ABSTRACT
Skeletal muscle, a critical component of the mammalian body, is essential for normal body movement. miRNAs are well documented in gene post-transcription regulation in many biological processes, including muscle development and maintenance. miR-92b-3p, which is often associated with tumorigenesis, has never been explored in myoblast development. Here, we used murine-derived C2C12 myoblasts to explore the potential functions of miR-92b-3p in skeletal muscle development. Our results demonstrated that miR-92b-3p mimics inhibited C2C12 cell proliferation and migration, whereas miR-92b-3p inhibitor promoted C2C12 cell proliferation and migration. C2C12 cell differentiation was not affected by miR-92b-3p mimics, according to immunofluorescence and qPCR results. Serum- and glucocorticoid-induced kinase 3 (SGK3) was predicted and validated as a target of miR-92b-3p. Overexpression of SGK3 promoted C2C12 cell proliferation. SGK3 and miR-92b-3p formed a regulatory pathway to modulate C2C12 cell proliferation. In conclusion, miR-92b-3p inhibited C2C12 cell proliferation by targeting SGK3 and impeded C2C12 cell migration.
Introduction
Myogenesis is the formation of skeletal muscle and consists of proliferation, migration, and differentiation. The development and maintenance of skeletal muscle is a complicated biological process that is orchestrated by a variety of cellular and molecular events [Citation1,Citation2]. Normal myogenesis maintenance is inseparable from the sophisticated regulation of the cell cycle [Citation3,Citation4]. Dysregulation of the cell cycle results in a series of diseases, such as cancer [Citation5].
miR-92b is generally considered as an oncogene or tumor suppressor. miR-92b-3p is the dominant form during miRNA formation. It manipulates the proliferation, metastasis, invasion, and apoptosis of various cancer positively or negatively, such as gastric cancer [Citation6], clear cell renal cell carcinoma [Citation7], cervical cancer [Citation8], pancreatic cancer [Citation9], and glioma [Citation10]. miR-92b also participates in many biological events, such as promoting neurite growth [Citation11], maintaining neuroblast self-renewal [Citation12], inhibiting cardiomyocytes hypertrophic growth [Citation13], modulating osteogenic differentiation [Citation14], restraining the production of intermediate cortical progenitors in the murine brain [Citation15], and manipulating the G1/S checkpoint gene p57 in human embryonic stem cells [Citation16]. Although miR-92b serves as a crucial factor, its role in skeletal muscle development has never been explored.
SGK3, a downstream effector of phosphoinositide-3-kinase regulatory subunit 1 (PI3K), belongs to the SGK family and has been shown to function in many biological processes. In Xenopus oocytes, SGK3 participates in numerous ion channel control [Citation17]. In hepatocellular carcinoma cells, SGK3 promotes cell migration and invasion, leading to a poor prognosis [Citation18].
In this study, we sought to illuminate the role of miR-92b-3p in skeletal muscle myoblast development and the molecular mechanism involved. Our results show that miR-92b-3p bound to the 3′UTR of SGK3. miR-92b-3p mimics induced cell cycle arrest and inhibited C2C12 cell proliferation and migration. Overexpression of SGK3 facilitated C2C12 cell proliferation by promoting cell cycle progression. miR-92b-3p mimics abated the promotion effect elicited by SGK3 on C2C12 cell proliferation. Our data have established the role of miR-92b-3p in regulating skeletal muscle myoblast development, providing new insight into skeletal muscle myogenesis.
Materials and methods
Cell culture
Mouse C2C12 cells were cultured in the growth medium, made up of Dulbecco’s modified Eagle’s medium (DMEM) (Hyclone, Utah, USA), supplemented with 20% heat-inactivated fetal bovine serum (FBS) (Hyclone, Utah, USA), and 1% penicillin/streptomycin (Gibco, Maryland, USA) under a humidified air containing 5% CO2 at 37°C for proliferation and DMEM contains 2% heated-inactivated horse serum (Hyclone, Utah, USA) and 1% penicillin/streptomycin for differentiation at the same condition.
Plasmid construction and transfection
For the construction of SGK3 overexpression plasmid, the coding sequence (CDS) of SGK3 was amplified and cloned into pcDNA3 to produce pcDNA3.1-SGK3. The recombinant plasmid was confirmed by sequencing. The primers involved were shown in . miR-92b-3p mimics, miR-92b-3p inhibitor, and three Si-SGK3s (Si-SGK3-1:5′- TCACGGTTTATAAAGTTCT-3′, Si-SGK3-2: 5′AGTACTTCGAAGCCACATT-3′, Si-SGK3-3: 5′-GAAGAAACGGTTCCCTATT-3′) were purchased from RiboBio Co., Ltd. (Guangzhou, China). The plasmids, siRNAs, miR-92b-3p mimics, and inhibitor were transfected into C2C12 cells by Lipofectamine 3000 (Invitrogen, California, USA) according to the manufacture’s instruction.
