ABSTRACT
RBM10 is the RNA-binding protein often absent or mutated in lung adenocarcinoma, rendering it as a potential biomarker or even therapeutic target to prolongate survival time. In this study, we investigated the involvement of RBM10 mutation in the pathogenesis and tumorigenesis of lung adenocarcinoma and identified the differentials in relative signal pathways, aiming to provide the new therapeutic approaches. By performing the systematic TCGA analysis, our results demonstrated that RBM10 mutation was identified in 6% lung adenocarcinoma patients, meanwhile 113 functional genes were identified as significant expression among these patients. Further gene ontology and KEGG analysis were employed to identify the most relative 10 genes and signal pathways. Moreover, four members of the 5-acyl-6, 7-dihydrothiophene [3, 2-c] pyridine (known as “ru-ski”)-ru-ski 43 were identified as the potential drugs for RBM10 mutation lung adenocarcinoma therapy, investigated by the GDSC database. Meanwhile there were 157 genes that were more frequently mutated in the RBM10 mutation group than the wild-type group (p value<0.05). KEGG analysis showed that these genes were enriched in various cancer development pathways and cell proliferation. Finally, our investigations provided the glance at the differential genes and cellular signaling pathways related to RBM10 mutation and identified series of potential drugs for personalized RBM10 mutation lung adenocarcinoma therapy.
Introduction
Lung carcinoma is the most common tumor counting 11.6% percentage in all cancer cases and occupied the 18.4% in death cases causing by cancers [Citation1]. Two main lung cancer, based on the proliferation rate and spreading mechanism, can be classified as small cell lung carcinoma (SLC) or non-small cell lung carcinoma (NSCLC). Among these different type of lung cancers, NSCLC has the highest cancer incidence and most mortality rate, with the 18.2% of 5-year overall survival rate [Citation2]. So far, the major therapeutic methods for NSCLC contains operation, chemotherapy, targeted therapies [Citation3], especially for the targeted therapies. In the past decade, therapeutic drugs to target ordinary gene mutations, for example EGFR, ALK, and ROS, have significantly improve the survival of NSCLC patients [Citation4,Citation5]. These great achievements in development of targeted therapies in NSCLC reveals that the precise therapy to target specific genes or proteins can significantly improve the survival of patients. Therefore, it is important to identify latent biomarkers for the progression and prognosis of lung adenocarcinoma, which may simultaneously represent potential targets for molecular directed therapy.
RBM10 is one RNA recognition motif-containing component of the spliceosome complex that is involved in regulation of alternative splicing of pre-mRNA [Citation6,Citation7] and mRNA stabilization [Citation8,Citation9]. Alterations in RBM10 are frequently associated with selectively exon skipping [Citation10]. In excess of 90% of human genes, including tumorigenesis-related genes, are produced by alternative splicing [Citation11]. Changes in mRNA alternative splicing, particularly in exon skipping [Citation10] or exon-inclusion of the affected proteins [Citation11,Citation12], can lead to abnormal intracellular protein expression. RBM10 gene knockdown has been shown to lead to alterations in 10–20% of all pre-mRNA splicing events [Citation13], and previous studies have suggested that RBM10 may therefore act as a tumor suppressor gene [Citation14]. Preliminary functional research has shown that RBM10 promotes programmed cell death [Citation11] and inhibits cell proliferation [Citation12,Citation13]. Furthermore, it has been confirmed that a knockdown of RBM10 V354E mutation can reduce tumor proliferation in xenograft animal model [Citation14], while over-expression of RBM10 has been shown to inhibit malignant proliferation of lung adenocarcinoma, including cell viability and cell cycle progression [Citation15]. To summarize, there is an emerging body of literature suggesting that RBM10 may represent a tumor suppressor gene.
In this investigation, we performed the enrichment of RNA-seq data, GSEA, KEGG signal pathway to analyze and identify the differentially expressed genes and relative genetic functions. Then, we employed the GDSC to be mined for further analysis, obtaining the potential clinical therapeutic drugs.
Materials and methods
Bioinformatics resources
Bioinformatic analysis was performed on the CNGrid with ScGrid environment (http://www.cngrid.org/). All the images were generated by utilizing R Software (version 4.0.2).
