ABSTRACT
Cardiovascular disease (CVD) has been identified as the leading cause of premature deaths in rheumatoid arthritis (RA), accounting for about 40 to 50% of all deaths. Macrophage inflammation is regarded as a key point to link to the two diseases. Recently, long non-coding RNAs (lncRNAs) have acknowledged as a regulator of inflammation significantly. Here, we firstly found that lncRNA myocardial infarction associated transcript (lncRNA MIAT), a crucial lncRNA to regulate CVD, expressed increasingly in synovium and myocardial tissues of collagen-induced arthritis (CIA) mice. Besides, we also verified that the increased infiltration of macrophage occurred in those tissues of the CIA. In vitro, we found that macrophage inflammation induced by LPS could up-regulate lncRNA MIAT expression. LncRNA MIAT seemed to inhibit the expression of IL-1β, TNF-ɑ and be suppressed by ATP-induced NLRP3 inflammasome activation pathway. Therefore, these data indicated an anti-inflammatory effect of lncRNA MIAT in macrophage and an original research direction for high cardiovascular risk in RA.
Introduction
Rheumatoid arthritis (RA) is a typically inflammatory autoimmune disease, with substantial disability and mortality [Citation1,Citation2]. The high cardiovascular (CV) risk is the main contributor to 40–50% death population of RA [Citation3–6] with uncertain pathogenesis. Of note, more evidence states that the spontaneous degeneration of inflammation leads to the development of high CV risk throughout a RA patient’s life [Citation6–8]. One of the hallmarks of persistent inflammation is the increased number of macrophage that derives from mature monocyte arising from bone marrow [Citation9,Citation10].
As is known, various cytokines (IL-6, IL-1β, TNF-α, and so on) in macrophage are induced by inflammatory factors such as LPS, contributing to the cause of RA and CVD [Citation11–13]. Besides, nucleotide-binding oligomerization domain, leucine-rich repeat and pyrin domain-containing protein 3 (NLRP3) inflammasome activation is recently pointed out as one of the effective pathways to generate amounts of pro-inflammatory cytokines in macrophage via a large, rapid release of IL-1famaily cytokines in macrophage via a large, rapid release of mature IL-1β formed by the cleavage effect of activated caspase-1 [Citation14–17]. The activation of NLRP3 inflammasome has been linked to the onset of RA and CVD [Citation18,Citation19]. Consequently, untying the pathways for inflammation of macrophage is the major interest to better understand the pathogenesis of RA with cardiovascular disease (CVD) and to explore a new approach to achieve low CV risk in RA patients.
Long non-coding RNAs (LncRNAs) are known as a class of transcripts longer than 200 nucleotides to potentially and limitedly regulate all levels of the gene expression [Citation20]. Numerous studies emerge that lncRNAs have an obvious effect on the biologic process, including organic inflammatory reaction and immune defense [Citation21]. LncRNAs relating to RA are gradually recognized by advanced technique [Citation22,Citation23]. For example, the study found that lnc0640 had a significantly increased expression in peripheral blood mononucleosis (PBMC) of RA patients, which might be linked to serum C reactive protein (CRP) levels [Citation24]. LncRNA myocardial infarction associated transcript (MIAT), expressing in cardiac cells and several retinal cells, has already been verified to have a decisive effect on CVD [Citation25]. Among all CV risk factors, the expression level of MIAT in patients with myocardial infarction (MI) is linked to hypertension and smoking [Citation26]. Besides, the expression level of MIAT is a meaningful single variable predictor of left ventricular dysfunction [Citation27]. However, no former studies put MIAT on the research of RA.
In our study, MIAT was verified that exiting higher expression in the knee joint synovium and the myocardial tissues of collagen-induced arthritis (CIA) mice than in these tissues of the control. Besides, our study demonstrated that MIAT might be an inhibitor for inflammation due to the increased expression of IL-1β, TNF-α when it was knocked down in vitro. The data also showed that MIAT was suppressed by ATP-induced NLRP3 inflammasome activation pathway.
1. Methods and materials
1.1 Mice
The Institutional Animal Care and Use Committee of the Affiliated Hospital of Qingdao University authorized mice experiments. DBA/1 mice (15 males, 6 −8 weeks old) were bought from Shanghai slack laboratory and bred in the Laboratory Animal Center of the Affiliated Hospital of Qingdao University.
1.2 Collagen-induced arthritis models and assessment of disease
Bovine type II collagen (2 mg/ml; Chondrex, Cat#20,022) was mixed into complete Freund’s complete adjuvant (4 mg/ml; Chondrex, Cat#7001) and injected into the skin of mice tail on day 0. A second injection with the same volume was finished on day 21. The arthritis scores was graded from 0 to 4 for each paw, based on the swollen and red situation [Citation28]. Mice serum was collected in the last day to detect the levels of cytokines secreted by ELISA.
