 h in 96-well plates, and cells treated with the same volume of PBS were regarded as blank controls.
Cell viability
After treatment and/or transfection as described, A549 and H1299 cells grown to 75–80% confluence were seeded in a 96-well plate; 24 h later, each well was co-incubated with 10 μL cell counting kit-8 (CCK-8) solution (Beyotime, Shanghai, China) for 4 h. Finally, the optical density (OD) at 450 nm was analyzed by a microplate reader.
DNA synthesis capacity by 5-bromo-2-deoxyuridine (BrdU)
A549 and H1299 cells at 80% confluence were seeded on each well of 96-well plates and transfected and/or treated as described. After 24 h, per well were added with 10 μM BrdU (RIBOBIO) and incubated for another 2 h. After discarding the medium, cells were fixed by 4% paraformaldehyde for 30 min, followed by interaction with peroxidase-coupled BrdU antibody (Sigma) for 60 min. Then cells were washed with PBS and incubated with peroxidase substrate. Finally, the absorbance values were analyzed at 450 nm.
Transwell assay
Cells cultured to 60% confluence were transfected and/or treated as described. For migration assay, cells with serum-free RPMI 1640 medium were placed in the upper chamber (8 µm pore size), then the lower chamber was filled with medium fixed with FBS. After incubation for 48 h at 37°C, the migratory cells on the lower face of the membranes were fixed and stained, and then were counted under a microscope. For cell invasion, the upper chamber membranes were pre-coated with matrigel and other philosophy of measurement was similar to the steps of cell migration.
Glucose consumption and lactate production assay
Cells in 70–80% confluences were transfected and/or treated as described, and then maintained in a 6-well plate and incubated for 48 h. After that, supernatants of cell culture media were collected to determine the levels of glucose and lactate by a glucose and lactate assay kit (Sigma) in line with the protocol of manufacturer. The relative glucose uptake and lactate production were assessed ground on the standard curve and normalized to protein content using the Pierce BCA Protein assay.
Measurement of extracellular acidification (ECA)
Cells were cultured to 70–80% confluences, and transfected and/or treated as described. Then A549 and H1299 cells were seeded in a XF24 cell culture microplate for 24 h. Subsequently, the culture medium was replaced with XF assay medium containing 2 mM l-glutamine. Finally, ECA was determined with the Seahorse XF24 Extracellular Flux Analyzer (Agilent Technologies, Santa Clara, CA, USA) in accordance with the recommendations of manufacturer after normalization to protein content.
Western blot
Whole-protein was prepared from transfected and/or treated cells using the RIPA buffer (Beyotime). Then all protein was separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (40 V, 4~5 h), transferred onto a polyvinylidene fluoride membranes (60 V, 2 h), and blocked with 5% nonfat-milk for 1 h. Afterward, western blot analysis was conducted using primary antibodies against glucose transporter 1 (GLUT1) (1:10,000, ab40084, Abcam, Cambridge, MA, USA), hexokinase-2 (HK2) (1:5000, ab104836, Abcam), phosphoglycerate kinase 1 (PGK1) (1:5000, ab113687, Abcam), lactate dehydrogenase A chain (LDHA) (1:3000, ab135366, Abcam), β-actin (1:2000, 4967, Cell Signaling Technology, Boston, MA, USA) and the secondary HRP-conjugated antibody (1:1000, ab9482, Abcam). Finally, protein bands were detected using electrochemiluminescence.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted with the TRIzol reagent (Invitrogen) following the standard procedure. After interaction with Rnase R (Epicentre, Madison, WI, USA) and incubation with RNeasy MinElute Cleanup Kit (Qiagen), extracted RNA was reversely transcribed into complementary DNA (cDNA) using the PrimeScript RT reagent kit (Takara, Dalian, China). Then qRT-PCR was carried out by SYBR™ Green Master Mix (Qiagen). The relative expression was detected using 2−ΔΔCt method and normalized by glyceraldehyde 3-phosphate dehydrogenase (GADPH) and U6 small nuclear B noncoding RNA (U6). The primers were listed as followed: circTADA2A: F 5’-AATGTGCACCAAGACCAAGG-3’, R 5’- CCA AAGCCACAG TCCATCAC-3’; miR-455-3p: F 5’-TAAGACGTCCATGGGCAT-3’, R 5’- GTGCAGGGTCCGAGGT-3’. FOXM1: F 5’-TTATCAGTGCTGCTAGCTGAGG-3’, R 5’-TGATGGGTCTCGCTAAGTGT-3’, GADPH: F 5’-GATATTGTTGCCATCAATGAC-3’, R 5’-TTGATTTTGGAGGGATCTCG-3’; U6: F 5’-GCTTCGGCAGCACATATACTAAAAT-3’, R 5’- CGCTTCACGAATTTGCGTGTCAT-3’.