RNA-seq data
RNA-Seq datasets of 561 lung adenocarcinoma patients with corresponding clinical profiles were obtained from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). RBM10 expression information was extracted from the TCGA seq file using R software. Survival information, including total survival and disease-free survival was extracted from clinical data of patients with RBM10 mutations from the TCGA database, including 33 patients with RBM110 mutation and 470 patients with wild-type RBM110. Furthermore, we carried out Kaplan-Meier analysis on survival data using the log-rank test with p value cutoff < 0.05. These data were visualized using the maftools package of EdgeR and cbioportal (https://www.cbioportal.org/).
Data mining and analysis of the GDSC database
Using the Genomics of Drug Sensitivity in Cancer Project (GDSC, https://www.cancerrxgene.org/), compounds targeting to RBM10 mutations were identified and filtered by sensitivity for lung adenocarcinoma (effect size<0, p value<0.001, FDR<0.01). Subsequently, volcano diagram, elastic network and a scatter diagram were generated by using R software. Moreover, Mann-Whitney-Wilcoxon (MWW) analysis was carried out (MWW p value<0.01).
Gene set enrichment analysis (GSEA)
GSEA v3.0 (http://software.broadinstitute.org/gsea/downloads. JSP) was used to identify the differentially expressed genes and enrich for biological functions and signal pathways among the lung adenocarcinoma patients with or without RBM10 mutation. The number of permutations was set to 5 and related enrichment statistic method was set to weighted with Signal2Noise metric method. Other parameters were set as default. Enrichment results with a false discovery rate (FDR q value<0.25 and p value<0.05) were considered as statistically significant.
Identification of differentially expressed genes
EdgeR (http://bioconductor.org/) was utilized to identify differential gene expression between lung cancer patients with or without RBM10 mutation [Citation16,Citation17]. The identification criteria of DEG was as follows: p value<0.05 and FDR q value<0.25, |log2FC|≥1. The differentially expressed genes were used for next bioinformatics analysis.
Functional annotation and pathway enrichment analysis of differentially expressed genes
Gene Ontology (GO) and KEGG enrichment analysis were carried out using the R cluster profiler package. To avoid high error discovery rates (FDR) originated from multiple tests, q value was calculated and relative cutoff values are. FDR q values<0.25 and p values<0.05, respectively. In addition, R cluster profiler package to compare clusters was used in order to automatically enrich for functional categories of each gene cluster, and visualize data.
Integration of protein–protein interaction (PPI) network and module analysis
All the protein-protein interactions were generated by using Retrieval of Interacting Genes (STRING, http://strin g-db.org) [Citation18]. The PPT networks was analyzed by utilizing Cytoscape software (Version 3.0.6, http://cytoscape.org) and relative images was generated by Cytoscape. The mutual impact relationship of the encoded proteins of the differentially expressed genes was analyzed. Significant interactions were considered at a score of > 0.4. In addition, KEGG pathway enrichment and GO enrichment of differentially expressed genes in the top two rankings was carried out.
KEGG pathway enrichment analysis of genes with differential mutation rates
We identified the genes that were more frequently mutated in the RBM10 mutation group than the wild-type group. A KEGG analysis was carried out using the cluster profiler package to automatically determine the enriched pathways for each gene cluster, and to visualize the data. The relative cutoff values considered statistically significant were set to FDR-adjusted q value <0.25 and p value <0.05, respectively.
Statistical analysis
GraphPad prism and R 4.0.2 were used to conduct all statistical analysis. To analyze mRNA levels of RBM10 between tissues with or without RBM10 mutation, Student’s t-tests was performed. To analyze the survival curve of patients suffering RBM10 mutation, Kaplan–Meier log-rank tests between RBM10 mutation groups were carried out with GraphPad prism. The FDR derived from edgeR and GSEA were adjusted for multiple testing using the Benjamini–Hochberg procedure. Enrichment results satisfying a p value<0.05, with a false discovery rate (FDR q value) <0.25 were considered statistically significant.
Results
Data information
By analyzing the RNA-seq information of TCGA database with 561 patients, there is 35 lung adenocarcinoma patients (6% in all collected patients) carried the RBM10 mutation.
Clinical impact of RBM10 mutation in lung adenocarcinoma progression and prognosis
The impact of RBM10 mutations on the progression of lung adenocarcinoma was evaluated. Firstly, the RBM10 mRNA levels in the wild-type group and in RBM10 mutation patients was assessed ()). In comparison of RBM10 wild-type patients, the expression of RBM10 was significantly decreased (p < 0.001). Then, we systematically investigated the relationship between patient survival and RBM10 conditions. As shown in ), the mutation condition of RBM10 is negative related with patient survival (p = 0.02), which indicated that patients carrying the RBM10 mutation will lead the poorer survival conditions. Meanwhile, higher expression of RBM10 mRNA in patient tissue also lead the poor survival compared with lower expression of RBM10. The progression-free survival rate of disease is shown in ). These data suggest that RBM10 mutation may contribute to the development of lung adenocarcinoma and early prevention may be advantageous in these patients.
