ABSTRACT
We have recently reported that transgenic mice overexpressing the HMGA1-pseudogene7 develop hematological neoplasia marked by monoclonal B-cell populations, and diagnosed as Diffuse Large B-cell Lymphoma. These findings prove the HMGA1-pseudogene7 oncogenic role in vivo.
Background
Pseudogenes are residues deriving from primordial genes following the deprivation of their encoding function by mutations in encoding regions and/or regulatory elements [Citation1]. Indeed, for a long time, many genes may not have worked as protein-coding genes, generating several mutations and degenerating into pseudogenes [Citation2].
Since their finding, pseudogenes have been ignored and viewed as defective versions of coding genes since they have lost their ability to code for proteins, remaining without an evident function [Citation3], even representing a considerable portion of the human transcriptome and proteome. Only recently, it has been reported that thousands of them are transcribed and hundreds are also translated [Citation4,Citation5]. To date, pseudogenes are classified among the group of long non-coding RNAs (lncRNAs) given that their mRNAs can produce non‐coding RNAs and anti‐sense RNAs [Citation6–8]. Notably, pseudogenes have frequently miRNA response elements (MREs) along their nucleotide sequences that, by a competing endogenous RNA (ceRNA) mechanism, can sequester microRNAs (miRNAs), then modulating the expression of the parental genes and other genes with the same MREs [Citation9].
Pseudogenes and ceRNA mechanisms in cancer
The importance of pseudogenes in cell physiology has become recently evident thanks to the study of Poliseno et al. who demonstrated that PTENP1 pseudogene can upregulate the parental PTEN gene expression levels by sequestering miR-21 (which is able to target PTEN) and, then, breaking the repression exerted by PTEN, thus generating a growth-suppressive signal [Citation3]. Consistently, it has been shown that patients affected by clear renal cell carcinoma, which do not express PTENP1, displayed a shorter overall survival compared to those expressing PTENP1 [Citation3].
Consequently, it is plausible to think that pseudogenes are not passively endured during evolution, but it is likely that they have been positively selected to remain and play some roles in gene expression and regulation.
Then, in agreement with the role of pseudogene expression in carcinogenesis, it has been described that the expression levels of 309 pseudogenes, out 440 analyzed, were extensively altered in breast cancer [Citation10].
To date, the genetically engineered mouse models have permitted to demonstrate that abnormally expressed pseudogenes are enough to cause cancer by ceRNA mechanisms. Indeed, mice ubiquitously overexpressing the Braf-rs1, the processed pseudogene of the mouse Braf kinase, which shares three miRNAs (miR-134, miR-543, and miR-653) with its primordial genes, were generated [Citation11]. These mice developed a symptomatology characterized by splenomegaly and enlarged lymph nodes, that together with the flow cytometric analysis of the neoplastic cells allowed the diagnosis of an aggressive form of Diffuse Large B-cell Lymphoma (DLBCL) [Citation11]. Consistently with the ceRNA theory, increased Braf and pErk levels have been found in these Braf-rs1-generated tumors [Citation11].
In humans, the processed pseudogene of human BRAF is BRAFP1: they share 40 miRNAs, being together involved in a ceRNA pathway each other. As expected by its potential oncogenic activity, the amplification of BRAFP1 genomic locus was found in most of the human malignancies, including DLBCL, where the expression levels of BRAFP1 and BRAF were associated by a positive correlation [Citation11].
Therefore, other than the BRAF mutations, in particular, that at Val600Glu residue, it can be affirmed that BRAF activation in cancer may also depend on its enhanced expression sustained by its parental pseudogene BRAFP1 [Citation12].
