ABSTRACT
The function of a new long non-coding RNA GAS6-AS2 in non-small cell lung cancer (NSCLC) is not fully understood. In this study, GAS6-AS2 was identified, and its roles as well as mechanisms in regulating proliferation of NSCLCs cells were investigated. qRT-PCR was used to analyze GAS6-AS2, miR-144-3p, and MAPK6 expression. Protein expression was detected by Western blotting. Cell Counting Kit-8 (CCK8) assay was used to examine the cell proliferation ability. The interaction between GAS6-AS2 and miR-144-3p was confirmed by dual-luciferase reporter assay and RNA pull down assay. A xenograft model was constructed to monitor the mice NSCLC tumor growth in vivo. GAS6-AS2 was up-regulated, while miR-144-3p was suppressed in NSCLC cells compared with normal lung cells. GAS6-AS2 suppression could inhibit the progression of NSCLC cells, and miR-144-3p could attenuate the effect. GAS6-AS2 could function as a competitive endogenous RNA (ceRNA) via direct sponging miR-144-3p-3p, which further regulating the expression of MAPK6. The knockdown of GAS6-AS2 could greatly suppress the tumor growth of NSCLC in vivo. GAS6-AS2 up-regulated MAPK6 by sponging miR-144-3p in NSCLC tissues and cells. Thus, GAS6-AS2 is an effective therapeutic target in NSCLC.
1. Introduction
Lung cancer is a very common human cancer and the top cause of cancer-related deaths all over the world [Citation1], with non-small cell lung cancer (NSCLC) constituting as the major type in lung cancer patients [Citation2,Citation3]. Although there exist surgeries, chemotherapies, and targeted drugs, the 5-year survival rates and recurrence rates still need to be improved [Citation4]. The key to novel and effective treatments is the accurate understanding of the pathological mechanism underlying the initiation, development, and progression of NSCLC [Citation5]. Therefore, it is quite important to understand their molecular regulatory mechanisms to explore novel therapeutic strategies.
In recent years, accumulating studies have been shown that non-coding RNAs such as long non-coding RNAs (lncRNAs) [Citation6–8] and micro RNAs (miRNAs) [Citation9,Citation10], as well as their corresponding messenger RNAs (mRNAs) [Citation11,Citation12] involve in the regulation of NSCLC cell viability, progression, and metastasis. LncRNA GAS6-AS2 (GAS6-AS2), a newly discovered tumor-related lncRNA, was mapped to 13q34 with a transcript length of 12,361 bp. Up to date, the studies on the expression and function of GAS6-AS2 in tumors were limited. miRNAs are a kind of endogenous small noncoding RNA, with a length of 22 nt, which negatively control the gene expression levels through binding to their complementary target mRNAs. By bioinformatics tools, miR-144-3p, the mature form of miR-144, was predicted to be sponged by GAS6-AS2. Although miR-144-3p was not only detected in lung cancer but also in breast and colorectal cancer, the regulatory mechanism between GAS6-AS2 and miR-144-3p remains unknown.
Moreover, it has been known that ERK3/4‐MK5 signaling pathway play an important role in cancer invasion and migration, including NSCLC. MAPK6 (mitogen‐activated protein kinase), a well-known member of the MAPK family, has been reported to be involved in the ERK3/4‐MK5 pathway in regulating lung cancers [Citation13,Citation14]. MAPK6 was mainly located in the nucleus and cytoplasm and in different organs [Citation15]. In lung cancer, MAPK6 signals can promote human lung cancer cell proliferation and invasion through SRC-3 coactivator and be upregulated by NEAT1/has-mir-98-5paxis [Citation13,Citation16].
In this study, we identified a novel lncRNA termed GAS6-AS2 for the first time, and our results demonstrated that GAS6-AS2 contributed to proliferation and metastasis of NSCLC cells via the GAS6-AS2/miR-144-3p/MAPK6 axis. The research broadened our insights into the underlying mechanisms in proliferation and metastasis, and provided a new therapeutic target for NSCLC cancer patients in clinic.
2. Materials and methods
2.1. Tissue samples
NSCLC tissues and matched adjacent normal lung tissues were obtained from 22 patients who were undergoing surgeries in Henan Provincial People’s Hospital. The patients had no therapies like radiotherapy, chemotherapy, or immunotherapy before surgery. All of them sent informed consent to us. The patient tissue specimens were obtained with written informed consents according to the established protocols approved by the Ethics Committee of First Affiliated Hospital of Henan Provincial People’s Hospital. Tissues were frozen in liquid nitrogen and maintained at −80°C. Ethics board of Henan Provincial People’s Hospital approved our research. Clinicopathological information is shown in .
Table 1. The relationship between lncRNA GAS6-AS2 expression and the clinicopathological features in NSCLC patients
2.2. Cell cultures
Human lung cancer cells (A549, H1299, H460, and H1975) and normal lung cell (BES-2B) were purchased from American Type Culture Collection, USA. A549 and H1299 were maintained in RPMI 1640 medium with 10% FBS, 100 U/ml penicillin, and 100 mg/ml streptomycin. H460 and BES-2B were maintained in DMEM, USA. The cells were incubated at 37°C in 5% CO2.
2.3. CCK8 assay
A549 and H1299 (50,000 cells/well) were plated in a 96-well plate with 50,000 cells/well. At 0, 2, 4, 6 days, 100 μL CCK8 (Dojindo, Japan) was utilized in every well. After 4 hours, the microplate reader (Bio Tek, US) was employed to measure the signals at 450 nm.