Lung adenocarcinoma cells with RBM10 mutations are sensitive to RU-SKI 43
To evaluate and identify the new therapeutic approaches for lung cancer with RBM10 mutation, we utilized the GDSC database to identify which drugs are sensitive for RBM10 mutation. As shown in –e), the statics results revealed that RU-SKI43 may have strong selectivity for lung adenocarcinoma patients with RBM10 mutations. However, exception lung cancer, we also observed that there is no significance between wild-type and mutation of RBM10, which is treated with RU-SKI43.
GSEA analysis
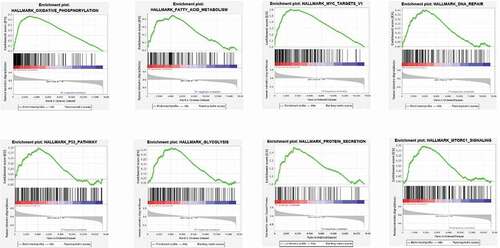
In order to identify the RBM10 mutation on the different functional signal pathway, we performed the GSEA for selected eight pathway, which are oxidative phosphorylation, fatty acid metabolism, MYC targets DNA repair, glycolysis, protein secretion, and MTORC1 signaling pathway. As shown in , the results indicated that the following pathways were markedly enriched: oxidative phosphorylation, fatty acid metabolism, MYC targets V1, DNA repair, glycolysis, p53 pathway, protein secretion, and mtorc1 signaling. To consider previous literature report [Citation19], This indicated that RBM10 mutations facilitate lung adenocarcinoma development by affecting various pathways involved in tumorigenesis, such as metastasis, cellular energy conversion, and cell proliferation.
Identification of differentially expressed genes
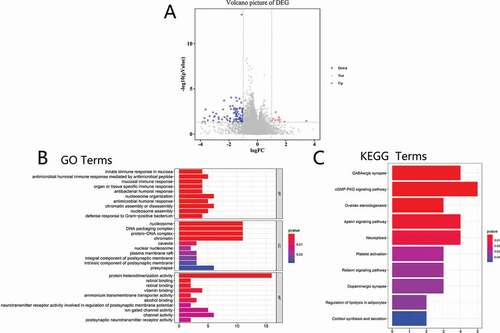
We identified differentially expressed genes to study altered genes and pathways caused by RBM10 mutations by querying RNA-Seq datasets from 35 patients with RBM10 mutation and 526 RBM10 wild-type group patients. Based on in silico parsing, 113 genes were identified as differentially expressed genes (log2FC ≥ 1.0 and p < 0.05). Among these genes, we found 10 genes that were upregulated and 103 genes that were downregulated. A volcano plot of differentially expressed genes is shown in ).
GO and KEGG analyses of differentially expressed genes
To study the functions of the 113 differentially expressed genes, GO was performed to analyze the relative biological functions. As shown in ), the enrichment of biological functions included nucleosome organization and assembly, chromatin assembly or disassembly, nucleosome, DNA packaging complex, protein-DNA complex, and protein heterodimerization activity. Moreover, KEGG pathway analysis was employed to enrich the differential genes to corresponding signal pathway: GABAergic synapse, cGMP-PKG signaling pathway, ovarian steroidogenesis, apelin signaling pathway, and necroptosis (as shown in )).
PPI network module screening
By investigate the protein–protein interaction (PPI) network, the key node information can clearly provide the role of node gene in genetic regulation process. Here, we utilized the STRING database to build the PPI network of differential genes. As shown in . We identified the top 10 genes as “central” genes. These node genes included HIST1H2BJ, HIST2H2BE, HIST3H2BB, HIST1H2BG, HIST1H2AG, HIST1H2BF, HIST1H4E, HIST1H2AL, HIST2H2BF, HIST1H2AE, and HIST1H2BJ, which has a score of 11 indicated the highest degree of nodes. We optimized PPI gene modules using the MCODE plug-in in Cytoscape and analyzed the top two signal pathway modules. The enrichment result revealed that genes in modules 1–2 were linked with the following functions: protein-DNA complex subunit organization, chromatin assembly or disassembly, nucleosome organization, protein heterodimerization activity, adenosine receptor signaling pathway, G protein-coupled chemoattractant receptor activity, growth hormone synthesis, secretion and action, and regulation of lipolysis in adipocytes.
