ABSTRACT
Osteosarcoma (OS) is a serious bone malignancy commonly occurred in childhood and adolescence. Circular RNA (circRNA) is a novel endogenous RNA that may be considered as a new biomarker for diseases’ diagnosis or prognosis. This study explored the roles and mechanism of circ_0001649 in OS. The qRT-PCR was performed to test circ_0001649 expression in OS tissues and cells. Luciferase was used to confirm the binding of circ_0001649 with miR-338-5p, miR-647 and miR-942. OS cells were stably transfected with pEX-circ_0001649 or miRNAs mimic, CCK-8 kit, colony formation, apoptosis and western blot analysis were used to detect the roles of circ_0001649. Circ_0001649 was low-expressed in OS tissues and cell lines. Circ_0001649 overexpression suppressed U2OS and HOS cell viability and survival fraction, and induced apoptosis presented as the increasing levels of Apaf-1, cleaved-caspase-3 and cleaved-caspase-9. Further, circ_0001649 worked as a sponge to absorb miR-338-5p, miR-647 and miR-942 to suppress cell proliferation, induce apoptosis and inhibit STAT pathway. Circ_0001649 suppressed OS cell proliferation and STAT pathway and induced apoptosis through sponging miR-338-5p, miR-647 and miR-942.
Introduction
Osteosarcoma (OS) is an osteoid-producing malignant sarcoma that usually occurs in pediatric sufferers. It is the most frequent original malignant tumor in the bone [Citation1]. The clinical features are pain at the tumor sites, limited joint activity and muscle atrophy. OS is the aggressive cancer, resulting in metastases in the lung to affect respiratory function and it is related to poor prognosis [Citation2,Citation3]. The treatments of OS include diverse combinations of surgical therapy and high toxicity chemotherapeutic agents, which are mainly doxorubicin, ifosfamide, methotrexate and cisplatin [Citation4,Citation5]. Although some achievements have been received through updated therapy especially the exploitation of effective chemotherapeutic agents, the effect of OS patients is still unsatisfactory. Over 20% of OS patients die of illness due to anti-tumor drugs, tumor metastasis and untreated tumor [Citation5]. The main factors affecting OS treatment are metastatic stage, not obvious detectable metastases clinically and chemotherapy resistance [Citation6]. Therefore, the development of therapeutic targets, the use of biomarkers to identify OS and the development of new methods for adjuvant therapy are new research direction [Citation7].
Circular RNAs (circRNAs) are a special type of endogenous non-coding RNAs, which are produced by genomic transcripts through joining the 5' and 3' ends closed loops and have scant or no protein coding potential [Citation8]. The latest data shows that circRNA is differentially expressed in many malignancies and may be considered as a new biomarker for diseases’ diagnosis or prognosis [Citation9,Citation10]. The key functions of circRNAs in various pathological processes have been observed, such as the regulation of proliferation and metastasis in cancer cells [Citation11]. Circ_0001649 is a novel identified circRNA and is reduced in cholangiocarcinoma [Citation12]. It is a transcription outcome of a tumor inhibitor gene, which is called Snf2 Histome Linker Phd Ring Helicase (SHPRH). Many researches have reported that circ_0001649 works as a tumor inhibitor in many malignancies, such as colorectal cancer and hepatocellular carcinoma [Citation13,Citation14]. But the roles of circ_0001649 in OS are still unknown.
It has been proved that some circRNAs are considered as miRNA sponges and negatively regulate miRNAs through absorbing and insulating miRNAs [Citation15]. For example, circRNA ciRS-7 exerts over 70 conventional miR-7-binding sites and is proved to be a miRNA suppressor [Citation16]. Circ_0001649 was predicted to exert a latent function in carrying some miRNAs, such as miR-1283, miR-182-3p, miR-4502, miR-4310 and miR-888-3p [Citation17]. In this study, we aimed to explore the roles of circ_0001649 in OS and its mechanism.
Materials and methods
Tissues
Twenty-six samples of OS and paired adjacent non-tumor tissues were got from Yantai Affiliated Hospital of Binzhou Medical University (Yantai, China). All experiments were approved by the ethics committee of Yantai Affiliated Hospital of Binzhou Medical University.. Written informed consent was got from every patient before the start of our research.
Cell
Human OS cell lines (MG63, U2OS, HOS, Saos-2) were bought from ATCC (Manassas, VA, USA) and were cultured in RPMI 1640 (Gibco, Grand Island, NY, USA) with 10% fetal bovine serum (FBS, HyClone, Logan, UT, USA) and 1% streptomycin/penicillin in 37°C. Normal cell line hFOB1.19 was also bought from ATCC and was kept in DMEM/Ham’s F-12 (Sigma-Aldrich, St. Louis, MO, USA) with 10% FBS and Geneticin (400 μg/ml) in 34°C with 5% CO2.