HMGA1-pseudogenes role in cancer
In this context, the recently identified HMGA1P6 and HMGA1P7 (HMGA1Ps) pseudogenes have been considered excellent candidates as pseudogenes with oncogenic activity. They are two processed pseudogenes of HMGA1 gene (located at chromosome 6p21.31), a well-studied human oncogene involved in cancer progression [Citation13,Citation14]. HMGA1P6 (located at chromosome 13q12.12) and HMGA1P7 (located at chromosome 6q23.2) are transcribed but not translated since the former yields in its sequence a mutation in the stop codon, whereas the latter carries a missense mutation of the initiator methionine codon [Citation15,Citation16]. However, HMGA1Ps perfectly conserve MREs for several miRNAs that can potentially target HMGA1 and also other related genes such as HMGA2, Enhancer of Zeste Homolog 2 (EZH2) and Vascular Endothelial Growth Factor (VEGF), whose involvement in carcinogenesis has been already widely reported [Citation15–17]. Thus, the pseudogenes of HMGA1 represent key regulators for a novel ceRNA oncogenic network. Indeed, it has been already reported that HMGA1P6 and HMGA1P7 are able to induce cell proliferation and increase the G1-S phase transition rate by enhancing HMGA1 protein expression. Interestingly, their expression has been found drastically upregulated in the undifferentiated anaplastic thyroid carcinomas (ATC), but not in the differentiated papillary (PTC) ones [Citation15], in stomach [Citation18], ovary [Citation15,Citation18], breast [Citation19] and in endometrial carcinomas (EEC) [Citation20], where their expression correlates with that of the parental gene. Moreover, in endometrial cancer HMGA1Ps expression represents a negative prognostic factor for EEC patients, since it is associated with increased histological grade and tumor size [Citation20]. More recently, it has been demonstrated that HMGA1P6 overexpression correlates with a shorter overall patient survival time compared with the low expressors in ovarian cancer. Interestingly, the same authors report that MYC controls the expression of HMGA1P6 by binding to its promoter, and that HMGA1P6 enhances sphere-forming efficiencies and invasiveness of ovarian cancer cells by the upregulation of both HMGA1 and HMGA2 through a ceRNA mechanism, then reinforcing HMGA1Ps critical role in cancer progression [Citation18].
Noteworthy, HMGA1P6 and HMGA1P7 expression have been found upregulated in growth hormone and nonfunctioning pituitary adenomas, where it significantly correlates with that of HMGA1 known to have, together with HMGA2, a pivotal role in the development of these neoplastic diseases [Citation21]. Moreover, in larynx squamous carcinomas, HMGA1Ps have been found overexpressed and a linear correlation between their expression levels and those of HMGA2 has been reported, confirming once more the upregulation of these pseudogenes in human cancer [Citation22].
In order to evaluate HMGA1P6 and HMGA1P7 oncogenic activity in vivo, our research group generated transgenic mice overexpressing these pseudogenes [Citation23] (). By a mean age of 12 months, about 50% of the transgenic mice died, developing splenomegaly and accumulation of lymphoid cells in spleens and also in other body compartments such as liver, kidney and lung [Citation24]. FACS and immunohistochemistry analyses showed that the expanded lymphoid population was positive for CD19 and CD45/B220 markers, respectively, and a clonal expansion was further confirmed by clonality assay, thus suggesting a diagnosis of B-cell lymphomas [Citation24] ().
Figure 1. Main features of HMGA1P7-transgenic mice. HMGA1P7-transgenic mice showed a high mortality rate, developing a diffuse large B-cell lymphoma through HMGA1-independent ceRNA mechanism
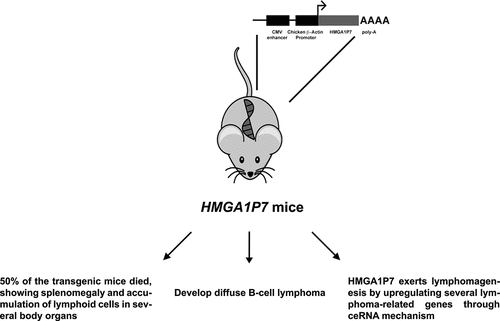
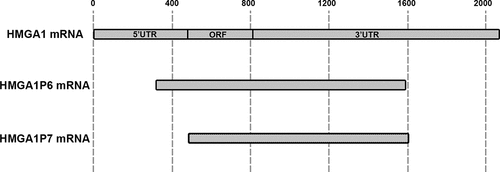
Figure 2. HMGA1-pseudogene family sequences. HMGA1 and its pseudogenes mRNA sequences are aligned and shown in gray
The transcriptomic analysis of HMGA1P7-explanted spleens compared with the WT ones revealed the upregulation of genes involved in inflammation, in the NFKB pathway and mTOR signaling. Conversely, the group of the downregulated genes included those decreased in post-germinal center (GC) BCL6 dependent B cell lymphomas and present in the GC B-cell type (GCB) Diffuse Large B cell Lymphoma (DLBCL) signature proposed a more specific diagnosis of DLBCL of the non-GCB type for these lymphoid proliferative lesions, in agreement also with the histological features [Citation24]. Noteworthy, a very similar pathology was also observed in the transgenic mice overexpressing HMGA1P6.