Table 1. The primers used in this study
RNA extraction and quantitative real-time PCR
Total RNA was extracted from transfected cells using TRIzol reagent (Vazyme, Nanjing, China). The expression of mRNA was detected by qPCR in a Bio-Rad CFX9600 Real-Time Detection System (Bio-Rad, California, USA) using qPCR SYBR® Green Master Mix (Yeasen, Shanghai, China). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used to normalize the tested mRNA expression. The 2−ΔΔCt method was performed to calculate the relative fold changes of candidate genes.
Western blot analysis
Cells were washed in phosphate-buffered saline (PBS) and lysed in RIPA buffer containing 1% (v/v) phenylmethanesulfonyl fluoride (PMSF). The western blotting was performed according to previously established procedures. The protein was quantified using the BCA Protein Assays Kit (Beyotime, Shanghai, China) and then separated in SDS-PAGE, followed by transfer onto PVDF membranes, and immunoblotted with following antibodies: anti-cyclin B1 (Bioss, Beijing, China), anti-cyclin D1 (Bioss, Beijing, China), anti-SGK3 (Bioss, Beijing, China), anti-β actin (Biodragon, Beijing, China), anti-P21 (Homemade). The protein bands were quantified using ImageJ software.
Cell proliferation assays
C2C12 cells were cultured in 48-well plates and transfected with SGK3 overexpression plasmid, siRNA, miR-92b-3p mimics, miR-92b-3p inhibitor according to the experimental needs. EdU staining was performed to evaluate the ability of cell proliferation using the EdU kit (RiboBio, Guangzhou, China). The EdU staining C2C12 cells were observed and recorded using a Nikon TE2000-U inverted microscope (Nikon Instruments, Tokyo, Japan). The EdU positive C2C12 cells were counted using Image Pro Plus (Media Cybernetics, MD, USA). And cells were harvested and treated with a cell cycle detection kit. Then C2C12 cells were dyed with propidium iodide (PI) and using flow cytometry to analyze the cell cycle status.
Wound healing assay
C2C12 cell migration after transfected with miR-92b mimics or inhibitor was detected with the wound healing assay. Briefly, C2C12 cells were cultured in 6-well plates 24 h before transfected with miR-92b-3p mimics or inhibitor. We used a sterile 200 μL plastic micropipette tip to scrape a homogeneous wound. Cell migration was monitored microscopically. Photographs were taken, and we used ImageJ software to calculate the migration area.
Immunofluorescence
After transfected with miR-92b-3p mimics, C2C12 cells were cultured in differentiation medium for 3, 5, 7, and 9 d, respectively, and stained. Briefly, cells were fixed with acetone at 4°C for 10 min, followed by washing with PBS three times, and 0.5% Triton X-100 for 3 min. Then, washed with PBS for three times. 5% BSA (Boster, Wuhan, China) was added and placed at 37°C for 1 h, followed by incubating with anti-myosin (Boster, Wuhan, China) at 37°C for 1 h. Afterward, cells were washed with PBS five times, incubated with goat anti-mouse IgG (Bioss, Beijing, China) for 1 h. Finally, washed with PBS for three times, and dyed with DAPI (Boster, Wuhan, China) for 10 min. Cells were photographed in Nikon TE2000-U inverted microscope (Nikon Instruments, Tokyo, Japan). And the myosin-positive cells were calculated with Image Pro Plus (Media Cybernetics, MD, USA).
miRNA target gene prediction
The potential targeted genes for miR-92b-3p were predicted using miR-Walk 2.0 (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/). miR-Walk 2.0 is a tool used for predicting and validating the interaction between miRNA and the potential targeted gene.
Dual-luciferase reporter assay
The putative sequences of the miR-92b-3p binding site in SGK3 3′UTR and the corresponding mutated sequences were cloned into a pmirGLO Dual-luciferase miRNA Target Expression Vector (Promega, Wisconsin, USA). The pmirGLO-SGK3-WT and pmirGLO-SGK3-Mut report vector was co-transfected with negative control (NC) mimics or miR-92b-3p mimics into HEK293 cells. After 24 h transfection, a Dual-Glo Luciferase Assay System (Promega, Wisconsin, USA) was used to detect the luciferase activity according to the manufacturer’s instruction.
Statistical analysis
Statistical analyses were performed using SPSS (IBM SPSS Statistic 24). All data were presented as the mean ± standard error of the mean (SEM.), and at least three replicates were used per group. Significant differences between treatment groups were determined by one-way ANOVA. Significant differences were achieved when P < 0.05.