Dual-luciferase reporter assay
The circTADA2A sequences containing the predicted potential miR-455-3p binding sites or mutant sites were amplified and cloned into the pmiR-RB-Report (Promega, Madison, WI, USA) to generate circTADA2A-WT and circTADA2A-MUT luciferase vectors. Similarly, the FOXM1 3’UTR-WT and FOXM1 3’UTR-MUT luciferase vectors were established. Subsequently, A549 and H1299 cells were transfected with these constructed vectors and miR-455-3p mimics or miR-NC for 48 h. Finally, a dual-luciferase reporter assay kit (Promega) was applied to analyze the relative luciferase activity.
Xenograft experiments in vivo
Female BALB/c nude mice (4–6 weeks old, N = 6) were purchased from Jinan Pengyue Animal Center (Jinan, China). A549 cells (1 × 107 cells) were subcutaneously injected into the flanks of the nude mice. When the volume of xenografts reached about 100 mm3, all mice were intratumorally injected with PBS or propofol. After 6 days following the inoculation, the tumor size was calculated every 3 days. On day 21, all mice were sacrificed, and tumor masses were weighed and harvested for further molecular analysis. All animal experiments were performed according to the guidelines permitted by the Animal Research Committee of The First Affiliated Hospital of Zhengzhou University.
Statistical analysis
Data from three independent experiments were presented as the mean ± standard deviation (SD). Significant differences were analyzed using the one-way analysis of variance (ANOVA) or Student’s t-test with GraphPad Prism 7 software (GraphPad Inc., San Diego, CA, USA). P-value less than 0.05 was suggested statistically significant.
Results
Propofol alleviates cell proliferation, migration, invasion, and aerobic glycolysis in lung cancer in vitro
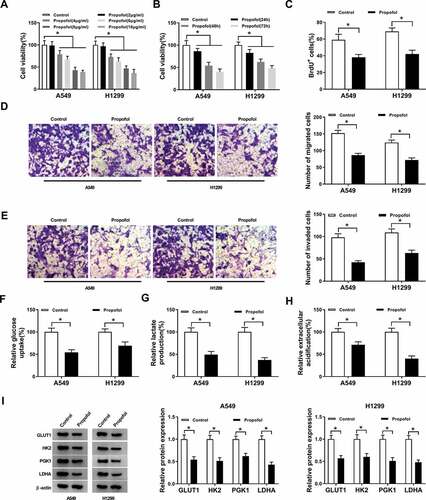
To investigate the effects of propofol treatment on cell tumorigenesis and aerobic glycolysis in lung cancer, A549 and H1299 cells were treated with PBS or exposed to different concentrations of propofol (2, 4, 6, 8, and 10 µg/mL). Results showed propofol decreased the viability of A549 and H1299 cells in a dose-dependent manner ()). Due to the decrease was about half at concentrations of 8 µg/mL, thus, we chose 8 µg/mL propofol for the following experiments. A549 and H1299 cells were treated with 8 µg/mL propofol for 24, 48, and 72 h, and we found the viability of A549 and H1299 cells was also decreased in a time-dependent manner ()). Meanwhile, BrdU assay exhibited propofol treatment inhibited the DNA synthesis capacity of A549 and H1299 cells ()). Additionally, transwell assay showed propofol exposure inhibited the migration and invasion of A549 and H1299 cells (,e)). Afterward, we started to investigate the effects of propofol treatment on glucose metabolism in lung cancer cells. Results suggested propofol exposure decreased glucose uptake ()), lactate production ()), and ECA ()). Furthermore, western blot analysis displayed propofol treatment remarkably down-regulated the protein expression levels of GLUT1, PKM2, HK2, and LDHA in A549 and H1299 cells ()), further indicating propofol decreased the rate of aerobic glycolysis in lung cancer cells. These results suggested that propofol inhibited cell tumorigenesis and aerobic glycolysis in lung cancer.
Figure 1. Propofol alleviates cell proliferation, migration, invasion and aerobic glycolysis in lung cancer in vitro. (a) Cell viability was detected using CCK-8 assay in A549 and H1299 cells treated with different concentrations of propofol (2, 4, 6, 8, and 10 µg/mL). (b) Cell viability was detected using CCK-8 assay in A549 and H1299 cells treated with 8 µg/mL propofol for 24, 48, and 72 h. A549 and H1299 cells were treated with 8 µg/mL propofol for 48 h. (c) Cell proliferation was analyzed using BrdU assay. (d,e) Cell migration and invasion were measured by transwell assay. (f,g) The glucose consumption and lactate production were determined by a glucose and lactate assay kit. (h) Extracellular acidification measurements were performed by Seahorse XF24 Extracellular Flux Analyzer. (i) Western blot was used to examine the protein expression of GLUT1, PKM2, HK2 and LDHA in A549 and H1299 cells. *P < 0.05.