Transfection
Amplified circ_0001649 sequences were cloned into pEX vector (GenePharma, Shanghai, China), referring as pEX-circ_0001649. Empty vector was worked as negative control (NC), referring as pEX. Si-circ_0001649, miR-338-5p mimic, miR-647 mimic, miR-942 mimic and related NC were synthesized by GenePharma. Transfections were done through Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA). 48 h post-transfection was considered as the harvest time in the subsequent experiments. The sequences of pEX-circ_0001649, miR-338-5p mimic, miR-647 mimic, miR-942 mimic and related NC are listed in .
Table 1. Sequences used in cell transfection
qRT-PCR
Total RNA was extracted from cells through Trizol Reagent (Invitrogen). For circ_0001649 detection, a script RT reagent kit and SYBR Premix Ex Taq II (all from TaKaRa, Dalian, China) were used to do loading; then, reactions were performed on a Roche LightCycler® 480II PCR instrument (Basel, Switzerland) according to producer’s instructions. GAPDH was worked as internal control. For miRNAs detection, cDNA was compounded through universal cDNA synthesis kit (Exiqon, Vedbaek, Denmark); then, qRT-PCR was done through ExiLERATE SYBR® Green Master Mix kit (Exiqon). U6 was worked as internal control.
Cell counting kit-8 (CCK-8) experiment
During the detection of cell viability, a CCK-8 assay kit (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was used according to the producer’s introductions. Cells at a density of 1000 cells/well were seeded in 96-well plates. 10 µl CCK-8 solution was added to every well, and cells were cultured in a 37°C wet environment with 95% air and 5% CO2 for 1 h. Absorbance at 450 nm was measured through a microplate reader (Tecan, Männedorf, Switzerland).
Colony formation experiment
Transfected U2OS and HOS cells, at a density of 1.5 × 102 cells/well, were seeded in 6-well plates and kept in medium with 10% FBS for 1 week. Then, phosphate-buffered saline (PBS) was used to clean the cells, which were fixed with 4% paraformaldehyde for 20 min and stained by 0.5% crystal violet solution for 15 min. Number of colonies was counted under an inverted microscope (Leica DMI3000B, Leica Microsystems CMS GmbH, Wetzlar, Germany).
Apoptosis experiment
After centrifuging, cells were cleaned by PBS, seeded in 6-well plates (4 × 105 per well) and kept in FBS-free medium for 12 h. When the degree of fusion was 80%, cells were digested with 0.25% trypsin into single cells. After centrifuging, cells were re-suspended through 1 × binding buffer. Then, cells were stained by FITC Annexin V Apoptosis Detection Kit (BD Biosciences, San Jose, CA, USA). Apoptosis cells were measured through Flow cytometry analysis (FACScan, BD Biosciences).
Dual-luciferase reporter experiment
Wild-type plasmid circ_0001649-WT and mutant-type plasmid circ_0001649-MUT were inserted into pGL3 promoter vector (GenePharma). 293 T cells were co-transfected 50 nM miRNAs mimics or NC and 80 ng plasmid constructed well through Lipofectamine 3000. Data were obtained by dual-luciferase reporter assay kit (Promega, Madison, WI, USA).
Western blot
RIPA Lysis Buffer (MA0151, Dalian Meilun Biotechnology Co., Ltd., China) with the protease inhibitor cocktails (FD1001, Fudebio, Hangzhou, China) on ice was used to get protein samples, which were isolated through SDS-PAGE and then the gel was transferred onto PVDF membranes. Then, 5% skim milk powder was used to block the membranes in the room for 1 h and then the membranes were combined with primary antibodies at 4°C all night. The next day, the membranes were washed and incubated with secondary antibodies carrying HRP-conjugates. Bands were examined through an enhanced chemiluminescence kit (FD8030, Fudebio).
Statistics
Graphpad 6.0 statistical software (GraphPad Software Inc., La Jolla, CA, USA) was employed for data analyses. t-test or one-way ANOVA was used to test the differences. A P < 0.05 was regarded statistically notable.