Results
miR-92b-3p mimics inhibited and miR-92b-3p inhibitor promoted C2C12 cell proliferation
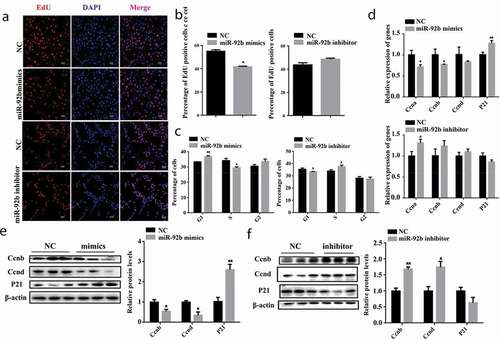
We synthesized miR-92b-3p mimics and inhibitor to assess the effect of miR-92b-3p on the proliferation of C2C12 cells. miR-92b-3p mimics effectively increased (p < 0.01) miR-92b-3p mRNA level and miR-92b-3p inhibitor decreased (p < 0.05) miR-92b-3p mRNA level (Figure S1(a,b)). EdU staining was performed 24 hours after C2C12 cells were transfected with either miR-92b-3p mimics or inhibitor. The EdU positive cells were significantly reduced (p < 0.05) when transfected with miR-92b-3p mimics. However, the miR-92b-3p inhibitor did not affect the cell numbers (). Flow cytometry results indicated that miR-92b-3p mimics increased (p < 0.01) the percentage of G1 phase cells and decreased (p < 0.05) the percentage of S phase cells compared to the negative control. Inhibition of miR-92b-3p increased (p < 0.05) the percentage of S phase cells and decreased (p < 0.05) the percentage of G1 phase cells ()). qPCR results showed miR-92b-3p mimics decreased (p < 0.05) the mRNA level of CCNA and CCNB, and increased (p < 0.01) the P21 mRNA level; miR-92b-3p inhibitor increased (p < 0.05) CCNA mRNA level, but did not change the mRNA level of CCNB, CCND, and P21 ()). Moreover, the CCNB and CCND protein level of C2C12 cells transfected with miR-92b-3p mimics decreased (p < 0.05), the P21 protein level increased significantly (p < 0.01) ()). Inhibition of miR-92b-3p increased CCNB (p < 0.01) and CCND (p < 0.05) protein level ()). These results indicate elevated level of miR-92b-3p induced cell cycle arrest, impeded CCNB and CCND expression level, and inhibited C2C12 cell proliferation.
Figure 1. miR-92b-3p mimics induced cell cycle arrest and inhibited C2C12 cell proliferation; miR-92b-3p inhibitor facilitated cell cycle progression and promoted C2C12 cell proliferation. (a) EdU staining of C2C12 cells 24 hours after transfected with miR-92b-3p mimics or inhibitor. miR-92b-3p mimics reduced EdU positive cells. miR-92b-3p inhibitor did not affect EdU positive cells. (b) The quantified data of EdU positive cells in Figure 1a. (c) Flow cytometry analysis after C2C12 cells transfected with miR-92b-3p mimics or inhibitor. miR-92b-3p mimics increased the percentage of G1 phase cells, decreased the percentage of S phase cells. miR-92b-3p inhibitor decreased the percentage of G1 phase cells, increased the percentage of S phase cells. (d) mRNA level of cell cycle genes measured by qPCR. miR-92b-3p mimics decreased CCNA, CCNB mRNA level, increased P21 mRNA level. miR-92b-3p inhibitor increased CCNA mRNA level. (e) Western blot results of C2C12 cells after transfected with miR-92b-3p mimics. miR-92b-3p mimics lowered the protein level of CCNB and CCND, increased the protein level of P21. (f) Western blot results of C2C12 cells after transfected with miR-92b-3p inhibitor. miR-92b-3p inhibitor increased the protein level of CCNB and CCND, decreased the protein level of P21. * p < 0.05, ** p < 0.01. All experiments were repeated three times, and results are presented as mean ± S.E.M
miR-92b-3p mimics inhibited and miR-92b-3p inhibitor promoted C2C12 cell migration
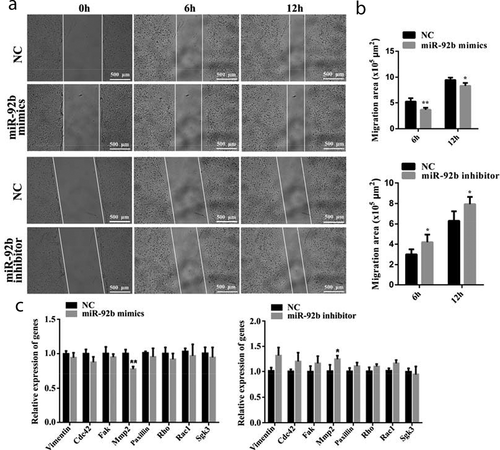
The motility of C2C12 cells was measured using a wound-healing assay. We found that miR-92b-3p mimics impeded the motility of C2C12 cells after 6 hours (p < 0.01) and after 12 hours (p < 0.05). miR-92b-3p inhibitor promoted C2C12 cell migration after 6 hours and 12 hours of treatment (p < 0.05) (). We also found that the mRNA level of migration-associated genes, including Vimentin, Cdc42, Fak, Paxillin, Rho, Rac1, and SGK3, was not affected by either miR-92b-3p mimics or inhibitor. Only MMP2 mRNA level was markedly decreased (p < 0.01) in C2C12 cells transfected with miR-92b-3p mimics and increased (p < 0.05) in C2C12 cells transfected with miR-92b-3p inhibitor ()). These results indicate that miR-92b-3p mimics inhibited C2C12 cell migration.