Propofol inhibits the expression of circTADA2A in lung cancer cell lines
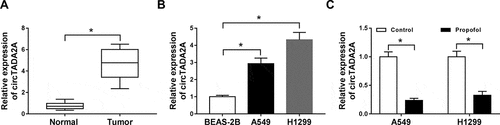
Immediately, we explored whether the expression of circTADA2A was impacted by propofol in lung cancer. First, qRT-PCR analysis showed that circTADA2A was significantly up-regulated in lung cancer tissues and cell lines compared with the controls (,b)). Next, we observed that propofol treatment markedly reduced the level of circTADA2A in lung cancer cells ()), indicating the potential involvement of circTADA2A in propofol-mediated regulation on lung cancer cell tumorigenesis and aerobic glycolysis.
Figure 2. Propofol inhibits the expression of circTADA2A in lung cancer cell lines. (a,b) The expression of circTADA2A in lung cancer tumor tissues and normal tissues, as well as lung cancer cell lines and normal BEAS-2B cell line was measured using qRT-PCR. (c) The expression of circTADA2A in A549 and H1299 cells treated with or without propofol was detected by qRT-PCR. *P < 0.05.
Propofol mitigates cell tumorigenesis and aerobic glycolysis in lung cancer by regulating circTADA2A
To confirm whether circTADA2A participated in propofol-induced inhibition on lung cancer progression, A549 and H1299 cells were transfected with pcDNA-NC or pcDNA-circTADA2A prior to propofol exposure; then a prominent up-regulation of circTADA2A expression was observed in pcDNA-circTADA2A groups ()). Subsequently, CCK-8 and BrdU assay showed that circTADA2A overexpression reversed propofol exposure-induced inhibition on the proliferation of A549 and H1299 cells (,c)). Meanwhile, transwell assay manifested that the migrated and invaded A549 and H1299 cells were decreased by propofol exposure, which was reversed by the up-regulation of circTADA2A (,e)). Besides that, circTADA2A up-regulation attenuated propofol-mediated decrease of glucose uptake ()), lactate production ()), ECA ()) as well as the protein levels of GLUT1, PKM2, HK2, and LDHA (,j)) in A549 and H1299 cells. Taken together, propofol inhibited cell tumorigenesis and aerobic glycolysis in lung cancer by regulating circTADA2A.
Figure 3. Propofol mitigates cell tumorigenesis and aerobic glycolysis in lung cancer by regulating circTADA2A. A549 and H1299 cells were transfected with pcDNA-NC or pcDNA-circTADA2A prior to propofol exposure. (a) The transfection efficiency was determined using qRT-PCR. (b,c) Cell proliferation was analyzed using CCK-8 and BrdU assay. (d,e) Transwell assay was applied to detect cell migration and invasion. (f,g) The glucose consumption and lactate production were assessed using a glucose and lactate assay kit. (h) Extracellular acidification measurements were conducted by Seahorse XF24 Extracellular Flux Analyzer. (I. J) The levels of GLUT1, PKM2, HK2, and LDHA protein in A549 and H1299 cells were examined by western blot. *P < 0.05.
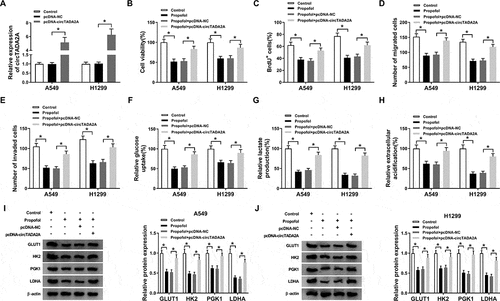
CircTADA2A is a sponge of miR-455-3p in lung cancer cells
Since the regulatory roles of circTADA2A in propofol-induced lung cancer cells were validated, the mechanisms underlying circTADA2A function were then investigated. Through searching online software program StarBase3.0, we found miR-455-3p had the potential to bind to circTADA2A ()). Subsequently, the reduction of luciferase activity in A549 and H1299 cells co-transfected with circTADA2A-WT and miR-455-3p confirmed the interaction between circTADA2A and miR-455-3p (,c)). Additionally, we also discovered that miR-455-3p expression was increased by circTADA2A deletion ()), but was decreased by circTADA2A overexpression ()) in A549 and H1299 cells. Therefore, circTADA2A targetedly repressed miR-455-3p in lung cancer cells. Moreover, qRT-PCR analysis also exhibited that miR-455-3p was decreased in lung cancer tissues ()) and cells ()); more importantly, miR-455-3p was up-regulated by propofol treatment in A549 and H1299 cells ()), indicating the possible involvement of miR-455-3p in the action of propofol on lung cancer progression.