1.3 Cell culture
J774A.1, a murine macrophage cell line (ATCC, Cat# TIB-67, RRID: CVCL_0358), was cultured in DMEM (HyClone, Cat#SH30022.01) with 10% fetal bovine serum (Gibco, Cat#10,100,147 C) and 0.1% penicillin-streptomycin liquid (Solarbio, Cat#P1400) at 37°C with 5% CO2. Because macrophage inflammation is a key for the pathogenesis of RA, we built different inflammation models in vitro by J774A.1.
J774A.1 was cultured by DMEM with 100 ng/mL LPS (Sigma, Cat#A0550)for 24 h to build inflammation models in contrast to group cultured by DMEM with the same volume of PBS (Gibco, Cat#20,012,027) [Citation29].
J774A.1 was cultured by DMEM with 100 ng/mL LPS for 4 h and 5 mM ATP (Sigma, Cat# A2383) for 0.5 h to build NLRP3 inflammasome activation models [Citation29] in contrast to DMEM with LPS group and LPS/ATP +Inhibitor group. ATP-related inhibitor- A-804,598 (MCE, Cat#HY-100,483) was used to block ATP-related receptor for inhibiting caspase-1 activation before NLRP3 inflammasome activation models were built. And there was no effective direct inhibitor for caspase-1 in vitro.
1.4 Vector construction and cell transfection
Short-hair RNA targeting to MIAT (sh-MIAT) was built by Gene pharma (Shanghai, China) to knock down MIAT expression in J774A.1. The sh-MIAT sequences are as follow:
5ʹ-3ʹ: GCTCCTTGTTCGGTTTATATC.
Sh-MIAT (KD group) and its scramble control vectors (NC group) were delivered into cells by transfection with Attractene Transfection Reagent (QIAGEN, German).
1.5 Histochemistry
Knee joints and myocardial tissues were collected in the last day. They were fixed in 4% paraformaldehyde and knee joints were decalcified in 10% EDTA-4Na, Then, they were used by paraffin embedding. Sections were observed under hematoxylin–eosin staining. At least 5 different regions were observed in each slice, and the cell count of macrophage in every region was performed by 3 people at the same time.
1.6 ELISA
Serum and cell supernatants were collected to measure the levels of cytokines secreted. The levels of IL-1β, TNF-ɑ and IL-6 (Abcam, Cambridge, MA, USA) were measured by ELISA on the basis of the protocols. All the assays were performed in triplicate.
1.7 RNA isolation and quantitative real-time PCR
Total RNA collected respectively by knee joints,myocardial tissues and J774A.1 cell were isolated using TRIzol Reagent (Invitrogen, Cat#15,596,018). Quantitative real time PCR was achieved using PrimeScript™ RT Reagent Kit (Takara, Cat#RR073A) on LightCycler® 96 System (Roche Diagnostics International). GAPDH was regarded as the reference gene to control in this study. All primer sequences are showed in Table 1.
2−ΔΔCt method was used to calculate the relative expression of genes.
1.8 Detection of caspase-1 activity
The FAM-FLICA® Caspase-1 Assay Kit (ImmunoChemistry Technologies, Cat#98) was to access caspase-1 activation. J774A.1 cells were gently digested with Trypsin (2.5%)- no EDTA (Gibco, Cat#15,090,046). Then, they were stained with FAM-FLICA for approximately 1 h at 37°C and were washed 3 times by apoptosis wash buffer. Finally, flow cytometry was used to analyze.
1.9 Western blotting
Cells were lysed by RIPA buffer (Cell signaling technology, Cat#9806). The proteins of lysates were separated by 12% SDS-PAGE and transferred to nitrocellulose filter (NC) membrane (Solarbio, Cat#YA1710). The membranes were incubated with anti-GAPDH antibody (Cell signaling technology, Cat# 5174S, RRID: AB_10622025), anti-Caspase-1 antibody (Sant Cruz biotechnology, Cat# sc-56,036, RRID: AB_781816), next to horseradish peroxidase (HRP)-conjugated appropriate secondary antibody. FluorChem Q (ProteinSimple, California, USA) was used to visual signals on NC membranes. GAPDH was used as the reference protein in our study.
1.10Statistical analysis
MEV software (version 4.9.0) was used to analysis data from GSE13,071 in the GEO DataSets (https://www.ncbi.nlm.nih.gov/gds/), a public microarray or high-throughput sequencing database.
Statistical analysis was performed with the GraphPad 9.0. One‐way analysis of variance (ANOVA) was used for statistical analysis among groups. Student’s t test was performed for two-group samples. Mean ± S.E.M was used to present all results, P < 0.05 was considered statistically significant.