Figure 2. miR-92b-3p mimics inhibited C2C12 cell migration; miR-92b-3p inhibitor promoted C2C12 cell migration. (a) Photomicrographs of wound-healing assay of C2C12 cells after transfected with miR-92b-3p mimics or inhibitor. miR-92-3p mimics impeded C2C12 cell migration. miR-92b-3p inhibitor promoted C2C12 cell migration. (b) The quantified data of C2C12 cell migration area in Figure 2a. (c) qPCR results of migration-related genes. miR-92b-3p mimics decreased the mRNA level of MMP2. miR-92b-3p inhibitor increased the mRNA level of MMP2. * p < 0.05, ** p < 0.01. All experiments were repeated three times, and results are presented as mean ± S.E.M
miR-92b-3p mimics did not affect C2C12 cell differentiation
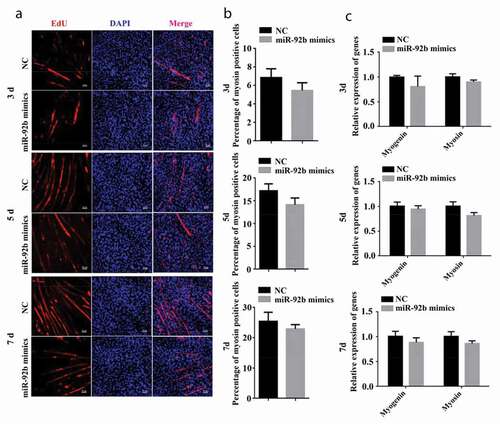
Immunofluorescent staining was performed 3, 5, and 7 d after C2C12 cells were cultured in differentiation medium and transfected with miR-92b-3p mimics. miR-92b-3p mimics did not change the number of myosin-positive cells (). We also performed qPCR to measure the mRNA level of myogenin and myosin after transfection. The mRNA level of myogenin and myosin were not affected by miR-92b-3p mimics compared to the negative control ()). These results indicated that miR-92b-3p mimics and inhibitor did not affect C2C12 cell differentiation.
Figure 3. miR-92b-3p mimics did not affect C2C12 cell differentiation. (a) Immunofluorescence of C2C12 cells 3, 5, and 7 d after transfected with miR-92b-3p mimics. miR-92b-3p mimics did not change the number of myosin-positive cells. (b) The quantified data of myosin positive cell in Figure3a. (c) qPCR results of myogenin and myosin. miR-92b-3p mimics did not change myogenin and myosin mRNA level. All experiments were repeated three times, and results are presented as mean ± S.E.M
SGK3 is a target gene of miR-92b-3p
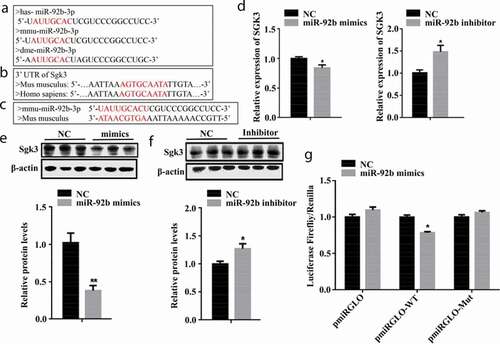
Through bioinformatic analysis, we found that miR-92b-3p is highly conserved ()), and humans and mice have the same miR-92b-3p complementary region in the 3′UTR of SGK3 ()). To elucidate the mechanism through which miR-92b-3p modulates C2C12 cell proliferation, we used miR-Walk 2.0 (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/) to predict the target gene of miR-92b-3p and found that miR-92b-3p might bind to the 3′UTR of SGK3 ()). We measured the mRNA level of SGK3 in C2C12 cells after transfected with either miR-29b-3p mimics or miR-92b-3p inhibitor. miR-92b-3p mimics decreased (p < 0.05) the mRNA level of SGK3; miR-92b-3p inhibitor increased (p < 0.05) the mRNA level of SGK3 ()). Consistently, the protein level of SGK3 was significantly decreased (p < 0.01) when transfected with miR-92b-3p mimics ()) and increased (p < 0.05) when transfected with miR-92b-3p inhibitor ()). To confirm that SGK3 is a target gene of miR-92b-3p, we constructed a luciferase reporter gene containing either SGK3 3′UTR binding site for miR-92b-3p (pmirGLO-WT) or mutated SGK3 3′UTR binding site for miR-92b-3p (pmirGLO-Mut), and co-transfected with either miR-92b-3p mimics or NC mimics into HEK293 cells. The luciferase activity was reduced (p < 0.05) when co-transfected with pmirGLO-SGK3-WT and miR-92b-3p mimics. Whereas the luciferase activity was not affected by miR-92b-3p mimics when co-transfected with pmirGLO-Mut ()). These results indicated that SGK3 is the genuine target of miR-92b-3p.