2.4. Flow cytometry (FCM)
The flow cytometry was performed using CytoFLEX (Beckman Coulter, Inc., USA). Cells in logarithmic growth phase of each group was digested with pancreatic enzymes and centrifuged. The supernatant was removed and cells collected. After cells were washed with pre-cold PBS, centrifugation was performed at 1000 rpm for 5 min twice. Then, cell density was adjusted into 106/ml and a total volume of 200 μl cells were washed with 1 ml PBS twice before centrifugation. Cells were re-suspended in 100 μl of binding buffer, added with 2 μl of Annexin-V-FITC (20 μg/ml) and mixed at ice without light exposure. After 15 min, cells were transferred into the testing tube and 300 μl of PBS was added. Each sample was added with 1 μl PI (50 μg/ml) before detection within 30 min. Using AnnexinV as horizontal axis, PI as vertical axis, cells in upper left area were mechanical damaged cells, upper right area were late apoptotic cells or necrotic cells, lower left area were negative normal cells and lower right area were early apoptotic cells.
2.5. qRT-PCR
Total RNA was extracted via RNAiso Plus (TaKaRaBio, China), and reversely transcribed via Prime Script TM RT Master (TaKaRa, China). qPCR was conducted via SYBR Premix ExTaqII (TaKaRaBio, China) on the RealTime PCR System (Applied Biosystems, USA). The relative expression levels of miRNA was normalized to GAS6-AS2 expression and calculated with 2−ΔΔCq method. The primer sequences for qPCR were as follows: GAS6‐AS2 forward: 5ʹ- AAGGAGGACGCAATACC-3ʹ; and reverse: 5ʹ-ATCCTGGCTAACACGGT; GAPDH forward: 5ʹ-GTCTCCTCTGACTTCAACAGCG-3ʹ; and reverse: 5ʹ-ACCACCCTGTTGCTGTAGCCAA-3ʹ. miR-144-3p forward: 5ʹ-GCGCGCTACAGTATAGATGATG‐3′, and reverse: 5′‐GCTGTCAACGATACGCTACG‐3′ MAPK6 forward: 5′‐GGAGCGAGATCCCTCCAAAAT‐3′, and reverse 5′‐GGCTGTTGTCATACTTCTCATGG‐3′.
2.6. Transfection
The sequence si-GAS6-AS2-1 was 5′‐CTGTATGTACACTTTTTTGTC‐3′ and si-GAS6-AS2-2 was 5′‐CTGGGAATGATCTTCAAGGAG‐3′, which were perchased from RiboBio (Guangzhou, China). GAS6-AS2 cDNA was cloned to vector pcDNA3.1 (Invitrogen, USA). miR-144-3p mimics, inhibitors or the negative controls was obtained from RiboBio, China. They were transfected to A549 and H1299 cells by Lipofectamine 2000 (Invitrogen, US).
2.7. Luciferase activity
GAS6-A2-WT (wild-type) or GAS6-A2-MUT (mutant) were synthesized and cloned to pGL3 Basic vector (Promega, USA). A549 cells were plated on 24-well plates for 2 days. miR-144-3p-mimics or miR-144-3p-inhibitors were co-transfected with 10 μg GAS6-A2-WT or GAS6-A2-MUT via Lipofectamine 2000. The MAPK6-WT or MAPK6-MUT was co-transfected with miR-144-3p-mimics or miR-144-3p-inhibitor by Lipofectamine 2000. Dual-Luciferase Reporter Assay System was used to detect the luciferase activities (Promega, USA).
PCR was performed to amplify the partial sequences of MAPK6 which contained the putative binding sites of miR-144-3p. The sequences were then cloned into the pmirGLO Dual Luciferase miRNA Target Expression Vector (Promega Corp., Fitchburg, WI, USA). GeneArtTM Site-Directed Mutagenesis system (Thermo Fisher Scientific, Inc.) was used to induce site-directed mutagenesis of miR-144-3p complementary bases in the sequences of MAPK6. Then, A549 cells were transfected with the constructed wild-type (WT) and mutant (MUT) reporter vectors, respectively, in the presence of miR-144-3p mimics or miRNC. The activity of luciferase was measured using the Dual Luciferase Assay system (Promega) and then normalized to Renilla luciferase activity based on the manufacturer’s instructions. The assay was performed in triplicate experiments.
2.8. RIP assay
RIP assay was performed via Magna RIP Kit (Millipore, US). A549 cells were lysed and maintained with human anti-Ago2 antibodies or control IgG antibodies, both conjugated with magnetic beads (Millipore, USA). qRT-PCR was carried out to measure GAS6-AS2 and miR-144-3p expressions.
2.9. RNA pull-down assay
The RNAs were biotin-labeled with Pierce RNA 3ʹEnd Desthiobiotinylation Kit (Thermo Fisher, US). MiR-144-3p-WT, miR-144-3p-Bio, miR-144-3p-MUT, miR-144-3p-Bio-MUT) and NC-Bio were treated via A549 lysates with magnetic beads. QRT-PCR was used to detect RNA.
2.10. Colony formation assay
400 cells/well were plated in 6-well plates. The culture medium was refreshed frequently. After 14 days, adherent cells were washed and fixed by 4% paraformaldehyde for 30 min at 25°C. Giemsa solution dyed the cells for 15 min. After washing and drying, cell colonies were counted.
2.11. EdU assay
The cell proliferation rate was detected through EdU assay via EdU Kit (RiboBio, China). Cells (1x104) were seeded in each well of 96-well plates. After 2 days at 37°C and 5% CO2, cells were added by 50 mM EdU for 2 hours. We fixed the cells with 4% paraformaldehyde and stained them by Apollo Dye Solution. Nucleic was stained by Hoechst 33,342. Cell proliferation rates were calculated, and the photos were recorded by fluorescence microscope (Olympus FSX100, Japan).