Interestingly, the development of DLBCL in HMGA1P7 mice does not seem to be dependent on HMGA1 induction, since its expression has not been found upregulated. However, the HMGA1P7 oncogenic potential lies likely in its ability to overexpress several cancer-related genes that share the same MREs with HMGA1P7 [Citation24]. Indeed, through the transcriptomic analysis, genes involved in lymphomagenesis as Cebpd, Ccl24, Fos, Bcl2l1 and Il1a have been found upregulated in HMGA1P7 transgenic mouse spleens compared to the WT ones, and, intriguingly, all these transcripts were regulated by HMGA1P7-induced ceRNA mechanism [Citation24]. Interestingly, the HMGA1P7 transgenic mice developed also sarcoma and two cases of renal cell carcinomas and one case of liver carcinoma, that have not been observed in the littermates and never reported in the WT C57BL/6 N. Then, it is reasonable to hypothesize that they are caused by HMGA1P7 overexpression by a ceRNA mechanism, however, by competition with unknown genes. A transcriptomic analysis of the HMGA1P7-solid tumors might give more information about the mechanisms underlying the development of these malignancies. It is likely that the early death of the HMGA1P7 mice has hidden the frequency of these solid tumors since it is well known that the malignant transformation of epithelial cells requires more genetic alterations in comparison with hematological malignancies.
Therefore, taken together all the data reported by De Martino et al. clearly support that HMGA1P7 and HMGA1P6 (unpublished data) represent the first human pseudogenes that show oncogenic activity in vivo.
Then, studies have been undertaken to define the role of HMGA1 and its pseudogenes in human DLBCL. Preliminary results of our research group show HMGA1 overexpression in almost all the cases of DLBCL analyzed, and, interestingly, these expression levels significantly correlate with those of EZH2, which plays a central role in human lymphomagenesis [Citation25]. Interestingly, even though no HMGA1Ps overexpression has been detected in DLBCL, the analysis of 40 samples of DLBCL available in the Cancer Genome Atlas (TGCA) revealed the overexpression of HMGA1P1, another HMGA1 pseudogene, whose locus is located on chromosome Xp21.3, and it has been also found amplified in these lymphomas (manuscript in preparation). Interestingly, only few point mutations have been found in HMGA1P1 sequence compared to HMGA1a one, and, importantly, all these mutations seem do not affect the translation ability of this pseudogene [Citation26]. Indeed, some HMGA1P1 mutations hit amino acidic residues that are frequently modified at post-translational level along the HMGA1 protein, controlling its functions. In this manner, HMGA1P1 overexpression might contribute to carcinogenesis by enhancing HMGA1 activity, which in turn may induce EZH2 expression by binding to its promoter (manuscript in preparation), and it may also have some oncogenic activity per se, that needs to be validated by further studies.
In conclusion, all these results seem to validate a critical role of the HMGA1 pseudogenes as oncogenes in cancer development.
Disclosure statement
No potential conflict of interest was reported by the author.
References
- An Y, Furber KL, Ji S. Pseudogenes regulate parental gene expression via ceRNA network. J Cell Mol Med. 2017;21(1):185–192.