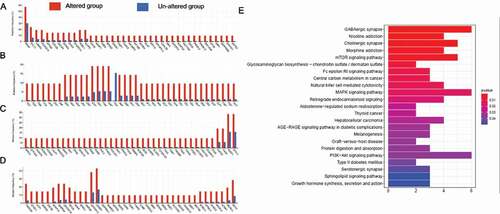
KEGG analysis of mutated genes in the RBM10 mutation group
To study the genetic function of 157 differential genes, KEGG analysis was performed and top 10 related pathways were enrichment with the cutoff value (p < 0.05). (–d)). The KEGG analysis indicated enrichment includes the GABAergic synapse, cholinergic synapse, mTOR signaling pathway, central carbon metabolism in cancer, MAPK signaling pathway, natural killer cell mediated cytotoxicity ()).
Discussion
RBM10 is a RNA-binding protein, as a variable splicing regulator, and the mutations always were observed in lung adenocarcinoma [Citation14]. RNA-binding proteins played a crucial role in tumorigenesis and always are marked dysfunctions compared with normal tissues [Citation20,Citation21]. For several types of RNA-binding proteins, the expression level is always associated with copy number variations, which highly promoted the tumorigenesis[Citation22]. As a consequence, to identify the role of RBM10 in lung cancer is very helpful for developing new therapeutic approaches or even drugs.
By performing to analyze the TCGA database of lung cancer, we found that 6% of 561 lung adenocarcinoma cases carried a variety RBM10 mutation, including frameshit mutations, truncations, and missense mutations (as shown in –b)). By analysis of RBM10 expression between wild-type and mutation, we also observed that expression of RBM10 in mutation patients is lower than wild-type patients ()). Furthermore, patients who carried RBM10 mutations had poorer survival rates and higher disease recidivation than wild-type RBM10 patients (–c)). Therefore, our results provided evidences that RBM10 mutations render proliferation and metastasis in patient with lung adenocarcinoma, which indicated a clinical significance for RBM10 mutations in lung adenocarcinoma patients and were consistent with previous literature reports [Citation23]. Moreover, our results also indicated that to apply the clinical interventions on the RBM10 mutation patients may significantly improve the survival conditions.
Figure 1. Mutation frequency (a) and types (b) of RBM10 in lung adenocarcinoma reproduced from The Cancer Genome Atlas (TCGA) database
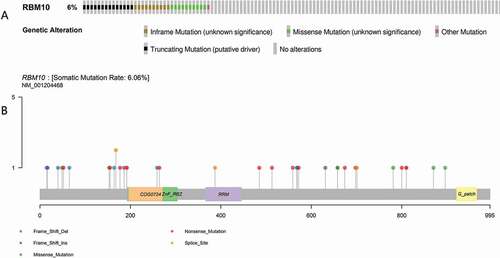
Figure 2. Mutations of RBM10 are associated with lung adenocarcinoma prognosis and drug selection. (a) Correlation between the RBM10 mutation and mRNA expression. (b,c) Kaplan–Meier survival and disease recurrence curves for lung adenocarcinoma patients stratified by the RBM10 mutation. (d,e) Reproduction of the GDSC database by excluding cancer of other types showed that lung adenocarcinoma cells with the RBM10 mutation was also significantly inhibited by RU-SKI43(p = 0.0044200). (f) Scattered plot and volcano plot show that multiple cancer cell types with the RBM10 mutation were not inhibited by RU-SKI43 (p > 0.05)
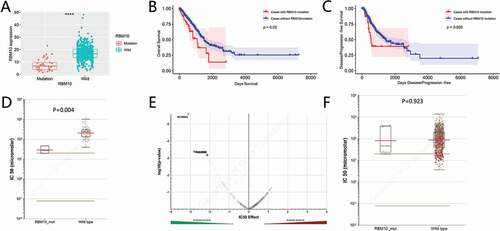
In clinical, to identify the genetic condition of Patients with or without RBM10 mutations in lung adenocarcinoma required the tissue section or genetic sequencing. After identification of genetic conditions, to precisely apply the drug interventions is helpful to improve the patients’ survival period. Here, performing the screening process in GDSC database, we identified the RU-SKI 43 series compounds, which displayed the higher sensitivity to lung adenocarcinoma harboring the RBM10 mutations (–f)). RU-SKI 43 was firstly reported as Hedgehog Acyltransferase inhibitor for Sonic hedgehog (Shh) signaling pathway [Citation24]. So far, no literature reported the relationship between Sonic hedgehog (Shh) signaling pathway and RBM10. But Sonic hedgehog signal pathway plays a critical role in lung cancer [Citation25].