2. Results
2.1 MIAT up-regulation in collagen-induced arthritis (CIA)
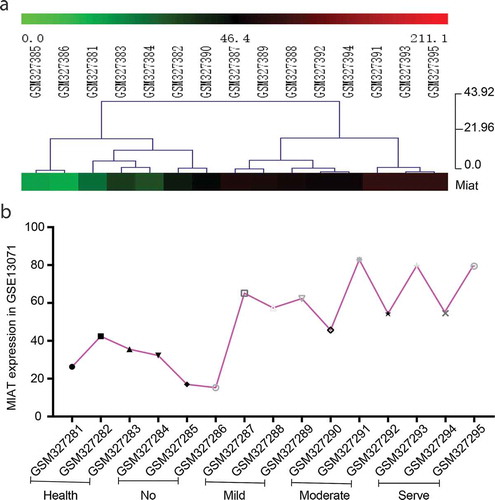
Public data uploaded in GEO DataSets was used to make preliminary confirmation that MIAT has an abnormal expression in arthritis. GSE13071 is a microarray for the whole genome RNA of knee joint synovium from different grades of inflammation in CIA (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi). Based on data from GSE 13071, lncRNAs expression was analyzed after standardization which former researchers did not accomplish. The color of MIAT in the heat map changed from the green to the red with the degree of joint inflammation. The heat map showed us MIAT absolute expression in joints of CIA increases with inflammation exacerbation (. A). The curve of gene expression also showed the same change (. B). These results suggest that up-regulated expression of MIAT might be involved in joints damage in arthritis.
Figure 1. MIAT expression in a microarray for the whole genome RNA of knee joint synovium from different grades of inflammation in collagen-induced arthritis (CIA) (GSE13071). A. Heat map of MIAT expression. The color from green to red means the change of MIAT expression from low expression to high expression. B. The curve of MIAT expression. All 15 samples in GSE13071 were divided into five groups (GSM327381-83: the healthy control; GSM327384-96: no joint inflammation; GSM327387-89: mild joint inflammation; GSM327390-92:moderate joint inflammation; GSM327393-95: severe joint inflammation). Each sample represents an individual mouse (n = 15)
2.2 Macrophage increased in knee joint synovium and myocardial tissues of CIA
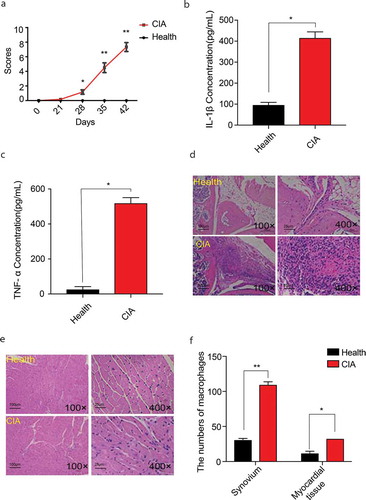
Firstly, CIA models were built. The arthritis scores showed that the scores of the model group were significantly higher than of the control group at the beginning of day28 ( A). IL-1β, TNF-ɑ level in CIA serum were significantly increased ( B-C). The HE staining of knee joint synovium in mice of the last day showed that the infiltration of inflammatory cells in the knee joint synovium was significantly increased in CIA mice ( D). To prove that arthritis can also affect the inflammatory state of the myocardial tissues, we treated the heart muscles of mice with HE staining, and the results showed that the infiltration of inflammatory cells in the myocardial tissues of CIA was significantly increased ( E). Moreover, the number of macrophages in both tissues of CIA increased than in the same tissues of the control ( F).
Figure 2. Assessments of CIA and the number of macrophages in knee joint synovium and myocardial tissues of CIA. A. The disease scores of CIA mice (n = 7) and the healthy control (n = 5). From day28, the redness and swallow of paw, knee or ankles in CIA mice appeared. B. C. IL-1β, TNF-ɑ levels in serum of CIA mice (n = 7) and the healthy control (n = 5). D. E. HE staining for inflammatory infiltration in knee joint synovium and myocardial tissue of CIA (n = 7) and of the healthy control (n = 5) (including microscopes of 100× and 400×) in the last day. F. The number of macrophages of synovium and myocardial tissues in CIA mice (n = 7) and the healthy control (n = 5) under the microscopes of 400 × . At least 5 different regions were observed in each slice, and the cell count of macrophages in every region was performed by 3 people at the same time. One‐way analysis of variance (ANOVA) was used for statistical analysis among groups. Student’s t test was performed for two-group samples, *P < 0.05, **P < 0.01
2.3 MIAT overexpression links to inflammation in macrophage
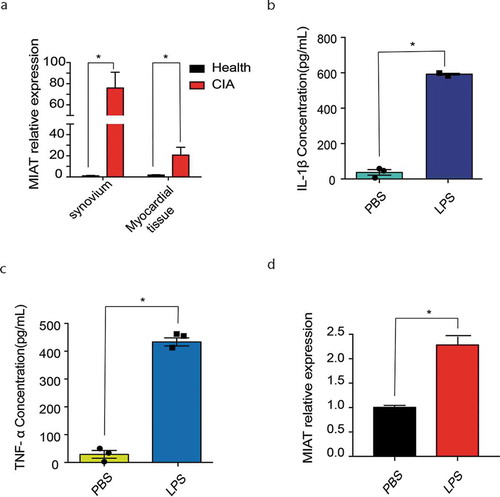
Then, knee joint synovium and myocardial tissues were collected for analysis of the MIAT relative expression. The result showed that the relative expression of MIAT increases significantly in CIA group than in the healthy group ( A). The result showed that MIAT high expression might have a relationship to increased NLRP3 inflammasome activation.