Results
Circ_0001649 was low-expressed in OS
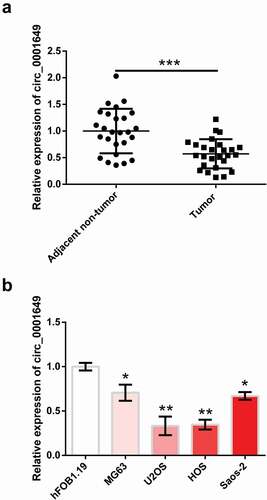
First, the relationship between circ_0001649 and OS was examined through qRT-PCR. ) shows that level of circ_0001649 was observably reduced in OS tissues contrasted with that in adjacent normal tissues (P < 0.001). Besides, circ_0001649 level was also notably reduced in OS cell lines MG63 (P < 0.05), U2OS (P < 0.01), HOS (P < 0.01) and Saos-2 (P < 0.05, )) compared with hFOB1.19. U2OS and HOS cells were selected as follow-up study cells because the difference was most significant.
Circ_0001649 overexpression suppressed OS cells growth
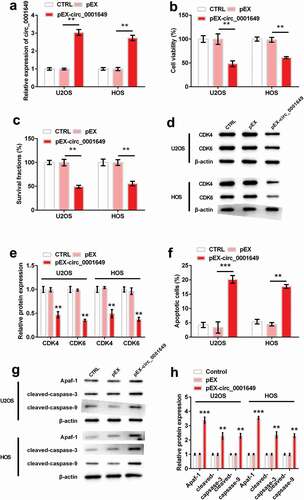
Circ_0001649 expression vector (pEX-circ_0001649), which could stably up-regulated the expression of circ_0001649, was employed to study the role of circ_0001649 in OS cells U2OS and HOS. ) showed the notable overexpression of circ_0001649 in both cells after transfection with pEX-circ_0001649 (both P < 0.01). The viabilities of both cells were significantly reduced by pEX-circ_0001649 transfection (both P < 0.01, )). And the same trends were occurred for survival fraction (both P < 0.01, )). Similarly, levels of cyclin-dependent kinase 4 (CDK4) and CDK6 were notably reduced when circ_0001649 was overexpressed in both cells (all P < 0.01, ). Besides, for apoptosis, circ_0001649 overexpression significantly increased apoptosis (P < 0.001 and P < 0.01, )), and levels of apoptosis-related factors (Apaf-1, cleaved-caspase-3 and cleaved-caspase-9) were also increased in both cells (both P < 0.001, P < 0.01 and P < 0.01, )).
Figure 2. Circ_0001649 overexpression inhibited the growth of U2OS and HOS cells, which were transfected with pEX-circ_0001649. (a) Expression of circ_0001649 was detected by qRT-PCR. Viability (b) and survival fraction (c) were detected by CCK-8 and colony formation assay. (d–e) Levels of CDK4 and CDK6 were detected by western blot analysis. (f) Apoptosis was detected by flow cytometry. (g–h) Levels of apoptosis-related factors were detected by western blot analysis. * P < 0.05, ** P < 0.01 and *** P < 0.001 contrasted with indicated group
Circ_0001649 could sponge multiple miRNAs
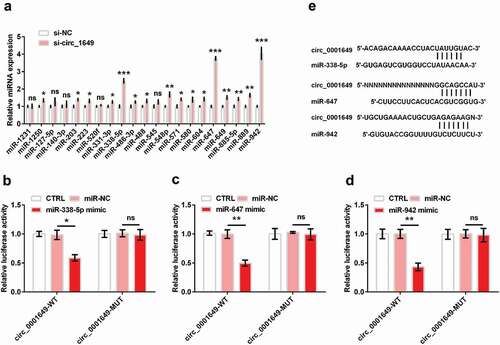
Many studies have found that circRNAs can sponge miRNAs and regulate the roles of miRNAs [Citation18,Citation19]. According to Circular RNA Interactome database (https://circinteractome.nia.nih.gov/), 21 top miRNAs were predicted to be the target gene of circ_0001649, including miR-1231, miR-1250, miR-127-5p, miR-140-3p, miR-203, miR-223, miR-520 f, miR-331-3p, miR-338-5p, miR-486-3p, miR-488, miR-545, miR-548p, miR-571, miR-580, miR-604, miR-647, miR-649, miR-885-5p, miR-889, miR-942. To assess the possible interaction between circ_0001649 and these miRNAs, we used qRT-PCR to detect the expression of miRNAs when circ_0001649 was knocked down in U2OS cell. We found that si-circ_0001649 could up-regulate part of the miRNAs expression ()). We chose 3 miRNA (miR-338-5p, miR-647 and miR-942, all P < 0.001) with expression folds more than 2 fold for subsequent assays. Then, we performed luciferase reporter assays to validate the target relationship in 293 T cell. showed that luciferase activities were significantly decreased in 293 T cell co-transfected with circ_0001649-WT and miRNA (miR-338-5p, miR-647 and miR-942) mimic (P < 0.05, P < 0.01 and P < 0.01), while the circ_0001649-MUT groups were no notable change of luciferase activity, suggesting the binding relationship between circ_0001649 and these three miRNAs was established. And ) displays the potential binding sequences.