Figure 4. CircTADA2A is a sponge of miR-455-3p in lung cancer cells. (a) The potential binding sites between circTADA2A and miR-455-3p were predicted by StarBase3.0 program. (b,c) Luciferase activity of A549 and H1299 cells co-transfected with circTADA2A-WT or circTADA2A-MUT and miR-455-3p mimics or miR-NC was examined by dual-luciferase reporter assay. (d,e) qRT-PCR analysis of miR-455-3p expression in A549 and H1299 cells transfected with si-NC, si-circTADA2A, pcDNA-circTADA2A, or pcDNA-NC was conducted. (f,g) The expression of miR-455-3p in lung cancer tumor tissues and normal tissues, as well as lung cancer cell lines and normal BEAS-2B cell line was measured using qRT-PCR. (h) The level of miR-455-3p was examined using qRT-PCR in A549 and H1299 cells treated with propofol. *P < 0.05.
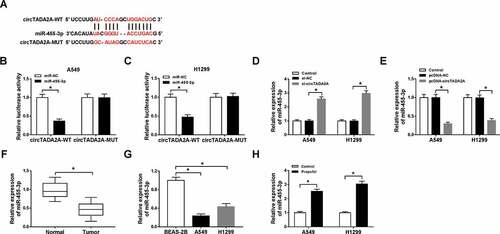
Propofol performs anti-tumor effects by regulating circTADA2A/miR-455-3p axis in lung cancer cells
Further experiments were conducted to probe the impact of circTADA2A/miR-455-3p axis on propofol-stimulated inhibition of lung cancer progression. First, A549 and H1299 cells were transfected with pcDNA-NC, pcDNA-circTADA2A, pcDNA-circTADA2A + miR-NC, or pcDNA-circTADA2A + miR-455-3p, and we found miR-455-3p expression was decreased by circTADA2A overexpression, while was rescued by miR-455-3p up-regulation after transfection ()). Then transfected cells were treated with propofol and rescue assay was conducted. Data in e) showed miR-455-3p re-expression rescued circTADA2A up-regulation-induced promotion on the proliferation (,c)), migration and invasion (,e)) of propofol-treated A549 and H1299 cells. Meanwhile, glucose metabolism analysis indicated miR-455-3p re-expression attenuated circTADA2A overexpression-induced increase of glucose uptake ()), lactate production ()), ECA ()) and the protein levels of GLUT1, PKM2, HK2, and LDHA (,j)) in propofol-exposed A549 and H1299 cells. Altogether, propofol suppressed cell tumorigenesis and aerobic glycolysis by regulating circTADA2A/miR-455-3p axis in lung cancer.
Figure 5. Propofol performs anti-tumor effects by regulating circTADA2A/miR-455-3p axis in lung cancer cells. A549 and H1299 cells were transfected with pcDNA-NC, pcDNA-circTADA2A, pcDNA-circTADA2A + miR-NC, or pcDNA-circTADA2A + miR-455-3p before propofol treatment. (a) The transfection efficiency was analyzed using qRT-PCR. (b,c) Cell proliferation was analyzed using CCK-8 and BrdU assay. (d,e) Cell migration and invasion were detected by transwell assay. (f,g) The glucose consumption and lactate production were examined with a glucose and lactate assay kit. (h) Seahorse XF24 Extracellular Flux Analyzer was applied to measure extracellular acidification. (i,j) The levels of GLUT1, PKM2, HK2, and LDHA protein in A549 and H1299 cells were examined by western blot. *P < 0.05.
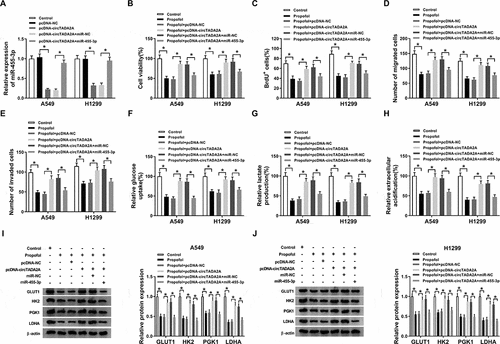
MiR-455-3p is a target of FOXM1 in lung cancer cells
By searching the StarBase3.0 program, we identified that miR-455-3p comprised the binding sites in FOXM1 3’UTR ()). The next dual-luciferase reporter assay proved the direct interaction between miR-455-3p and FOXM1 due to the decline of luciferase activity in A549 and H1299 cells co-transfected with FOXM1 3’UTR-WT and miR-455-3p (,c)). After that, expression analysis showed FOXM1 expression was elevated in lung cancer tissues (,e)) and cell lines (,g)) both at protein and mRNA levels. Moreover, propofol treatment decreased FOXM1 expression in A549 and H1299 cells (,i)), suggesting FOXM1 might also participate in the action of propofol on lung cancer progression. Additionally, A549 and H1299 cells were tran sfected with anti-miR-NC, anti-miR-455-3p, anti-miR-455-3p + si-NC, anti-miR-455-3p + si-FOXM1 before propofol treatment, and we found miR-455-3p inhibition increased FOXM1 expression, while this increase was reversed by FOXM1 deletion in A549 and H1299 cells (,k)), indicating miR-455-3p negatively regulated FOXM1 expression.