- Poliseno L. Pseudogenes: newly discovered players in human cancer. Sci Signal. 2012;5(242):re5.
- Poliseno L, Salmena L, Zhang J, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465(7301):1033–1038.
- Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108.
- Kim MS, Pinto SM, Getnet D, et al. A draft map of the human proteome. Nature. 2014;509(7502):575–581.
- Groen JN, Capraro D, Morris KV. The emerging role of pseudogene expressed non-coding RNAs in cellular functions. Int J Biochem Cell Biol. 2014;54:350–355.
- Milligan MJ, Lipovich L. Pseudogene-derived lncRNAs: emerging regulators of gene expression. Front Genet. 2014;5:476.
- Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199–208.
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–358.
- Welch JD, Baran-Gale J, Perou CM, et al. Pseudogenes transcribed in breast invasive carcinoma show subtype-specific expression and ceRNA potential. BMC Genomics. 2015;16:113.
- Karreth FA, Reschke M, Ruocco A, et al. The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell. 2015;161(2):319–332.
- Lin JD, Fu SS, Chen JY, et al. Clinical manifestations and gene expression in patients with conventional papillary thyroid carcinoma carrying the BRAF(V600E) mutation and BRAF pseudogene. Thyroid. 2016;26(5):691–704.
- Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat Rev Cancer. 2007;7(12):899–910.
- De Martino M, Fusco A, Esposito F. HMGA and cancer: a review on patent literatures. Recent Pat Anticancer Drug Discov. 2019;14(3):258–267.
- Esposito F, De Martino M, Petti MG, et al. HMGA1 pseudogenes as candidate proto-oncogenic competitive endogenous RNAs. Oncotarget. 2014;5(18):8341–8354.
- Esposito F, De Martino M, Forzati F, et al. HMGA1-pseudogene overexpression contributes to cancer progression. Cell Cycle. 2014;13(23):3636–3639.
- De Martino M, Palma G, Azzariti A, et al. The HMGA1 pseudogene 7 Induces miR-483 and miR-675 upregulation by activating Egr1 through a ceRNA mechanism. Genes (Basel). 2017;8:11.
- Tian X, Song J, Zhang X, et al. MYC-regulated pseudogene HMGA1P6 promotes ovarian cancer malignancy via augmenting the oncogenic HMGA1/2. Cell Death Dis. 2020;11(3):167.
- De Martino M, Forzati F, Marfella M, et al. HMGA1P7-pseudogene regulates H19 and Igf2 expression by a competitive endogenous RNA mechanism. Sci Rep. 2016;6:37622.
- Palumbo Junior A, de Sousa VPL, Esposito F, et al. Overexpression of HMGA1 figures as a potential prognostic factor in endometrioid endometrial carcinoma (EEC). Genes (Basel). 2019;10(5):372.
- Esposito F, De Martino M, D’Angelo D, et al. HMGA1-pseudogene expression is induced in human pituitary tumors. Cell Cycle. 2015;14(9):1471–1475.
- Palumbo A Jr., De Martino M, Esposito F, et al. HMGA2, but not HMGA1, is overexpressed in human larynx carcinomas. Histopathology. 2018;72(7):1102–1114.
- De Biase D, Esposito F, De Martino M, et al. Characterization of inflammatory infiltrate of ulcerative dermatitis in C57BL/6NCrl-Tg(HMGA1P6)1Pg mice. Lab Anim. 2018;23677218815718. DOI:10.1177/0023677218815718
- De Martino M, De Biase D, Forzati F, et al. HMGA1-pseudogene7 transgenic mice develop B cell lymphomas. Sci Rep. 2020;10(1):7057.
- Lue JK, Amengual JE. Emerging EZH2 inhibitors and their application in lymphoma. Curr Hematol Malig Rep. 2018;13(5):369–382.
- De Martino M, Forzati F, Arra C, et al. HMGA1-pseudogenes and cancer. Oncotarget. 2016;7(19):28724–28735.