2.12. Western blotting
The proteins were loaded and separated by 10% SDS polyacrylamide gels. They were transferred to PVDF membranes (Millipore, USA) and treated by first antibody at 4°C for a night. Next, they were maintained with secondary antibody for 1 hour at 25°C. The primary antibodies were: anti-MAPK6 (1:500, Abcam, UK) and anti-GAPDH (1:500, Abcam, UK). (Viber Lourmat, France).
2.13. Xenograft model
Male BALB/c nude mice (20–22 g) were purchased from Animal Center of the Chinese Academy of Science. 5 × 106 A549 cells transfected with sh-GAS6-AS2-2 or sh-GAS6-AS2-2+ anti-miR144 were inoculated into the left flank in the dorsal of the nude mice (n = 5 per group) for in vivo xenograft assay. Tumor sizes and mice weight were detected every week. The tumor volumes were calculated according to (length × width2)/2. Mice were sacrificed at 28 days after the injection, mice were sacrificed. Tumor xenografts were harvested, weighed, and preserved in liquid nitrogen. Animal Ethics Committee of Henan Provincial People’s Hospital proved all the experiments, and performed in accordance with the NIH guidelines for the care and use of laboratory animals.
2.14. Bioinformatics analysis
The prediction of target genes of miR-144-3p was performed by Starbase [Citation17] (http://starbase.sysu.edu.cn), TargetScan [Citation18] (http://www.targetscan. org/) and miRDB [Citation19] (mirdb.org/miRDB/) bioinformatics analysis, followed by further analysis with the KEGG system. Twenty predicted targets were identified during first screening. Next, the secondary screening was performed by selection of genes with functional change that induced any changes in GAS6-AS2 and MAPK6 in previous studies (Data not shown), which were selected priority over other candidates.
2.15. Statistical analysis
Results were shown as mean ± SD. Comparisons within groups were analyzed by t-test or one-way ANOVA. Pearson’s correlation coefficient was used for the correlation analysis between plasma levels of GAS6-AS2 and miR-144-3p. P < 0.05 was considered as statistically significant. SPSS 18.0 and GraphPad Prism v6. were used for data analysis.
3. Results
3.1. GAS6-AS2 was enhanced, and miR-144-3p was reduced in NSCLC tissues and cell lines
To determine the expression of GAS6-AS2 in NSCLC tissues, RT-qPCR was performed to detect GAS6-AS2 expression in 22 pairs of NSCLC and adjacent normal tissues. In NSCLC tissues, the expression of GAS6-AS2 was elevated compared with adjacent normal lung tissues (p < 0.05, )), which hinted that GAS6-AS2 might attend to the modulation of NSCLC cancer cell growth and proliferation. Pearson’s correlation coefficient was used for the correlation analysis between expression of GAS6-AS2 and miR-144-3p. ) illustrates that the expression of GAS6-AS2 had a negative relationship with the expression of miR-144-3p (R2 = 0.578).. However, when we further to explore whether GAS6-AS2 is related to tumor development, the clinical data gave us a negative result. Statistical analysis showed that in different groups of tumor patients (age <55, female, tumor size <3 cm, TNM III–IV, lymph node metastasis, poorly differentiation, distant metastasis), GAS6-AS2 expression was not statistically different (P > 0.05, ).
Figure 1. GAS6-AS2 was enhanced, and miR-144-3p was reduced in NSCLC tissues and cell lines. (a) qRT-PCR for GAS6-AS2 expression in NSCLC and adjacent normal tissues, n = 22. (b) The negative correlation between GAS6-AS2 and miR-144-3p in NSCLC tissues. (c and d) qRT-PCR for GAS6-AS2 and miR-144-3p expression in NSCLC cell lines (A549, H460, H1299 and H1975) compared with normal cells (BEAS-2B). *compared with adjacent normal tissues, p < 0.05; *compared with BEAS-2B, p <0.05. n = 3
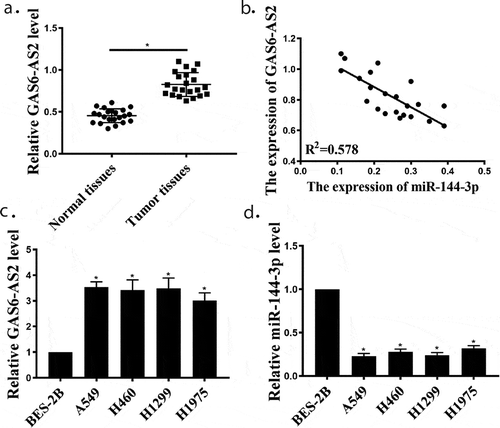
Additionally, we also measured the expression of GAS6-AS2 and miR-144-3p in four NSCLC cell lines (A549, H460, H1299 and H1975) and human lung normal epithelial cells (BES-2B). ) showed that the expression of GAS6-AS2 in NSCLC cells was significantly enhanced than that in BESA-2B cells (p < 0.05). In addition, ) demonstrates that the expression of miR-144-3p in NSCLC cells were significantly down-regulated compared with that in BES-2B cells (p < 0.05). Our results pointed out that GAS6-AS2 might be an oncogene via suppressing miR-144-3p in NSCLC. These results suggest that GAS6-AS2 may be related to the occurrence of tumors, but may not have statistical significance with the development of tumors.