Figure 6. MiR-455-3p is a target of FOXM1 in lung cancer cells. (a) The potential binding sites between FOXM1 and miR-455-3p were predicted through searching StarBase3.0 program. (b,c) Luciferase activity of A549 and H1299 cells co-transfected with FOXM1 3’UTR-WT or FOXM1 3’UTR-MUT and miR-455-3p mimics or miR-NC was determined by dual-luciferase reporter assay. (dg) The expression of FOXM1 in lung cancer tumor tissues and normal tissues, as well as lung cancer cell lines and normal BEAS-2B cell line was measured using qRT-PCR and western blot. (h,i) The level of FOXM1 was examined using qRT-PCR and western blot in A549 and H1299 cells treated with propofol. (j,k) The level of FOXM1 was examined using qRT-PCR and western blot in A549 and H1299 cells transfected with anti-miR-NC, anti-miR-455-3p, anti-miR-455-3p + si-NC, anti-miR-455-3p + si-FOXM1. *P < 0.05.
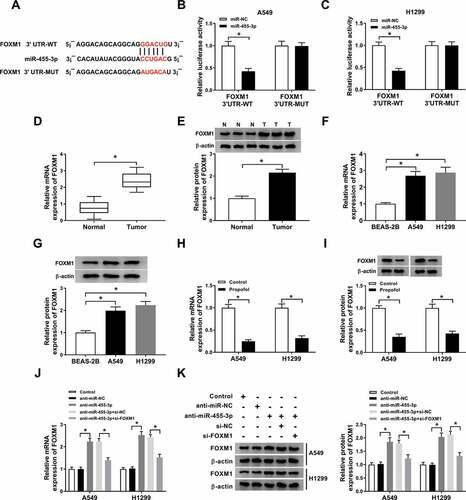
Propofol inhibits cell tumorigenesis and aerobic glycolysis by regulating miR-455-3p/FOXM1 axis in lung cancer
According to the regulatory relationship between propofol and miR-455-3p or FOXM1 expression, we wanted to know whether the miR-455-3p/FOXM1 axis affected the effects of propofol on the inhibition of cell tumorigenesis and aerobic glycolysis. Results showed miR-455-3p inhibition reversed propofol treatment-induced inhibition on A549 and H1299 cell proliferation (,b)), migration and invasion (,d)), while these reverses were abated by following FOXM1 silence (d)). After that, we found that the reduction of glucose uptake ()), lactate production ()), ECA ()) and the protein levels of GLUT1, PKM2, HK2 and LDHA (,i)) mediated by propofol treatment in A549 and H1299 cells was overturned by miR-455-3p inhibition, which was rescued by following FOXM1 deletion. These data revealed that propofol affected cell tumorigenesis and aerobic glycolysis through regulating miR-455-3p/FOXM1 axis in lung cancer.
Figure 7. Propofol inhibits cell tumorigenesis and aerobic glycolysis by regulating miR-455-3p/FOXM1 axis in lung cancer. A549 and H1299 cells transfected with anti-miR-NC, anti-miR-455-3p, anti-miR-455-3p + si-NC, anti-miR-455-3p + si-FOXM1 before propofol treatment. (a,b) The analysis of A549 and H1299 cell proliferation was conducted by CCK-8 and BrdU assay. (c,d) The migrated and invaded A549 and H1299 cells were measured using transwell assay. (e,f) The glucose consumption and lactate production were examined with a glucose and lactate assay kit. (g) Extracellular acidification measurements were carried out by Seahorse XF24 Extracellular Flux Analyzer. (h,i) Western blot analysis of GLUT1, PKM2, HK2, and LDHA protein in A549 and H1299 cells was conducted. *P < 0.05.
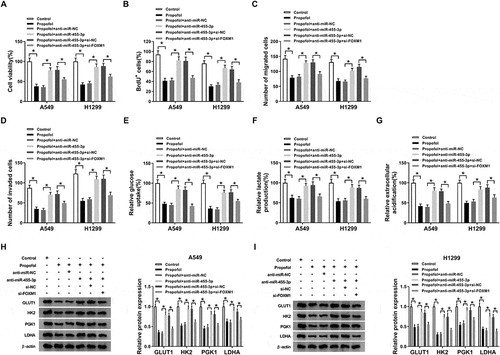
CircTADA2A indirectly regulates FOXM1 expression by binding to miR-455-3p in lung cancer cells
In view of the interaction between miR-455-3p and circTADA2A or FOXM1, we speculated that circTADA2A could affect FOXM1 expression through the regulation of miR-455-3p. A549 and H1299 cells were transfected with miR-NC, miR-455-3p, miR-455-3p + pcDNA-NC, pcDNA-circTADA2A, then western blot analysis exhibited that the expression of FOXM1, both at protein and mRNA levels, was inhibited by miR-455-3p overexpression, while this condition was restored by circTADA2A up-regulation (,b)). Therefore, we considered that circTADA2A could regulate FOXM1 expression through miR-455-3p in lung cancer cells.