Figure 3. Circ_0001649 could sponge multiple miRNAs. (a) MiRNA expression in si-circ_0001649 transfected U2OS cell was detected by qRT-PCR. Luciferase activity experiment detected target relationship between circ_0001649 and miR-338-5p (b), miR-647 (c) and miR-942 (d), respectively. (e) The potential sequences of circ_0001649 and miR-338-5p, miR-647 and miR-942, respectively. * P < 0.05, ** P < 0.01 and *** P < 0.001 contrasted with indicated group
Circ_0001649 suppressed OS cells proliferation through sponging multiple miRNAs
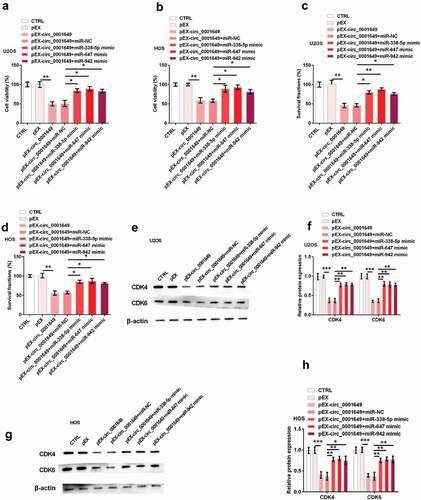
To investigate whether circ_0001649 functions by interacting with miR-338-5p, miR-647 and miR-942, we did co-transfection assays. U2OS and HOS cells were both co-transfected with pEX-circ_0001649 and miRNA (miR-338-5p, miR-647 and miR-942, respectively) mimic. showed that cell viabilities in both cells were notably inhibited by pEX-circ_0001649 (both P < 0.01), while were significantly restored by the adding of miR-338-5p mimic, miR-647 mimic and miR-942 mimic (all P < 0.05). Similarly, the reducing survival fractions of both cells were notably raised after co-transfection with miR-338-5p mimic (both P < 0.05), miR-647 mimic (P < 0.01 and P < 0.05) and miR-942 mimic (both P < 0.05), respectively (). And the decreasing levels of CDK4 and CDK6 were also significantly raised in U2OS (CDK4: all P < 0.01; CDK6: all P < 0.01) and HOS (CDK4: P < 0.01, P < 0.01 and P < 0.05; CDK6: all P < 0.01) cells by these miRNAs mimic (). These results indicated that circ_0001649 inhibited OS cell proliferation through sponging miR-338-5p, miR-647 and miR-942.
Figure 4. Circ_0001649 inhibited U2OS and HOS cells proliferation through sponging miR-338-5p, miR-647 and miR-942 after cells were co-transfected with pEX-circ_0001649 and miRNAs mimic. Cell viability was detected through CCK-8 in U2OS (a) and HOS (b) cells. Survival fraction was tested through colony formation in U2OS (c) and HOS (d) cells. Expression of CDK4 and CDK6 was detected through western blot and western blot quantitation in U2OS (e, f) and HOS (g, h) cells. * P < 0.05 and ** P < 0.01 contrasted with indicated group
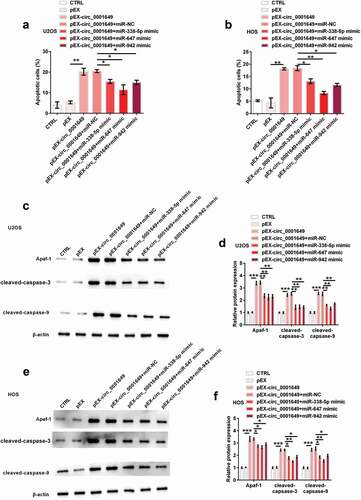
Circ_0001649 promoted OS cells apoptosis through sponging multiple miRNAs
Besides, indicated that apoptosis was significantly increased by pEX-circ_0001649 (both P < 0.01), while was obviously reduced after co-transfection with miR-338-5p mimic (both P < 0.05), miR-647 mimic (P < 0.01 and P < 0.05) and miR-942 mimic (both P < 0.01), respectively, in U2OS and HOS cells. Similarly, the increasing levels of Apaf-1 (U2OS: all P < 0.01; HOS: all P < 0.05), cleaved-caspase-3 (U2OS: all P < 0.01; HOS: P < 0.05, P < 0.05 and P < 0.05) and cleaved-caspase-9 (U2OS: all P < 0.01; HOS: P < 0.05, P < 0.05 and P < 0.05) were significantly reduced by the adding of miR-338-5p mimic, miR-647 mimic and miR-942 mimic in both cells (). These results indicated that circ_0001649 promoted OS cell apoptosis through sponging miR-338-5p, miR-647 and miR-942.