3.2. GAS6-AS2 promoted the viability and proliferation of A549 and H1299
Among the five NSCLC cell lines examined, there is no statistical difference in the expression of GAS6-AS2. A549 and H1299 that the most commonly used in NSCLC study were therefore selected for further experiments. GAS6-AS2 expression was silenced in A549 and H1299cells via the transfection of si-GAS6-AS2-1 and si-GAS6-AS2-2 . The silencing efficiency was verified using RT-qPCR. The results demonstrated the decreased expression of GAS6-AS2 in A549 and H1299 cells after the introduction of si-GAS6-AS2-1 or si-GAS6-AS2-2 (p < 0.05,)). ) shows that silence of GAS6-AS2 notably reduced the viabilities of A549 and H1299 cells (p < 0.05 andp<0.05). The percentages of positive cells cells and colony number of A549 and H1299 cells were both reduced after silencing GAS6-AS2 (,), p < 0.05). As cell cycle arrest was one of the most important manners that contributes to proliferation suppression, we further examined the cell cycle after GAS6‐AS2 silence. ) shows that more cells were arrested in G0/G1 phase after GAS6-AS2 silence (p < 0.05 and p < 0.05). The observations uncovered that silence of GAS6-AS2 could repress A549 and H1299 cell proliferation. GAS6-AS2-2 exhibited the highest efficiency in knocking down GAS6-AS2 expression, although there is no statistical difference, and was therefore used in subsequent functional experiments.
Figure 2. GAS6-AS2 promoted the growth and proliferation of A549 and H1299 cells (a) qRT-PCR for GAS6-AS2 expression in A549 and H1299 cells, transfected with GAS6-AS2 siRNA1 or GAS6-AS2 siRNA2 for 2 days. (b) Cell viabilities of A549 and H1299 cells transfected by si-GAS6-AS2-1 or si-GAS6-AS2-2. (c) Cell proliferations of A549 and H1299 cells transfected by si-GAS6-AS2-1 or si-GAS6-AS2-2. (d) Colony formation of A549 and H1299 cells transfected by si-GAS6-AS2-1 or si-GAS6-AS2-2. (e) Flow cytometry analysis of A549 and H1299 cells transfected by si-GAS6-AS2-1 or si-GAS6-AS2-2. *compared with si-NC, p <0.05
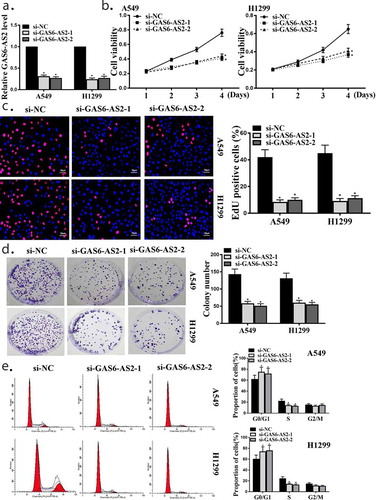
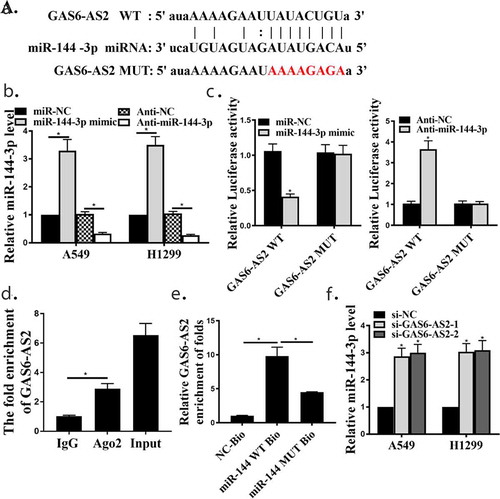
Figure 3. GAS6-AS2 functioned as a ceRNA via directly sponging of miR-144-3p. (a) Online software showed binding relationship between miR-144-3p and GAS6-AS2. (b) increased expression of miR-144-3p in miR-144-3p mimic group and reduced expression of miR-144-3p in mir-144-3p inhibitor group. (c) The co-transfection assay show the relationship of GAS6-AS2 with miR-144-3p in both A549. (d) RIP assay showed that the relationship of GAS6-AS2 with miR-144-3p in both A549. (e) RNA pull-down assay showed that the relationship of GAS6-AS2 with miR-144-3p in both A549 and H1299 cells. (f) The expression of miR-144-3p after GAS6-AS2 was silenced. *compared with NC group, p <0.05. n = 3
3.3. GAS6-AS2 functions as a ceRNA via directly sponging of miR-144-3p
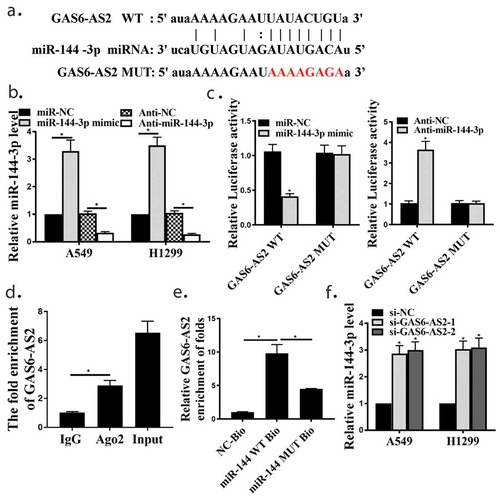
To further explore the in-depth mechanisms of GAS6-AS2 involved in NSCLC proliferation. Bioinformatics prediction showed GAS6-AS2 might directly bind with the seed sequence (2–8 nucleotides) of miR-144-3p ()). To identify the roles of GAS6-AS2 in NSCLC cells, A549 and H1299 cells were transfected with miR-144-3p mimic and anti-mir-144-3p to generate miR-144-3p overexpressing and silence cells. qRT-PCR analysis was performed to confirm the transfection efficiency, indicating that the down-regulation and up-regulation of miR-144-3p was appropriate ()).