To well-understand the underlying biology functions of the RBM10 mutations in lung adenocarcinoma tumorigenesis, we performed the RNA-Seq aligning and analysis of lung adenocarcinoma patients derived from the TCGA database to identify alterations in genes and pathways related to RBM10 mutations. To enrich the most relative signaling pathways, GSEA results () showed that RBM10 mutations were linked with multiple cancer-related pathways, including oxidative phosphorylation, fatty acid metabolism, MYC targets V1, DNA repair, glycolysis, p53 pathway, protein secretion, and mtorc1 signaling. p53 pathway anticipated and regulated amount key factors in cell proliferation and tumorigenesis process [Citation19]. By GSEA enrichment, the enrichment of p53 pathway indicated that the p53 pathway played the key role in poor survival period of RBM10 mutation lung cancer patients. These results suggest that RBM10 mutation may promote lung adenocarcinoma progression by influencing pathways in cellular proliferation, metabolism and DNA repair. These pathways are highly associated with the recently research which shows significantly the RBM10 mutation can promote tumor cell proliferation and invasion capacity [Citation23].
To identify the impact of genes on lung cancer, we found 113 differentially expressed genes (DEGs) between the RBM10 mutation group and wild-type group. Up- and downregulation DEGs are 10 and 103 ()), respectively. By further GO and KEGG enrichment (–c)), the corresponding results implied that DEGs were involved in various biological procedures, including GABAergic synapses, cGMP-PKG signaling pathways, regulation of lipolysis in adipocytes, nucleosome organization, and assembly. These results revealed that patients with RBM10 mutations may have great impact on the cellular signaling pathways and protein or organelle assembly for cell proliferation which is consistent with the recently reports [Citation14,Citation23].
Figure 4. Cluterprofiler enrichment results of differentially expressed genes. (a) Volcano plot for differentially expressed genes. (b) The GO enrichment terms of differentially expressed genes. (c) The KEGG pathway analysis of differentially expressed genes
By performing the protein–protein interaction network analysis ( and )), we identified 16 differentially expressed genes, which played the key role in the maintenance of tumorigenesis. The first key genetic clusters are histone protein family (as shown in )), which are related to nucleosome assembly and negative regulation of tumor necrosis factor-mediated signaling pathways. KEGG and GO analysis ( and )) of the top two modules in the PPI network showed an enrichment of genes involved in protein-DNA complex subunit organization, protein heterodimerization activity, regulation of lipolysis in adipocytes, growth hormone synthesis, cholinergic synapse, G protein-coupled chemoattractant receptor activity, adenosine receptor signaling pathway, showing again that lung adenocarcinomas with the RBM10 mutation tend to be highly active in a variety cellular biological processes.
Figure 5. Top two modules from the PPI network. (a,b) PPI network and GO and KEGG analyses of module 1. (c,d) PPI network and GO and KEGG analyses of module 2

Meanwhile, we also investigated the genetic level conditions of lung cancer patients harboring RBM10 mutation. After alignment, 157 genes were identified as more frequent mutation in the RBM10 mutation group than the wild-type group (p value<0.05). The subsequent KEGG analysis indicated enrichment regarding GABAergic synapse, cholinergic synapse, mTOR signaling pathway, central carbon metabolism in cancer, MAPK signaling pathway, natural killer cell mediated cytotoxicity. These enrichment signal pathways () indicated that numerous specific signaling pathways are in operation during the development of LUAD with RBM10 mutations. Moreover, these pathways demonstrated that numerous specific signaling pathways are in operation during the development of LUAD with RBM10 mutations.
Figure 6. (a,b,c,d) Comparisons of the mutation frequencies of genes between the RBM10 mutation and wild-type groups of lung adenocarcinoma (LUAD) patients (p value<0.01). (e) KEGG analysis of the genes with differential mutation rates
There are, however, limitations to our study. We were only able to investigate whether RBM10 mutations in lung adenocarcinoma affect disease development and prognosis. The exact underlying mechanisms of RBM10 mutations in lung adenocarcinoma development remain to be more thoroughly investigated in both in vitro biological studies and clinical research.