Figure 8. CircTADA2A indirectly regulates FOXM1 expression by binding to miR-455-3p in lung cancer cells. A549 and H1299 cells were transfected with miR-NC, miR-455-3p, miR-455-3p + pcDNA-NC, pcDNA-circTADA2A. (a,b) The protein and mRNA expression of FOXM1 was detected using qRT-PCR and western blot. P < 0.05.
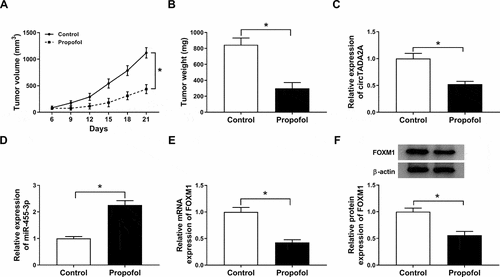
Propofol inhibits tumor growth in vivo
We further established mouse xenograft models to determine the role of propofol in tumor growth in vivo. We found propofol injection inhibited tumor growth in vivo, reflected by the decrease of tumor volume and weight in propofol treatment groups (,b)). Subsequently, molecular analysis displayed that propofol treatment down-regulated circTADA2A ()) and FOXM1 (,f)) expression, while up-regulated miR-455-3p expression ()) in the tumor masses. Collectively, propofol might suppress tumor growth in vivo by regulating circTADA2A/miR-455-3p/ FOXM1 axis.
Discussion
Growing evidence discloses that propofol has neuroprotective, anti-anxiety, anti-oxidation and anti-cancer roles in pathogenic situations and significantly affects the process of cancer progression [Citation11,Citation29]. In lung cancer, the involvement of propofol in tumor progression has been identified. For example, propofol inhibited cell invasion in lung cancer through the down-regulation of matrix metalloproteinase-9 and aquaporin-3 [Citation30]. Propofol restrained the proliferation, migration and invasion of lung cancer cells by decreasing the expression of miR-372 [Citation31]. In this study, propofol was found to inhibit cell proliferation, migration and invasion in lung cancer in vitro, which was consistent with previous studies [Citation30,Citation31]. Additionally, propofol treatment also inhibited tumor growth in vivo.
Glucose metabolism is a major source of energy, tumor cells have been reported to preferably take adenosine triphosphate (ATP) by aerobic glycolysis rather than oxidative phosphorylation despite sufficient oxygen [Citation32]. This metabolic switch has been demonstrated in lung cancer contexts and shown to be regulated by many oncoproteins to impact the proliferation, migration and metastasis of lung cancer cells [Citation33]. More importantly, a recent study exhibited that propofol could repress aerobic glycolysis in colorectal cancer cells via inactivating the NMDAR-CAMKII-ERK pathway [Citation34]. Thus, it is imperative to uncover the role of propofol in aerobic glycolysis of lung cancer cells. This study demonstrated that the glucose uptake, lactate production and extracellular acidification was remarkably down-regulated by propofol treatment in lung cancer cells. Meanwhile, all three glycolysis enzymes, HK2, PKM2, and LDHA and a glucose transporter GLUT1 were also decreased by propofol treatment in lung cancer cells. All these results indicated propofol treatment suppressed cell aerobic glycolysis in lung cancer.
Propofol often modulates cancer malignant in direct and indirect methods, propofol influences key RNAs and signaling pathways to conduce to cancer development, or controls host immune function to affect the degree of immunosuppression [Citation35]. Thus, it is of great clinical significance to detect the effects of propofol on lung cancer progression, including the regulation of circRNAs. CircTADA2A has been found to serve as an oncogene in NSCLC tumorigenesis [Citation28]. In this review, circTADA2A was elevated in lung cancer, while we observed propofol treatment decreased the expression of circTADA2A. Besides that, circTADA2A re-expression reversed propofol treatment-induced suppression on cell tumorigenesis and aerobic glycolysis in lung cancer. Strikingly, circRNAs can function as sponges for microRNA (miRNA) and modulate gene expression [Citation36]. In this study, we confirmed that miR-455-3p directly interacted with circTADA2A and FOXM1, and circTADA2A could regulate FOXM1 expression through binding to miR-455-3p in lung cancer cells. MiR-455-3p was demonstrated to act as a tumor suppressor in NSCLC progression through interacting with HOXB5 and was an unfavorable prognostic factor to predict poor overall survival in lung cancer [Citation37]. This study showed miR-455-3p was increased by propofol treatment, and miR-455-3p overexpression suppressed the carcinogenesis and aerobic glycolysis of cells under propofol treatment plus circTADA2A up-regulation. FOXM1 is a key transcription factor of FOX family, emerging evidence has shown FOXM1 expression is aberrantly elevated in various cancers and acts as a critical regulator in many cancers by involving in tumor cell metastasis, angiogenesis, cell cycle acceleration, and drug resistance [Citation38–40]. In lung cancer, FOXM1 was also found to be up-regulated and acted as an oncogene to contribute to cell tumorigenesis [Citation41,Citation42]. In this study, FOXM1 was elevated in lung cancer, which was consistent with the previous study; immediately, we observed propofol treatment decreased FOXM1 expression, and FOXM1 knockdown reversed miR-455-3p inhibition-mediated promotion on cell tumorigenesis and aerobic glycolysis in propofol treated-lung cancer cells.