Figure 5. Circ_0001649 promoted U2OS and HOS cells apoptosis through sponging miR-338-5p, miR-647 and miR-942 after cells were co-transfected with pEX-circ_0001649 and miRNAs mimic. Apoptosis was detected through flow cytometry in U2OS (a) and HOS (b) cells. Expression of apoptosis-related factors was detected through western blot and western blot quantitation in U2OS (c, d) and HOS (e, f) cells. * P < 0.05, ** P < 0.01 and *** P < 0.001 contrasted with indicated group
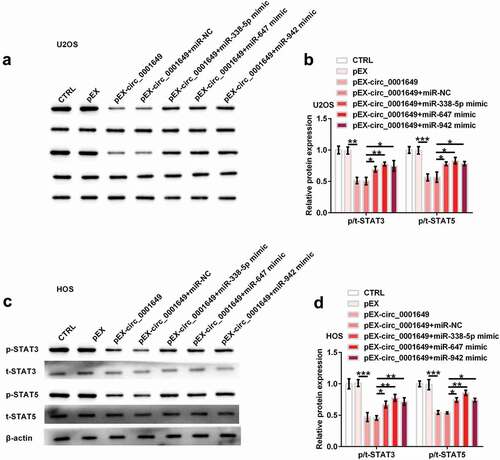
Circ_0001649 suppressed signal transducers and activators of transcription (STAT) signal pathway through sponging multiple miRNAs
STAT signal pathway takes important roles in cell proliferation and differentiation [Citation20,Citation21]. STAT3 and STAT5 mediate various cellular procedures and regulate gene expression [Citation22]. We detected the effects of circ_0001649 on STAT signal pathway through western blot analysis. As shown in , levels of STAT3 and STAT5 were significantly reduced by pEX-circ_0001649 (both P < 0.01), while were notably raised after co-transfection with miR-338-5p mimic (both P < 0.05), miR-647 mimic (P < 0.01 and P < 0.05) and miR-942 mimic (both P < 0.05), respectively, in U2OS cell. Similar trends occurred in HOS cell (). So, circ_0001649 inhibited STAT signal pathway through sponging miR-338-5p, miR-647 and miR-942.
Figure 6. The inhibition of STAT signal pathway by circ_0001649 was detected after co-transfected with pEX-circ_0001649 and miRNAs mimic in U2OS and HOS cells. Expression of STAT3 and STAT5 was detected through western blot analysis (a, b) in U2OS cell. Expression of STAT3 and STAT5 was detected through western blot analysis (c, d) in HOS cell. * P < 0.05, ** P < 0.01 and *** P < 0.001 contrasted with indicated group
Discussion
Recently, increasing evidence has shown the emerging vital function of circRNAs in human diseases, especially, cancers. With the development of RNA-seq, many circRNAs have been proved to be incvolved in lung cancer, hepatocellular carcinoma and colorectal cancer, offering prospective molecular biomarkers and treating goals for human diseases [Citation23–26]. OS is the most frequent primary bone tumor with high incidence in children and brings out poor prognosis [Citation27]. Though OS patients are cured through multi-agent chemotherapy and surgery, overall survival has not improved significantly, and the 5-year survival rates remain low [Citation28]. Thus, it is important to explore key biomolecules, therapeutic targets or tumor-related pathways. In our research, we observed that circ_0001649 could sponge miR-338-5p, miR-647 and miR-942 to inhibit OS cell growth and the activation of STAT signal pathway, which a possible method for OS treatment.
In tumor study areas, according to high-throughput sequencing and microarray analysis, circRNA has been gradually discovered and reported [Citation29]. Recently, many studies have indicated the key roles of circRNA in various biological procedures, such as the progression and metastasis of cancers [Citation30,Citation31]. Liu et al. found that lots of circRNAs were expressed differentially in OS cell lines and relative non-tumor cells [Citation32]. But the roles of these circRNAs are largely unclear. Here, we focused on circ_0001649, which was reported to down-regulate in some malignancies. For example, Xu et al. reported that circ_0001649 was down-regulated in cholangiocarcinoma tissues, being related to the unhealthy phenotype and influencing the growth capacities of cholangiocarcinoma cells [Citation12]. Wang et al. found that expression of circ_0001649 was reduced in glioma tissues and cells, which was associated with larger tumor size and poor overall survival in glioma patients, suggesting the inhibition roles of circ_0001649 in glioma [Citation33]. As we expected, circ_0001649 was low-expressed in OS tissues and cells. Further experiments indicated that circ_0001649 decelerated viability, survival fraction and induced apoptosis and activated STAT pathway. What is more, circ_0001649 played its anti-tumor roles through sponging miRNAs. These findings revealed that circ_0001649 might work as a tumor inhibitor in OS.