Conclusion
In summary, by performing the GSEA enrichment and further analysis, our results identified prime genes and pathways highly associated with RBM10 mutations in lung adenocarcinoma. Through our studies, our results may provide the new therapeutic approaches, i.e. RU-SKI 43 to target Shh pathway, and be helpful to improve the survival rate of patients with RBM10 mutations in lung adenocarcinoma.
Data availability
Publicly available datasets were analyzed in this study. This data can be found here: https://portal.gdc.cancer.gov/.
Disclosure statement
The authors declare no conflicts of interest.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
- DeSantis CE, Lin CC, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64:252–271.
- Camidge DR, Doebele RC, Kerr KM. Comparing and contrasting predictive biomarkers for immunotherapy and targeted therapy of NSCLC. Nat Rev Clin Oncol. 2019;16:341–355.
- Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500.
- Kwak EL, Bang Y-J, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non–small-cell lung cancer. N Engl J Med. 2010;363:1693–1703.
- Glisovic T, Bachorik JL, Yong J, et al. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986.
- Zhao J, Sun Y, Huang Y, et al. Functional analysis reveals that RBM10 mutations contribute to lung adenocarcinoma pathogenesis by deregulating splicing. Sci Rep. 2017;7:1–11.
- Mueller CF, Berger A, Zimmer S, et al. The heterogenous nuclear riboprotein S1-1 regulates AT1 receptor gene expression via transcriptional and posttranscriptional mechanisms. Arch Biochem Biophys. 2009;488:76–82.
- Sun X, Jia M, Sun W, et al. Functional role of RBM10 in lung adenocarcinoma proliferation. Int J Oncol. 2019;54:467–478.
- Wang Y, Gogol‐Döring A, Hu H, et al. Integrative analysis revealed the molecular mechanism underlying RBM 10‐mediated splicing regulation. EMBO Mol Med. 2013;5:1431–1442.
- Pan Q, Shai O, Lee LJ, et al. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415.
- David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 2010;24:2343–2364.
- Ji Y, Xie S, Jiang L, et al. Increased cell apoptosis in human lung adenocarcinoma and in vivo tumor growth inhibition by RBM10, a tumor suppressor gene. Oncol Lett. 2017;14:4663–4669.
- Hernández J, Bechara E, Schlesinger D, et al. Tumor suppressor properties of the splicing regulatory factor RBM10. RNA Biol. 2016;13:466–472.
- Cao B, Fang Z, Liao P, et al. Cancer-mutated ribosome protein L22 (RPL22/eL22) suppresses cancer cell survival by blocking p53-MDM2 circuit. Oncotarget. 2017;8:90651.
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140.
- McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297.
- Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9. 1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2012;41:D808–D15.
- Jung JH, Lee H, Cao B, Liao P, Zeng SX, Lu H. RNA-binding motif protein 10 induces apoptosis and suppresses proliferation by activating p53. Oncogene. 2020;39(5):1031–1040. doi:10.1038/s41388-019-1034-9
- Blackinton JG, Keene JD. Post-transcriptional RNA regulons affecting cell cycle and proliferation. Semin Cell Dev Biol. 2014;44–54. DOI: 10.1016/j.semcdb.2014.05.014.
- Bonnal S, Vigevani L, Valcárcel J. The spliceosome as a target of novel antitumour drugs. Nat Rev Drug Discov. 2012;11:847–859.
- Wang ZL, Li B, Luo YX, et al. Comprehensive genomic characterization of RNA-binding proteins across human cancers. Cell Rep. 2018;22:286–298.
- Yin LL, Wen XM, Li M, et al. A gene mutation in RNA-binding protein 10 is associated with lung adenocarcinoma progression and poor prognosis. Oncol Lett. 2018;16:6283–6292.
- Petrova E, Rios-Esteves J, Ouerfelli O, et al. Inhibitors of hedgehog acyltransferase block sonic hedgehog signaling. Nat Chem Biol. 2013;9:247–249.
- Abe Y, Tanaka N. The hedgehog signaling networks in lung cancer: the mechanisms and roles in tumor progression and implications for cancer therapy. Biomed Res Int. 2016;2016:7969286.