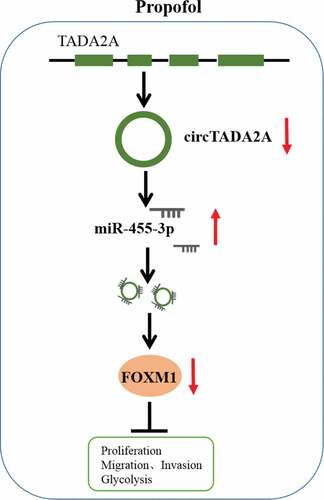
In conclusion, propofol down-regulated circTADA2A expression through miR-455-3p/FOXM1 axis to inhibit cell carcinogenesis and aerobic glycolysis in lung cancer (), suggesting an effective clinical implication for propofol to prevent the malignant behavior of lung cancer cells.
Highlight
Propofol alleviates lung cancer cell proliferation, migration, invasion, and aerobic glycolysis in vitro as well as inhibits tumor growth in vivo.
Propofol increases the level of miR-455-3p, but decreases the levels of circTADA2A and FOXM1.
MiR-455-3p directly interacts with circTADA2A and FOXM1 in lung cancer cells.
CircTADA2A indirectly regulates FOXM1 expression through binding to miR-455-3p in lung cancer.
Propofol performs anti-tumor functions by regulating circTADA2A/miR-455-3p/FOXM1 in vitro and in vivo.
Authors’ contribution
All authors made substantial contribution to conception and design, acquisition of the data, or analysis and interpretation of the data; take part in drafting the article or revising it critically for important intellectual content; gave final approval of the revision to be published; and agree to be accountable for all aspect of the work.
Ethics approval and consent to participate
The present study was approved by the ethical review committee of The First Affiliated Hospital of Zhengzhou University.
Availability of data and materials
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
Disclosure statement
The authors declare that they have no competing interests.
Additional information
Funding
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29.
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132.
- Yang B, Zhang L, Cao Y, et al. Overexpression of lncRNA IGFBP4-1 reprograms energy metabolism to promote lung cancer progression. Mol Cancer. 2017;16(1):154.
- Cassinello F, Prieto I, Del Olmo M, et al. Cancer surgery: how may anesthesia influence outcome? J Clin Anesth. 2015;27(3):262–272.
- Bajwa SJ, Anand S, Kaur G. Anesthesia and cancer recurrences: the current knowledge and evidence. J Cancer Res Ther. 2015;11(3):528–534.
- Chidambaran V, Costandi A, D’Mello A. Propofol: a review of its role in pediatric anesthesia and sedation. CNS Drugs. 2015;29(7):543–563.
- Oren-Ziv A, Hoppenstein D, Shles A, et al. Sedation methods for intra-articular corticosteroid injections in Juvenile Idiopathic Arthritis: a review. Pediatr Rheumatol Online J. 2015;13:28.
- Kushida A, Inada T, Shingu K. Enhancement of antitumor immunity after propofol treatment in mice. Immunopharmacol Immunotoxicol. 2007;29(3–4):477–486.
- Song J, Shen Y, Zhang J, et al. Mini profile of potential anticancer properties of propofol. PLoS One. 2014;9(12):e114440.
- Yu B, Gao W, Zhou H, et al. Propofol induces apoptosis of breast cancer cells by downregulation of miR-24 signal pathway. Cancer Biomark. 2018;21(3):513–519.
- Zhang W, Wang Y, Zhu Z, et al. Propofol inhibits proliferation, migration and invasion of gastric cancer cells by up-regulating microRNA-195. Int J Biol Macromol. 2018;120(Pt A):975–984.
- Yang N, Liang Y, Yang P, et al. Propofol inhibits lung cancer cell viability and induces cell apoptosis by upregulating microRNA-486 expression. Braz J Med Biol Res. 2017;50(1):e5794.