Increasing studies have shown that circRNAs work as miRNA sponges to regulate the transcription and post-transcription level [Citation34]. For example, circ-ITCH inhibited Wnt/β pathway through sponging miR-17, miR-214 and miR-7 [Citation35]. Here, we firstly found 21 miRNAs to circ_0001649 via online prediction ()). Among them, miR-338-5p, miR-647 and miR-942 were significantly expressed. The tumor promotion roles of these three miRNAs have been reported in the previous studies. For example, miR-338-5p could elevate melanoma progression through regulating CD82 expression [Citation36]. MiR-647 was demonstrated to elevate the proliferation and migration of colorectal cancer cells through targeting nuclear factor I/X [Citation37]. MiR-942 was proved to elevate the development of esophageal squamous cell carcinoma tumors, and the incidence of esophageal squamous cell carcinoma was positively correlated with miR-942 expression [Citation38]. Therefore, it will be interesting to explore the regulatory mechanism between circ_0001649 and these miRNAs. For the first time, we found thatcirc_0001649 suppressed OS development through sponging miR-338-5p, miR-647 and miR-942, which were consistent with the tumor-promoting roles of these three miRNAs. But, miR-338-5p and miR-647 were also reported to play tumor inhibitor roles in some tumor types, such as glioblastoma [Citation39] and glioma [Citation40]. This phenomenon might depend on different cell situations. More work is needed to explore the functional mechanism between circ_0001649 and miR-338-5p/miR-647 in diverse cells to provide more evidence for the regulation of biological processes by circ_0001649.
Dysregulation of cell cycle mechanisms is a basic process of tumor growth [Citation41,Citation42]. CDKs are participated in controlling the progression of the whole cell cycle through their association with cyclins. The binding of CDK4 and CDK6 to cyclin D is essential for G1 progression [Citation43]. Some inhibitors of CDK4 and CDK6 can be employed as potential cancer therapeutics [Citation44]. It has been reported that lncRNA HOTTIP could up-regulate the expression of cyclin D1 and CDK4 and β-catenin in OS cell line MG63 cells to play tumor-promoting roles [Citation45]. Accumulating studies show that STAT pathway is involved in the progresses of many malignant tumors [Citation46–48]. STAT3 and STAT5 are of great attracted because they are not only actuated through various ligands controlling survival, proliferation and so on, but also their dysregulation elevates tumor progresses in human cancers [Citation49,Citation50]. In OS progression, the up-regulation of protein inhibitor of activated STAT (PIAsxα) played inhibitory roles in the progression of OS cells through the cyclin D and CDK pathway. PIAsxα inhibited the expression of CDK4, CDK6 and CDK8, which are cell cycle regulators through modulating DNA synthesis and the progression between G1 and S stages, suggesting that PIAsxα might be a potential novel clinical treatment for OS [Citation51]. CDK4, CDK6, STAT3 and STAT5 are interrelated in cancer progression (https://www.kegg.jp/kegg-bin/highlight_pathway?scale=1.0&map=map05200&keyword=CDK4%20CDK6). As we expected, circ_0001649 took anti-OS roles through inhibiting the activation of STAT3/5. Additionally, for the first time, the potential regulatory relationship among circ_0001649-miRNA and STAT was uncovered in this study. The activation of STAT pathway was suppressed by circ_0001649 through sponging miR-338-5p, miR-647 and miR-942, providing evidence for the anti-OS functions of circ_0001649. Additionally, JAK/STAT pathway is discovered as OS signal pathway and its activation has been reported to elevate OS development [Citation52]. Therefore, we guessed that the regulation of CDK4, CDK6, STAT3 and STAT5 might be through JAK/STAT pathway. We will verify this hypothesis and explore the mechanism of circ_0001649 on the target proteins of miRNAs (miR-338-5p, miR-647 and miR-942) in the subsequent experiments.
In conclusion, our data indicate that circ_0001649 expression is notably reduced in OS tissues and cell lines. We further functionally and mechanistically evaluated circ_0001649 that it could inhibit viability and survival, induced apoptosis and activated STAT signal pathway in U2OS and HOS cells through sponging miR-338-5p, miR-647 and miR-942. Our findings are first to demonstrate the effects of circ_0001649 in OS, which might provide a new latent therapeutic goal for monitoring and treating OS. We will verify these findings in vivo animal experiments in the next step to further support the potential application of circ_0001649 in OS.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This research was ratified via the Medical Ethics Committee of Yantai Affiliated Hospital of Binzhou Medical University (Yantai, China).