Figure 4. SGK3 is a target gene of miR-92b-3p. (a) The sequence of mature miR-92b-3p in different species. Humans, mice, and Drosophila have the same seed region (marked in red). (b) The sequence of SGK3 3′UTR of humans and mice. Humans and mice have the same complementary region (marked in red) in the 3′UTR of SGK3. (c) The complementary pairing (marked in red) of mmu-miR-92b-3p and SGK3 3′UTR of Mus musculus. (d) SGK3 mRNA level after transfected with miR-92b-3p mimics or inhibitor. miR-92b-3p mimics decreased SGK3 mRNA level. miR-92b-3p inhibitor increased the SGK3 mRNA level. (e) The protein level of SGK3 after transfected with miR-92b-3p mimics. miR-92b-3p mimics decreased the SGK3 protein level. (f) The protein level of SGK3 after transfected with miR-92b-3p inhibitor. miR-92b-3p inhibitor increased the SGK3 protein level. (g) Result of dual luciferase assay. miR-92b-3p mimics decreased the luciferase activity of pmirGLO-WT but did not affect the luciferase activity of pmirGLO-Mut. * p < 0.05, ** p < 0.01. All experiments were repeated three times, and results are presented as mean ± S.E.M
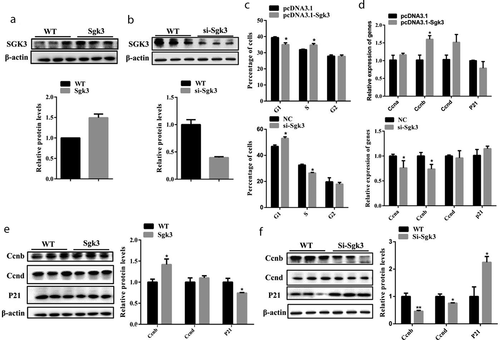
SGK3 overexpression promoted C2C12 cell proliferation
Given that SGK3 is a direct target of miR-92b-3p, we hypothesized that SGK3 also plays a role in C2C12 cell proliferation. We constructed the SGK3 overexpressing recombinant plasmid pcDNA3.1-SGK3 and transfected C2C12 cells cultured in the growth medium. qPCR results indicated pcDNA3.1-SGK3 significantly increased (p < 0.001) the mRNA level of SGK3 24 hours after transfection (Figure S2(a)). We also designed three siRNA oligos, Si-SGK3-1, Si-SGK3-2, and Si-SGK3-3 against mouse SGK3. qPCR results indicated that Si-SGK3-3 achieved the highest inhibition (p < 0.01) of the SGK3 mRNA level at 75 nM (Figure S2(b)). Consistent with qPCR results, pcDNA3.1-SGK3 increased (p < 0.01) the protein level of SGK3 ()), and Si-SGK3-3 decreased (p < 0.01) the protein level of SGK3 ()). We performed flow cytometry analysis to examine the effects of SGK3 overexpression and inhibition on C2C12 cell proliferation. Flow cytometry results indicated that C2C12 cells transfected with pcDNA3.1-SGK3 exhibited a higher (p < 0.05) percentage in S phase and lower (p < 0.05) percentage in G1 phase compared to the negative control. C2C12 cells transfected with Si-SGK3-3 exhibited a lower (p < 0.05) percentage in S phase and higher (p < 0.05) percentage in G1 phase compared to the negative control ()). qPCR results showed that pcDNA3.1-SGK3 increased (p < 0.05) CCNB mRNA level, but did not change the mRNA level of CCNA, CCND, and P21. Si-SGK3-3 decreased (p < 0.05) the mRNA level of CCNA and CCNB, but did not change the mRNA level of CCND and P21 ()). Moreover, overexpression of SGK3 increased (p < 0.05) the protein level of CCNB and decreased (p < 0.05) the protein level of P21, but did not change the protein level of CCND ()). Inhibition of SGK3 decreased the protein level of CCNB (p < 0.01) and CCND (p < 0.05), but increased the protein level of P21 ()). These results indicated that SGK3 overexpression accelerated cell cycle progression, facilitated CCNB expression, and promoted C2C12 cell proliferation.