As demonstrated using Pearson’s correlation coefficient, the dual luciferase reporter assay was performed in A549 cells cotransfected with both cloned vector and mir-144-3p mimics or anti-miR-144-3p. Compared with mir-NC mimics group, luciferase activities of pmir-Glo-GAS6-AS2-wt was significantly decreased while the luciferase activities in mut group showed no observable change in the mir-144-3p mimics group, on the contrary, luciferase activities of pmir-Glo-GAS6-AS2-wt was significantly increased in the anti-miR-144-3p group ()). Immunoprecipitation was performed and both GAS6-AS2 and miR-144-3p could be pulled down by AGO2 antibody ()), which indicated that GAS6-AS2 could combine with AGO2 and miR-144-3p and further function as a ceRNA. Moreover, we further analyzed the expression correlation between GAS6-A2 and miR-144-3p, and found that the expression of miR-144-3p was up-regulated when GAS6-AS2 was silenced, and similar results appeared after MiR-144-3p silenced (,), p < 0.05). It was then speculated that GAS6-AS2 functions as a ceRNA via directly sponging of miR-144-3p.
3.4. The relationship of miR-144-3p with cell proliferation in NSCLC
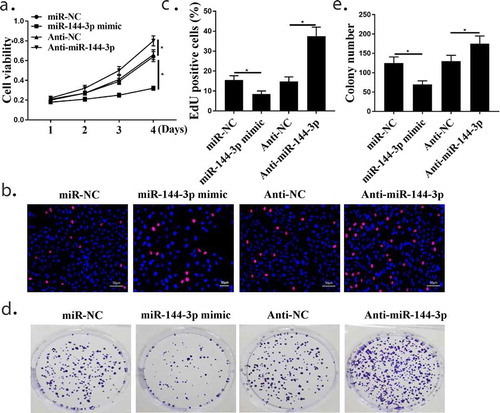
CCK-8 assay(), Edu test(,)) and Colony formation assay(,)) in A549 cells showed that cells transfected with miR-144-3p mimic inhibited cell proliferation, while transfection with anti-miR-144-3p promoted cell proliferation (both p < 0.05). These results collectively demonstrate that miR-144-3p plays cancer-inhibiting roles during NSCLC proliferation.
Figure 4. The relationship of miR-144-3p with cell proliferation in A549 cells. (a) CCK-8 assay showed that the relationship of miR-144-3p with cell proliferation in A549 cells. (Band C) Edu staining assay showed that the relationship of miR-144-3p with cell proliferation in A549 cells. (d and e) Colony formation assays showed that the relationship of miR-144-3p with cell proliferation in A549 cells. *compared with NC group, p <0.05. n = 3
3.5. MiR-144-3p mimic transfection weakened the influences of overexpression GAS6-AS2 in NSCLC cell proliferation
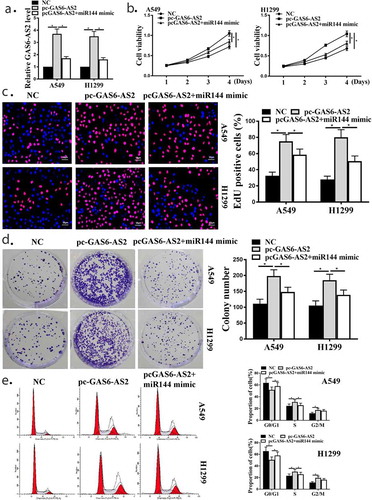
Next, we examined the biological functions of GAS6-AS2 in NSCLCs. GAS6-AS2 was overexpressed in A549 and H1299 cells through transfection with GAS6-AS2 expression vector pcDNA3.1/GAS6-AS2(pc-GAS6-AS2) and pc-GAS6-AS2+ miR144 mimic co-transfection vector, with the empty vector as the NC. ) shows that the transfection efficiency was measured at 48 h post-transfection. CCK-8 assay, Edu assay and colony formation assay in A549 and H1299 cells showed that enhanced proliferation ability was observed in pc-GAS6-AS2 transfected A549 and H1299 cells compared with NC-transfected cells. However, the effect of pc-GAS6-AS2 transfected was down-regulated by up-regulated mir-144-3p (), all p < 0.05). ) shows that less cells were arrested in G0/G1 phase after GAS6-AS2 overexpression in A549 and H1299 cells, and overexpression GAS6-AS2 and mir-144-3p showed that more cell were arrested in G0/G1 phase (), all p < 0.05). Altogether, these data demonstrated that GAS6-AS2 was involved in the A549 and H1299 cell proliferation through targeting miR-144-3p.