Disclosure statement
The authors declare that they have no conflict of interest.
Data availability statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Additional information
Funding
References
- Gitelis S. Bone and soft tissue tumors: clinical features, imaging, pathology and treatment. JBJS. 2001;83:801–802.
- Kansara M, Teng MW, Smyth MJ, et al. Translational biology of osteosarcoma. Nat Rev Cancer. 2014;14:722–735.
- Botter SM, Neri D, Fuchs B. Recent advances in osteosarcoma. Curr Opin Pharmacol. 2014;16:15–23.
- Luetke A, Meyers PA, Lewis I, et al. Osteosarcoma treatment – where do we stand? A state of the art review. Cancer Treat Rev. 2014;40:523–532.
- Ouyang Z, Li H, Zhai Z, et al. Zoledronic acid: pleiotropic anti-tumor mechanism and therapeutic outlook for osteosarcoma. Curr Drug Targets. 2018;19:409–421.
- Chan HS, Grogan TM, Haddad G, et al. P-glycoprotein expression: critical determinant in the response to osteosarcoma chemotherapy. J Natl Cancer Inst. 1997;89:1706–1715.
- Sizemore G, Lucke-Wold B, Rosen C, et al. Temporal lobe epilepsy, stroke, and traumatic brain injury: mechanisms of hyperpolarized, depolarized, and flow-through ion channels utilized as tri-coordinate biomarkers of electrophysiologic dysfunction. OBM Neurobiol. 2018;2:009.
- Ivanov A, Memczak S, Wyler E, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170–177.
- Abu N, Jamal R. Circular RNAs as promising biomarkers: a mini-review. Front Physiol. 2016;7:355.
- Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. 2016;32:309–316.
- Wang P, He X. Current research on circular RNAs associated with colorectal cancer. Scand J Gastroenterol. 2017;52:1203–1210.
- Xu Y, Yao Y, Zhong X, et al. Downregulated circular RNA hsa_circ_0001649 regulates proliferation, migration and invasion in cholangiocarcinoma cells. Biochem Biophys Res Commun. 2018;496:455–461.
- Ji W, Qiu C, Wang M, et al. Hsa_circ_0001649: a circular RNA and potential novel biomarker for colorectal cancer. Biochem Biophys Res Commun. 2018;497:122–126.
- Zhang X, Qiu S, Luo P, et al. Down-regulation of hsa_circ_0001649 in hepatocellular carcinoma predicts a poor prognosis. Cancer Biomarkers. 2018;22:135–142.
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388.
- Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215.
- Qin M, Liu G, Huo X, et al. Hsa_circ_0001649: a circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomarkers. 2016;16:161–169.
- Li Y, Zheng F, Xiao X, et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Reports. 2017;18:1646–1659.
- Liu H, Liu Y, Bian Z, et al. Circular RNA YAP1 inhibits the proliferation and invasion of gastric cancer cells by regulating the miR-367-5p/p27 (Kip1) axis. Mol Cancer. 2018;17:151.
- Quesnelle KM, Boehm AL, Grandis JR. STAT-mediated EGFR signaling in cancer. J Cell Biochem. 2007;102:311–319.
- Roberts LE, Fini MA, Derkash N, et al. PD98059 enhanced insulin, cytokine, and growth factor activation of xanthine oxidoreductase in epithelial cells involves STAT3 and the glucocorticoid receptor. J Cell Biochem. 2007;101:1567–1587.
- Linher-Melville K, Singh G. The complex roles of STAT3 and STAT5 in maintaining redox balance: lessons from STAT-mediated xCT expression in cancer cells. Mol Cell Endocrinol. 2017;451:40–52.
- You X, Conrad TO. Acfs: accurate circRNA identification and quantification from RNA-Seq data. Sci Rep. 2016;6:38820.
- Bolha L, Ravnik-Glavac M. Circular RNAs: biogenesis, function, and a role as possible cancer biomarkers. Int J Genom. 2017;2017:6218353.
- Chen BJ, Byrne FL, Takenaka K, et al. Analysis of the circular RNA transcriptome in endometrial cancer. Oncotarget. 2018;9:5786–5796.
- Lu C, Shi X, Wang AY, et al. RNA-Seq profiling of circular RNAs in human laryngeal squamous cell carcinomas. Mol Cancer. 2018;17:86.
- Rothermundt C, Seddon BM, Dileo P, et al. Follow-up practices for high-grade extremity osteosarcoma. BMC Cancer. 2016;16:301.
- Anderson ME. Update on survival in osteosarcoma. Orthop Clin North Am. 2016;47:283–292.