Figure 5. SGK3 overexpression accelerated cell cycle progression and promoted C2C12 cell proliferation; SGK3 inhibition impeded cell cycle progression and inhibited C2C12 cell proliferation. (a) The protein level of SGK3 24 hours after C2C12 cells were transfected with pcDNA3.1-SGK3. pcDNA3.1-SGK3 increased the protein level of SGK3. (b) The protein level of SGK3 24 hours after C2C12 cells transfected with Si-SGK3-3. Si-SGK3-3 decreased the protein level of SGK3. (c) Flow cytometry analysis after C2C12 cells were transfected with pcDNA3.1-SGK3 or Si-SGK3-3. pcDNA3.1-SGK3 lowered the percentage of G1 phase cells, increased the percentage of S phase cells. Si-SGK3-3 increased the percentage of G1 phase cells and decreased the percentage of S phase cells. (d) mRNA level of cell cycle genes measured by qPCR. pcDNA3.1-SGK3 increased the mRNA level of CCNB. Si-SGK3-3 decreased the mRNA level of CCNA and CCND. (e) Western blot results after C2C12 cells were transfected with pcDNA3.1-SGK3. pcDNA3.1-SGK3 increased the protein level of CCNB, decreased the protein level of P21. (f) Western blot results after C2C12 cells were transfected with Si-SGK3-3. Si-SGK3-3 decreased the protein level of CCNB and CCND, increased the protein level of P21. * p < 0.05, ** p < 0.01, *** p < 0.001. All experiments were repeated three times, and results are presented as mean ± S.E.M
SGK3 overexpression promoted C2C12 cell proliferation
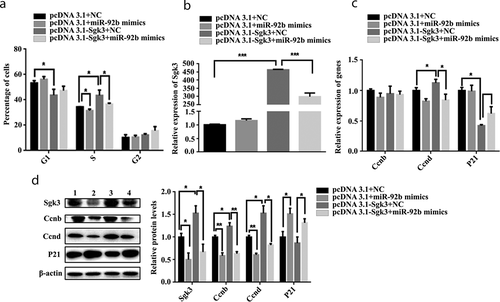
We wanted to confirm that the effect of miR-92b-3p on C2C12 cell proliferation was mediated through SGK3. Then we transfected C2C12 cells with four groups, G1: pcDNA3.1 and NC, G2: pcDNA3.1 and miR-92b-3p mimics, G3: pcDNA3.1-SGK3 and NC, and G4: pcDNA3.1-SGK3 and miR-92b-3p mimics, respectively, 24 hours after transfection. G2 versus G1 manifests the effect of miR-92b-3p; G3 versus G1 manifests the effect of SGK3; G4 versus G3 manifests the inhibiting effect of miR-92b-3p on SGK3. Flow cytometry analysis results showed that miR-92b-3p mimics weakened (p < 0.05) the promoting effect (p < 0.05) of pcDNA3.1-SGK3 on cell number in S phase, but did not weaken the inhibiting effect of pcDNA3.1-SGK3 on cell number in G1 phase ()). We also measured the mRNA level of SGK3, CCNB, CCND, and P21, 24 hours after transfection. The promoting effect (p < 0.001) induced by pcDNA3.1-SGK3 on SGK mRNA level was weakened (p < 0.001) when co-transfected with miR-92b-3p mimics ()). miR-92b-3p mimics weakened (p < 0.05) the promoting effect (p < 0.05) induced by pcDNA3.1-SGK3 on CCND mRNA level, but did not weaken the inhibiting effect (p < 0.05) induced by pcDNA3.1-SGK3 on P21 mRNA level, and the mRNA level of CCNB was not affected by either pcDNA3.1-SGK3 or miR-92b-3p mimics ()). Moreover, miR-92b-3p mimics weakened (p < 0.05) the promoting effect (p < 0.05) induced by pcDNA3.1-SGK3 on SGK3 protein level. The protein level of CCND was decreased (p < 0.01) when transfected with miR-92b-3p mimics, and miR-92b-3p mimics weakened (p < 0.05) the promoting effect (p < 0.05) induced by pcDNA3.1-SGK3 on CCND protein level. miR-92b-3p mimics increased the P21 protein level, either co-transfected with pcDNA3.1 or pcDNA3.1-SGK3. Inconsistent with CCNB mRNA level, miR-92b-3p mimics decreased (p < 0.01) the protein level of CCNB, and weakened (p < 0.01) the promoting effect (p < 0.05) induced by pcDAN3.1-SGK3 on CCNB protein level ()). These results indicated that miR-92b-3p mimics induced cell cycle arrest, impeded CCNB and CCND expression level, and inhibited C2C12 cell proliferation by targeting SGK3.
Figure 6. miR-92b-3p mimics inhibited C2C12 cell cycle progression and proliferation by targeting SGK3. (a) Flow cytometry analysis of C2C12 cells after transfection. pcDNA3.1-SGK3 decreased cell number in the G1 phase and increased cell number in the S phase. miR-92b-3p mimics decreased cell number in the S phase and weakened the promoting effect induced by pcDNA3.1-SGK3 on cell number in the S phase. (b) mRNA level of SGK3 after transfection measured by qPCR. pcDNA3.1-SGK3 increased the mRNA level of SGK3, miR-92b-3p mimics weakened the promoting effect induced by pcDNA3.1-SGK3 on SGK3 mRNA level. (c) mRNA level of cell cycle genes measured by qPCR. pcDNA3.1-SGK3 increased the mRNA level of CCND and decreased the mRNA level of P21. miR-92b-3p mimics weakened the promoting effect induced by pcDNA3.1-SGK3 on CCND mRNA level. (d) Western blot results of C2C12 cells after transfection. pcDNA3.1-SGK3 increased the protein level of SGK3, CCNB, and CCND. miR-92b-3p mimics decreased the protein level of SGK3, CCNB, and CCND; increased the protein level of P21; and weakened the promoting effects induced by pcDNA3.1-SGK3 on SGK3, CCNB, and CCND protein level. * p < 0.05, ** p < 0.01, *** p < 0.001. All experiments were repeated three times, and results are presented as mean ± S.E.M
Discussion
In the present study, we intended to explore the role and mechanism of miR-92b-3p in skeletal muscle development. Our data demonstrated that miR-92b-3p elicited cell cycle arrest and slowed the migration of C2C12 cells during proliferation. The inhibition effect of miR-92b-3p on C2C12 cell proliferation was mediated through SGK3. However, the differentiation of C2C12 cells was not affected by miR-92b-3p.