Figure 5. miR-144-3p mimic transfection weakened the influences of overexpression GAS6-AS2 in NSCLC cell proliferation. (a) qRT-PCR showed the function of pc-GAS6-AS2 in A549 and H1299 cells. (b) CCK-8 assay showed that the function GAS6-AS2 in A549 and H1299 cells. (c) Edu staining showed that the function GAS6-AS2 in A549 and H1299 cells. (d) Colony formation showed that the function GAS6-AS2 in A549 and H1299 cells. (e) (E) Flow cytometry showed that the function GAS6-AS2 in A549 and H1299 cells. *compared with NC group, p <0.05. n = 3
3.6. MAPK6 was directly targeted of miR-144-3p in NSCLC cells
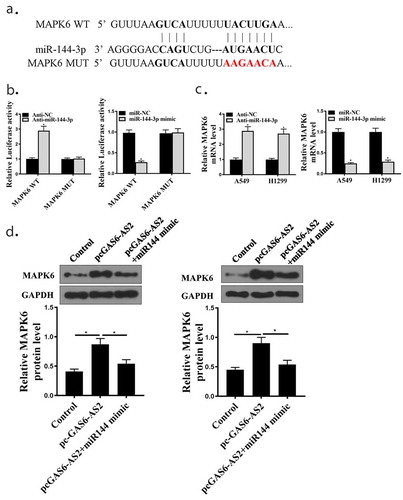
Previous studies revealed that MAPK6 could act as a target of miR-144-3p in tumor proliferation [Citation20]. MAPK6 was significantly overexpressed in NSCLC cells (Supplementary Figure). A previous study has identified MAPK6 ()) as a direct target of miR-144-3p in NSCLC cells. To confirm this observation, wt-MAPK6 and mut-MAPK6 reporter plasmids were constructed and transfected into in A549 and H1299 cells in the presence of miR-144-3p mimic or anti-miR-144-3p. In MAPK6-WT, the relative luciferase activity in the anti-miR-144-3p group was significantly promoted compared with that of the anti-NC group, and there was an opposite result in group of mir-144-3p mimic (), all p < 0.05). There was no significant difference between anti-NC and anti-miR-144-3p or mir-144-3p mimic group in MAPK6-MUT. Furthermore, qPCR assays elucidated that MAPK6 expression was upgraded in anti-miR-144-3p and downgraded in mir-144-3p mimic group (), all p < 0.05). In addition, we next applied western blot to evaluate the expression changes of MAPK6 in A549 and H1299 cells in pc-GAS6-AS2 group and pc-GAS6-AS2+ miR-144-3p mimic co-transfection group. When overexpression of GAS6-AS2, the MAPK6 protein was up-regulated, on the other side, co-expression group reduced MAPK6 with GAS6-AS2 up-regulated (), all p < 0.05). Although miR-144-3p could decrease MAPK6 expression, the levels of MAPK6 were remarkably restored by re-enhancing GAS6-AS2 expression. Collectively, these results demonstrate that GAS6-AS2 acts as a molecular sponge for miR-144-3p in NSCLC cells, thereby increasing the expression of its downstream target gene MAPK6.
Figure 6. Relationship between MAPK6 and miR-144-3p. (a) Putative binding sites of MAPK6 and miR-144-3p are shown. (b) Luciferase activities for cells co-transfected with MAPK6-WT or MAPK6-MUT with miR-NC or miR-144-3p in A549 and H1299 cells. (c) mRNA expression of MAPK6 in A549 and H1299 cells by qRT-PCR. (d) Protein expression of MAPK6 in A549 and H1299, transfected with GAS6-AS2 or GAS6+ miR-144-3p mimic. *compared with NC group, p <0.05.n = 3
3.7. shMAPK6 reversed the effect of down-regulation of miR-144-3p on the proliferation in NSCLC
To determine whether the biological activities in NSCLC cells are mediated by the regulation of the miR-144-3p/MAPK6 axis, A549 cells previously transfected with shMAPK6 were treated with miR-144-3p inhibitor or NC inhibitor. Transfection efficiency was assessed by qRT-PCR ()). CCK-8 assay, Edu test and Colony formation assay in A549 cells showed that cells transfected with miR-144-3p inhibited or promoted cell proliferation, while transfection with miR-144-3p inhibitor + shMAPK6 reduced cell proliferation by miR-144-3p inhibitor (). both p < 0.05). GAS6-AS2 silence inhibited NSCLC proliferation via miR-144-3p/MAPK6 in vivo
Figure 7. GAS6-AS2 promotes NSCLC cells proliferation by up-regulating MAPK6 via sponging miR-144-3p in A549 cells. (a) The transfection efficiency of MAPK6 was assessed by qRT-PCR. The cells proliferation were assessed via(b) CCK-8 assay. (c) Edu staining. (d) Colony formation. *compared with NC group, p <0.05. n = 3
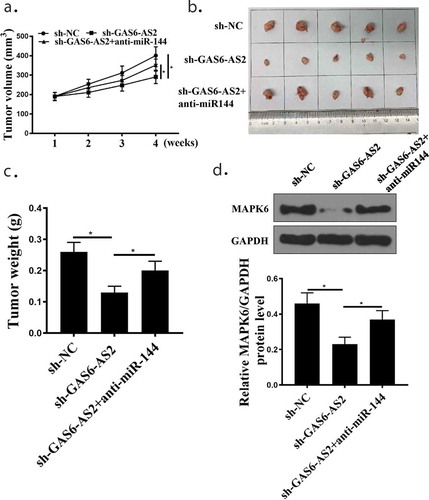
To address the NSCLC role of GAS6-AS2 in vivo, xenograft models were established by injecting A549 cells stably expressing sh-GAS6-AS2 or sh-NC into nude mice. The above results indicated that GAS6-AS2 promoted proliferation of NSCLC cells by miR-144-3p/MAPK6, we further evaluated its functions in vivo. Compared with the sh-NC group, the tumor volume and tumor weight of sh-GAS6-AS2 and sh-GAS6-AS2+ anti-miR-144 groups reduced significantly, while the si-GAS6-AS2+ anti-miR-144-3p group had significantly increased tumor weight compared with the sh-GAS6-AS2 group (), all p < 0.05). ) shows that MAPK6 protein expression was suppressed in cancer tissues in sh-GAS6-AS2 group (p < 0.05). Therefore, the interference of GAS6-AS2 inhibits the tumor growth of HCC due to altered miR-144-3p/MAPK6 expression.