- Peng N, Shi L, Zhang Q, et al. Microarray profiling of circular RNAs in human papillary thyroid carcinoma. PloS One. 2017;12:e0170287.
- Qiu M, Xia W, Chen R, et al. The circular RNA circPRKCI promotes tumor growth in lung adenocarcinoma. Cancer Res. 2018;78:2839–2851.
- Zhang J, Liu H, Hou L, et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16:151.
- Liu W, Zhang J, Zou C, et al. Microarray expression profile and functional analysis of circular rnas in osteosarcoma. Cell Physiol Biochem. 2017;43:969–985.
- Wang Y, Sui X, Zhao H, et al. Decreased circular RNA hsa_circ_0001649 predicts unfavorable prognosis in glioma and exerts oncogenic properties in vitro and in vivo. Gene. 2018;676:117–122.
- Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42–51.
- Li F, Zhang L, Li W, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6:6001–6013.
- Long J, Luo J, Yin X. MiR-338-5p promotes the growth and metastasis of malignant melanoma cells via targeting CD82. Biomed Pharmacothe. 2018;102:1195–1202.
- Liu S, Qu D, Li W, et al. miR647 and miR1914 promote cancer progression equivalently by downregulating nuclear factor IX in colorectal cancer. Mol Med Rep. 2017;16:8189–8199.
- Zhang J, Zhang Z, Sun J, et al. MiR-942 regulates the function of breast cancer cell by targeting FOXA2. Biosci Rep. 2019;39. DOI:https://doi.org/10.1042/BSR20192298.
- Lei D, Zhang F, Yao D, et al. MiR-338-5p suppresses proliferation, migration, invasion, and promote apoptosis of glioblastoma cells by directly targeting EFEMP1. Biomed Pharmacothe. 2017;89:957–965.
- Qin K, Tian G, Chen G, et al. miR-647 inhibits glioma cell proliferation, colony formation, and invasion by regulating HOXA9. J Gene Med. 2019. p. e3153.
- Mendrzyk F, Radlwimmer B, Joos S, et al. Genomic and protein expression profiling identifies CDK6 as novel independent prognostic marker in medulloblastoma. J Clin Oncol. 2005;23:8853–8862.
- Koontongkaew S, Chareonkitkajorn L, Chanvitan A, et al. Alterations of p53, pRb, cyclin D(1) and cdk4 in human oral and pharyngeal squamous cell carcinomas. Oral Oncol. 2000;36:334–339.
- Poomsawat S, Buajeeb W, Khovidhunkit SO, et al. Alteration in the expression of cdk4 and cdk6 proteins in oral cancer and premalignant lesions. J Oral Pathol Med. 2010;39:793–799.
- Zhu K, Liu L, Zhang J, et al. MiR-29b suppresses the proliferation and migration of osteosarcoma cells by targeting CDK6. Protein Cell. 2016;7:434–444.
- Li Z, Zhao L, Wang Q. Overexpression of long non-coding RNA HOTTIP increases chemoresistance of osteosarcoma cell by activating the Wnt/beta-catenin pathway. Am J Transl Res. 2016;8:2385–2393.
- Nam S, Xie J, Perkins A, et al. Novel synthetic derivatives of the natural product berbamine inhibit Jak2/Stat3 signaling and induce apoptosis of human melanoma cells. Mol Oncol. 2012;6:484–493.
- Wei R, Yang Q, Han B, et al. microRNA-375 inhibits colorectal cancer cells proliferation by downregulating JAK2/STAT3 and MAP3K8/ERK signaling pathways. Oncotarget. 2017;8:16633–16641.
- Yang N, Han F, Cui H, et al. Matrine suppresses proliferation and induces apoptosis in human cholangiocarcinoma cells through suppression of JAK2/STAT3 signaling. Pharmacol Rep. 2015;67:388–393.
- Hu H, Wang S, Shi D, et al. Lycorine exerts antitumor activity against osteosarcoma cells in vitro and in vivo xenograft model through the JAK2/STAT3 pathway. Onco Targets Ther. 2019;12:5377–5388.
- Yun HM, Park KR, Quang TH, et al. 4-parvifuran inhibits metastatic and invasive actions through the JAK2/STAT3 pathway in osteosarcoma cells. Arch Pharm Res. 2017;40:601–609.
- Wang J, Ni J, Yi S, et al. Protein inhibitor of activated STAT xalpha depresses cyclin D and cyclin D kinase, and contributes to the inhibition of osteosarcoma cell progression. Mol Med Rep. 2016;13:1645–1652.
- Angulo P, Kaushik G, Subramaniam D, et al. Natural compounds targeting major cell signaling pathways: a novel paradigm for osteosarcoma therapy. J Hematol Oncol. 2017;10:10.