Cell migration, proliferation, and differentiation play essential roles in skeletal muscle development. miRNAs are crucial regulators in skeletal muscle development, and many studies have demonstrated miRNAs’ post-transcriptional regulation in myogenesis. In pulmonary artery smooth muscle cells, miR-92b-3p was shown to facilitate abnormal cell proliferation by directly targeting TSC1 [Citation19]. In hepatocellular carcinoma (HCC), miR-92b-3p was shown to promote HCC cell proliferation by targeting Smad7 [Citation20].In the current study, we found that miR-92b-3p targeted SGK3 caused C2C12 cells to enter cycle arrest, reduced the expression of CCNB and CCND, and increased the expression of P21, led to the inhibition of C2C12 cell proliferation. These results indicate that miRNAs are closely related to skeletal muscle development, and miR-92b-3p is also involved in the regulatory mechanisms of myogenesis.
The motility of satellite cells during myogenesis is a crucial trait for skeletal muscle development. During this progression, the extracellular matrix (ECM) affects cell migration. Matrix metalloproteases (MMPs) are a class of biological enzymes that can precisely digest the components within ECM. In the present study, we found that miR-92b-3p mimics suppressed C2C12 cell migration and decreased the MMP2 mRNA level. Whereas other migration-associated genes, including Vimentin, Cdc42, Fak, Paxillin, Rho, and Rac1, were not affected, and neither was SGK3. These results, taken together, indicated that MMP2 might be involved in the miR-92b-3p-mediated migratory inhibition, but the specific mechanism remained to be established. Consistently, miR-92b suppressed cell migration by targeting Gabra3 in triple-negative breast cancer cells [Citation21]. In nasopharyngeal carcinoma cells, miR-92b inhibited cell migration by targeting Smad3 [Citation22]. Whereas in gastric cancer SGC‐7901 cells, miR-92b-3p increased MMP2/9 expression level and promoted cell migration [Citation23]. These studies indicated that miR-92b-3p might act as a versatile molecule in modulating cell migration. Taken together, the diversified mechanisms miR-92b-3p participate in many biological processes manifest its critical functions, and our results illuminated its role in C2C12 proliferation and migration.
After explored the functions of miR-92b-3p in C2C12 cell proliferation and migration, we examined the role of miR-92b-3p in C2C12 cell differentiation by transfecting miR-92b-3p mimics into differentiating C2C12 cells. Immunofluorescence and qPCR results showed that the expression of myogenin and myosin were not affected by miR-92b-3p mimics, which indicating miR-92b-3p mimics did not affect C2C12 differentiation. Whereas in Drosophila muscle development, miR-92b was of vital importance, it could be activated by myocyte enhancer factor 2 (MEF2) and, in turn, bind to the 3′UTR of MEF2 to inhibit the expression level of MEF2. miR-92b and MEF2 formed an orchestrated regulatory mechanism to modulate muscle development [Citation24]. Whether there are other mechanisms neutralize functions of miR-92b-3p in C2C12 cell differentiation remains to be explored.
In conclusion, miR-92b-3p inhibited skeletal muscle cell proliferation by targeting SGK3 and suppressed skeletal muscle cell migration. To our knowledge, this is the first study to investigate miR-92b-3p functions in skeletal muscle development. Results from our study expand our understanding of the mechanisms underlying skeletal muscle development, and miR-92b-3p could be a potential biomarker for modulating skeletal muscle development in humans and animals.
Author contribution
Chong Wang and Lianjie Hou conceptualized the project; Jia Shi performed the experiments; Zuocheng Ning analyzed the data; Zijian Ye validated the data and wrote the manuscript; Ching Yuan Hu and Chong Wang reviewed and edited the manuscript.
Supplemental Material
Download MS Word (2.7 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Cohen S, Goldberg A. Muscle wasting in disease: molecular mechanisms and promising therapies. Drug Discovery. 2015;14:58–74.
- Tajbakhsh S. Skeletal muscle stem cells in developmental versus regenerative myogenesis. J Intern Med. 2009;266:372–389.