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033.
- Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21(3):297–308.
- Vander Heiden MG, DeBerardinis RJ. Understanding the intersections between metabolism and cancer biology. Cell. 2017;168(4):657–669.
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, et al. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7(1):11–20.
- Cassim S, Pouyssegur J. Tumor microenvironment: a metabolic player that shapes the immune response. Int J Mol Sci. 2019;21(1):157.
- Cassim S, Raymond VA, Dehbidi-Assadzadeh L, et al. Metabolic reprogramming enables hepatocarcinoma cells to efficiently adapt and survive to a nutrient-restricted microenvironment. Cell Cycle. 2018;17(7):903–916.
- Enzo E, Santinon G, Pocaterra A, et al. Aerobic glycolysis tunes YAP/TAZ transcriptional activity. Embo J. 2015;34(10):1349–1370.
- Ganapathy-Kanniappan S, Geschwind JF. Tumor glycolysis as a target for cancer therapy: progress and prospects. Mol Cancer. 2013;12(1):152.
- Hamanaka RB, Chandel NS. Targeting glucose metabolism for cancer therapy. J Exp Med. 2012;209(2):211–215.
- Rybak-Wolf A, Stottmeister C, Glazar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58(5):870–885.
- Zhang J, Liu H, Hou L, et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16(1):151.
- Li P, Chen H, Chen S, et al. Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br J Cancer. 2017;116(5):626–633.
- Gao D, Zhang X, Liu B, et al. Screening circular RNA related to chemotherapeutic resistance in breast cancer. Epigenomics. 2017;9(9):1175–1188.
- Yu T, Wang Y, Fan Y, et al. CircRNAs in cancer metabolism: a review. J Hematol Oncol. 2019;12(1):90.
- Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94.
- Cui J, Li W, Liu G, et al. A novel circular RNA, hsa_circ_0043278, acts as a potential biomarker and promotes non-small cell lung cancer cell proliferation and migration by regulating miR-520f. Artif Cells Nanomed Biotechnol. 2019;47(1):810–821.
- Kurt M, Bilge SS, Kukula O, et al. Anxiolytic-like profile of propofol, a general anesthetic, in the plus-maze test in mice. Pol J Pharmacol. 2003;55(6):973–977.
- Ye HJ, Bai JJ, Guo PP, et al. [Propofol suppresses invasion of human lung cancer A549 cells by down-regulating aquaporin-3 and matrix metalloproteinase-9]. Nan Fang Yi Ke Da Xue Xue Bao. 2016;36(9):1286–1290.
- Sun H, Gao D. Propofol suppresses growth, migration and invasion of A549 cells by down-regulation of miR-372. BMC Cancer. 2018;18(1):1252.
- Xu XD, Shao SX, Jiang HP, et al. Warburg effect or reverse Warburg effect? A review of cancer metabolism. Oncol Res Treat. 2015;38(3):117–122.
- Li XB, Gu JD, Zhou QH. Review of aerobic glycolysis and its key enzymes - new targets for lung cancer therapy. Thorac Cancer. 2015;6(1):17–24.
- Chen X, Wu Q, Sun P, et al. Propofol disrupts aerobic glycolysis in colorectal cancer cells via inactivation of the NMDAR-CAMKII-ERK pathway. Cell Physiol Biochem. 2018;46(2):492–504.
- Jiang S, Liu Y, Huang L, et al. Effects of propofol on cancer development and chemotherapy: potential mechanisms. Eur J Pharmacol. 2018;831:(46–51.
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388.
- Gao X, Zhao H, Diao C, et al. miR-455-3p serves as prognostic factor and regulates the proliferation and migration of non-small cell lung cancer through targeting HOXB5. Biochem Biophys Res Commun. 2018;495(1):1074–1080.
- Li Q, Zhang N, Jia Z, et al. Critical role and regulation of transcription factor FoxM1 in human gastric cancer angiogenesis and progression. Cancer Res. 2009;69(8):3501–3509.
- Monteiro LJ, Khongkow P, Kongsema M, et al. The Forkhead Box M1 protein regulates BRIP1 expression and DNA damage repair in epirubicin treatment. Oncogene. 2013;32(39):4634–4645.
- Schimmel J, Eifler K, Sigurethsson JO, et al. Uncovering SUMOylation dynamics during cell-cycle progression reveals FoxM1 as a key mitotic SUMO target protein. Mol Cell. 2014;53(6):1053–1066.
- Zhang Y, Qiao WB, Shan L. Expression and functional characterization of FOXM1 in non-small cell lung cancer. Onco Targets Ther. 2018;11:3385–3393.
- Wei P, Zhang N, Wang Y, et al. FOXM1 promotes lung adenocarcinoma invasion and metastasis by upregulating SNAIL. Int J Biol Sci. 2015;11(2):186–198.