Figure 8. GAS6-AS2 silence inhibited NSCLC proliferation via miR-144-3p/MAPK6 in vivo. (a) Tumor volume was measured every week. (b) The image of tumor at the end. (c) Tumor weight was detected at the end. (d) Western blot examined the protein expression of MAPK6 in tumor tissues. *compared with NC group, p <0.05. n = 5
4. Discussion
NSCLC possesses as the most commonly known type of lung cancers, with more than 70% of lung patients are diagnosed as NSCLC in recent years [Citation21]. In NSCLC patients, many of them have lost the opportunities of surgeries at the time when the disease was diagnosed [Citation22]. This is due to the progression and metastasis of lung cancer, making it hard to perform surgeries [Citation21]. Despite chemotherapies and radiotherapies and even targeted drugs were used, the prognosis of NSCLC is still unsatisfied. In this study, we proposed a novel axis composed of lncRNA GAS6-AS2, miR-144-3p and MAK6, which was demonstrated to involve in the progression of NSCLC cells in vivo and in vitro.
Previous studies have shown that the dysregulation of lncRNAs and miRNAs involves in human cancers. For instance, W. Nie reported that the expression of lncRNA UCA1 was increased in human NSCLC tissues, and the expression of miR-193a-3p was suppressed in cancer tissue/cell compared with healthy lung tissues/cell [Citation6]. In NSCLC tissues, the qRT-PCR results showed that the mRNA expression of GAS6-AS2 was elevated in both NSCLC tissues and cell lines compared with normal ones. In addition, the expression of miR-144-3p in NSCLC cells was greatly down-regulated in contrast to healthy lung cells of BES-2B. Our results indicated that the expression of GAS6-AS2 and miR-144-3p was abnormal in NSCLC samples, which is in consistent with previous findings.
In 2017, Z. Zhao pointed out that lncRNA CCAT2 promoted the tumorigenesis of NSCLC, and over-expressions of CCAT2 increased rates of cancer cell survival and invasion [Citation23]. Our qRT-PCR revealed that GAS6-AS2 inhibition negatively impacted cell survival of A549 and H1299 cells, as well as the proliferation rate. Our colony formation assay confirmed that down-regulation of GAS6-AS2 suppressed NSCLC progression which further proved the oncogene role of lncRNAs in specific human cancers.
It was widely reported that lncRNAs and miRNAs could sponge to each other, and through the binding to miRNA, the specific lncRNA could exert promotion effect in the carcinogenesis [Citation24,Citation25]. Through the bioinformatics prediction, we found that miR-144-3p was a target of GAS6-AS2. The co-transfection of miR-144-3p mimics elevated mRNA expression of miR-144-3p, while anti-miR-144-3p reduced this expression. Co-transfection of the GAS6-AS2-WT with miR-144-3p mimics reduced activity compared with the control, while co-transfection of GAS6-AS2-WT with anti-miR-144-3p increased activity. miRNAs and Ago proteins could generate the RNA-induced silencing complex by inducing the degradation of suppression of mRNA [Citation26]. RIP assay revealed that GAS6-AS2 and miR-144-3p were remarkably richer in Ago2 pellets than that in IgG pellet. The miR-144-3p-biotin probe had a greater level of GAS6-AS2 than NC-bio or miR-144-3p-MUT-bio. All the results suggested that negative relationship existed between miR-144-3p and GAS6-AS2. Our results supplement a new pair of lncRNA and miRNA in the development of tumor gene biology.
Massive evidence has shown that miRNAs can regulate the cancer cell progression, and diminish the promoting effect from lncRNAs [Citation27]. According to S. Li, lncRNA MALAT1 contributed to the development of NSCLC through the targeting of miR-124 [Citation29]. In their study, miR-124 inhibited the NSCLC progression in vitro and reversed the promoting effect from MALAT [Citation29]. Our results demonstrated that miR-144-3p reduced GAS6-AS2 expression in A549 and H1299 cells. NSCLC cell proliferation was significantly reduced by GAS6-AS2 over-expression, but miR-144-3p had a reverse role. Supported by the results, we could conclude that GAS6-AS2 promoted NSCLC progressions by down-regulating miR-144-3p. As far as we know, we are the first to report this phenomenon Citation28.
Many miRNAs were proved to target MAPK6, including miR-26a in smooth muscle cell [Citation30], miR-499a in hepatocellular carcinogenesis, and miR-98-5p in lung cancers [Citation13]. From bioinformatics, we found that MAPK6 was a potential target of miR-144-3p. The co-transfection of MAPK6-WT with miR-144-3p induced reduced activities, but co-transfection of MAPK6-WT with miR-144-3p-inhibitors significantly increased the luciferase activity. Protein expression of MAPK6 were enhanced by GAS6, but this effect was relieved by a miR-144 mimic.
In a recent study, F. Wu et al. reported that NEAT1/miR-98-5p/MAPK6 axis effectively enhanced NSCLC growth, migration and invasion in vivo [Citation13]. In our in vivo experiments, the knockdown of GAS6-AS2 greatly inhibited tumor growth, but thes effects were attenuated by anti-miR-144-3p. MAPK6 protein expression were suppressed in cancer tissues by si-GAS6-AS2. It was obvious that the knockdown of GAS6-AS2 could suppress tumor growth in vivo. This information proved that GAS6-AS2 sponged miR-144-3p, miR-144-3p targeted MAPK6, and the whole GAS6-AS2/miR-144-3p/MAPK6 axis participated in the growth and proliferation of NSCLC cells.