- Calegari F, Huttner WB. An inhibition of cyclin-dependent kinases that lengthens, but does not arrest, neuroepithelial cell cycle induces premature neurogenesis. J Cell Sci. 2003;116:4947–4955.
- Calegari F, Haubensak W, Haffner C, et al. Selective lengthening of the cell cycle in the neurogenic subpopulation of neural progenitor cells during mouse brain development. J Neurosci. 2005;25:6533–6538.
- Abukhdeir AM, Park BH. p21 and p27: roles in carcinogenesis and drug resistance. Expert Rev Mol Med. 2008;10:e19.
- Ni QF, Zhang Y, Yu J, et al. miR‐92b promotes gastric cancer growth by activating the DAB2IP‐mediated PI3K/AKT signaling pathway. Cell Prolif. 2019;53:e12630.
- Wang C, Uemura M, Tomiyama E, et al. MicroRNA‐92b‐3p is a prognostic oncomiR that targets TSC1 in clear cell renal cell carcinoma. Cancer Sci. 2020;111:1146–1155.
- Sun Y, Feng Y, Zhang G, et al. The endonuclease APE1 processes miR-92b formation, thereby regulating expression of the tumor suppressor LDLR in cervical cancer cells. Ther Adv Med Oncol. 2019;11:1–20.
- Long M, Zhan M, Xu S, et al. miR-92b-3p acts as a tumor suppressor by targeting Gabra3 in pancreatic cancer. Mol Cancer. 2017;16:167.
- Wang K, Wang X, Zou J, et al. miR-92b controls glioma proliferation and invasion through regulating Wnt/beta-catenin signaling via Nemo-like kinase. Neuro Oncol. 2013;15:578–588.
- Chen Z, Li Z, Jiang C, et al. MiR‐92b‐3p promotes neurite growth and functional recovery via the PTEN/AKT pathway in acute spinal cord injury. J Cell Physiol. 2019;234:23043–23052.
- Yuva-Aydemir Y, Xu XL, Aydemir O, et al. Downregulation of the host gene jigr1 by miR-92 Is essential for neuroblast self-renewal in drosophila. PLOS Genet. 2015;11(5):e1005264.
- Hu ZQ, Luo JF, Yu XJ, et al. Targeting myocyte-specific enhancer factor 2D contributes to the suppression of cardiac hypertrophic growth by miR-92b-3p in mice. Oncotarget. 2017;8:92079–92089.
- Li Y, Feng C, Gao M, et al. MicroRNA‐92b‐5p modulates melatonin‐mediated osteogenic differentiation of bone marrow mesenchymal stem cells by targeting ICAM‐1. J Cell Mol Med. 2019;23:6140–6153.
- Nowakowski T, Fotaki J, Pollock V, et al. MicroRNA-92b regulates the development of intermediate cortical progenitors in embryonic mouse brain. PNAS. 2013;110:7056–7061.
- Sengupta S, Nie J, Wagner RJ, et al. MicroRNA 92b controls the G1/S checkpoint gene p57 in human embryonic stem cells. Stem Cells. 2009;27:1524–1528.
- Lang F, Böhmer C, Palmada M, et al. (Patho)physiological significance of the serum- and glucocorticoid-inducible kinase isoforms. Physiol Rev. 2006;86:1151–1178.
- Kong X, Liu F, Gao J. MiR-155 promotes epithelial-mesenchymal transition in hepatocellular carcinoma cells through the activation of PI3K/SGK3/β-catenin signaling pathways. Oncotarget. 2016;7:66051–66060.
- Lee J, Heo J, Kang H. miR-92b-3p-TSC1 axis is critical for mTOR signaling-mediated vascular smooth muscle cell proliferation induced by hypoxia. Cell Death Differ. 2019;26(9):1782–1795.
- Zhuang LK, Yang YT, Ma X, et al. MicroRNA-92b promotes hepatocellular carcinoma progression by targeting Smad7 and is mediated by long non-coding RNA XIST. Cell Death Dis. 2016;7:e2203.
- Li YY, Zheng XH, Deng AP, et al. MiR-92b inhibited cells EMT by targeting Gabra3 and predicted prognosis of triple negative breast cancer patients. Eur Rev Med Pharmacol Sci. 2019;23:10433–10442.
- Zhao C, Zhao F, Feng H, et al. MicroRNA-92b inhibits epithelial-mesenchymal transition-induced migration and invasion by targeting Smad3 in nasopharyngeal cancer. Oncotarget. 2017;8:91603–91613.
- Cheng L, Huo B, Wang Y, et al. Downregulation of microRNA‐92b‐3p suppresses proliferation, migration, and invasion of gastric cancer SGC‐7901 cells by targeting Homeobox D10. J Cell Biochem. 2019;120:17405‐17412.
- Chen Z, Liang S, Zhao Y, et al. miR-92b regulates Mef2 levels through a negative-feedback circuit during Drosophila muscle development. Development. 2012;139:3543–3552.