5. Conclusions
The GAS6-AS2/miR-144-3p/MAPK6 axis could promote the progress of NSCLC. GAS6-AS2 might be an important prognostic marker for NSCLC treatment.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Henan Provincial People’s Hospital. The research has been carried out in accordance with the World Medical Association Declaration of Helsinki. All patients and healthy volunteers provided written informed consent prior to their inclusion within the study.
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Disclosure statement
The authors declare that they have no competing interests.
References
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30.
- Graham MV, Purdy JA, Emami B, et al. Clinical dose–volume histogram analysis for pneumonitis after 3D treatment for non-small cell lung cancer (NSCLC). Int J Radiat Oncol* Biol* Phys. 1999;45(2):323–329. .
- Zhang J-G, Wang -J-J, Zhao F, et al. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC). Clin Chim Acta. 2010;411(11–12):846–852.
- Fukuoka M, Yano S, Giaccone G, et al. Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced non–small-cell lung cancer. J clin oncol. 2003;21(12):2237–2246. .
- Peters S, Adjei A, Gridelli C, et al. Metastatic non-small-cell lung cancer (NSCLC): ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2012;23(suppl_7):vii56–vii64. .
- Nie W, Ge H-J, Yang X-Q, et al. LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett. 2016;371(1):99–106. .
- Gibb EA, Vucic EA, Enfield KS, et al. Human cancer long non-coding RNA transcriptomes. PloS One. 2011;6(10):e25915. .
- Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochimica Et Biophysica Acta (Bba)-gene Regulatory Mechanisms. 2014;1839(11):1097–1109.
- Hu Z, Chen J, Tian T, et al. Genetic variants of miRNA sequences and non–small cell lung cancer survival. J Clin Invest. 2008;118(7):2600–2608. .
- Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64(11):3753–3756. .
- Tanabe H, Yagihashi A, Tsuji N, et al. Expression of survivin mRNA and livin mRNA in non-small-cell lung cancer. Lung Cancer. 2004;46(3):299–304.
- Sasaki H, Moriyama S, Nakashima Y, et al. Histone deacetylase 1 mRNA expression in lung cancer. Lung Cancer. 2004;46(2):171–178. .
- Wu F, Mo Q, Wan X, et al. NEAT1/hsa‐mir‐98‐5p/MAPK6 axis is involved in non–small‐cell lung cancer development. J Cell Biochem. 2019;120(3):2836–2846.
- Bian K, Muppani NR, Elkhadragy L, et al. ERK3 regulates TDP2-mediated DNA damage response and chemoresistance in lung cancer cells. Oncotarget. 2016;7(6):6665. .
- WEINHÄUSEL A Lung cancer diagnostic method and means. Google Patents; 2018.
- Long W, Foulds CE, Qin J, et al. ERK3 signals through SRC-3 coactivator to promote human lung cancer cell invasion. J Clin Invest. 2012;122(5):1869–1880. .
- Yang JH, Li JH, Shao P, et al. starBase: a database for exploring microRNA-mRNA interaction maps from argonaute CLIP-Seq and degradome-Seq data. Nucleic Acids Res. 2011;39(Databaseissue):D202–9.
- Agarwal V, Bell GW, Nam JW, et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4. DOI:10.7554/eLife.05005
- Chen Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48(D1):D127–D31.
- Wu J, Zhao Y, Li F, et al. MiR-144-3p: a novel tumor suppressor targeting MAPK6 in cervical cancer. J Physiol Biochem. 2019;75(2):143–152.
- Reck M, Rodríguez-Abreu D, Robinson AG, et al. Pembrolizumab versus chemotherapy for PD-L1–positive non–small-cell lung cancer. N Engl J Med. 2016;375(19):1823–1833. .
- Garon EB, Rizvi NA, Hui R, et al. Pembrolizumab for the treatment of non–small-cell lung cancer. N Engl J Med. 2015;372(21):2018–2028. .
- Zhao Z, Wang J, Wang S, et al. LncRNA CCAT2 promotes tumorigenesis by over-expressed Pokemon in non-small cell lung cancer. Biomed Pharmacother. 2017;87:692–697.
- Shao Y, Ye M, Li Q, et al. LncRNA-RMRP promotes carcinogenesis by acting as a miR-206 sponge and is used as a novel biomarker for gastric cancer. Oncotarget. 2016;7(25):37812. .
- Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochimica Et Biophysica Acta (Bba)-gene Regulatory Mechanisms. 2016;1859(1):169–176.
- Pillai RS, Artus CG, Filipowicz W. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. Rna. 2004;10(10):1518–1525.
- Ballantyne M, McDonald R, Baker A. lncRNA/MicroRNA interactions in the vasculature. Clin Pharmacol Ther. 2016;99(5):494–501.
- Li S, Mei Z, Hu HB, et al. The lncRNA MALAT1 contributes to non‐small cell lung cancer development via modulating miR‐124/STAT3 axis. J Cell Physiol. 2018;233(9):6679–6688.
- Tan J, Yang L, Liu C, et al. MicroRNA-26a targets MAPK6 to inhibit smooth muscle cell proliferation and vein graft neointimal hyperplasia. Sci Rep. 2017;7:46602.
- Xiang Z, Wang S, Xiang Y. Up-regulated microRNA499a by hepatitis B virus induced hepatocellular carcinogenesis via targeting MAPK6. PloS One. 2014;9(10):e111410.