Figure 3. MIAT relative expression in tissues of CIA and in inflammation models of J774A.1. A. MIAT relative expression in knee joint synovium and myocardial tissue of CIA (n = 7) and of the healthy control (n = 5). B. C. IL-1β, TNF-ɑ levels in cell supernatant of J774A.1 with PBS and LPS. D. MIAT relative expression of J774A.1 with PBS and LPS. Data of cells are collected from three independent experiments. One‐way analysis of variance (ANOVA) was used for statistical analysis among groups. Student’s t test was performed for two-group samples, *P < 0.05
To explore whether MIAT is related to macrophage inflammation, J774A.1, a murine macrophage cell line was used in vitro. J774A.1 was collected after 100ug/mL LPS stimulating for 24 h to build an inflammatory model [Citation29]. IL-1β, TNF-ɑ level in cell supernatant were significantly increased after LPS stimulation ( B-C). MIAT relative expression had a prominent augmentation after LPS stimuli ( D). These results confirmed that MIAT expression in macrophage could be up-regulated remarkably when inflammation reaction occurs.
2.4 MIAT inhibits IL-1β, TNF-ɑ expression but facilitates IL-6 expression
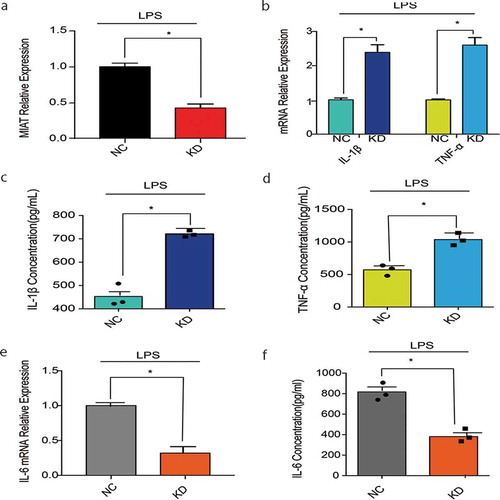
To probe the role of MIAT in macrophage inflammation, MIAT was knocked down in J774A.1 by shRNA. With LPS stimulating, MIAT relative expression in the shMIAT group is 37% of expression in the shNC control ( A). IL-1β, TNF-ɑ mRNA relative expression were detected to present a significant elevation in the shMIAT group than in the shNC control ( B). IL-1β, TNF-ɑ level in cell supernatant were significantly increased in shMIAT than in the shNC ( C-D). These results verified that MIAT might regulate IL-1β, TNFɑ-expression reduced negatively. Notably, IL-6 mRNA relative expression had a visible decrease in the shMIAT group than in the shNC control ( E). IL-6 level in cell supernatant was decreased as the mRNA level expression ( F). It showed that MIAT seemed to control IL-6 expression positively.
Figure 4. Cytokines in J774A.1 with MIAT-knocking down. A. MIAT relative expression with LPS stimuli in the negative control and MIAT-knocking down group. B. IL-1β, TNF-ɑ mRNA relative expression with LPS stimuli in the negative control and MIAT-knocking down group. C. D. IL-1β, TNF-ɑ levels in cell supernatant of J774A.1 with LPS stimuli in the negative control and MIAT-knocking down group. E. IL-6 mRNA relative expression with LPS stimuli in the negative control and MIAT-knocking down group. F. IL-6 levels in cell supernatant of J774A.1 with LPS stimuli in the negative control and MIAT-knocking down group. Data of cells are collected from three independent experiments. ANOVA was used for statistical analysis among groups. Student’s t test was performed for two-group samples, *P < 0.05
2.5 MIAT is inhibited by ATP-induced NLRP3 inflammasome activation
To demonstrate whether MIAT links to NLRP3 inflammasome activation, caspase-1 was used to assess. The increasing difference of activated caspase-1 expression in knee joint synovium and myocardial tissues of CIA had significant statistic importance than of the control ( A-B).
Figure 5. NLRP3 inflammasome activation in CIA. A. Activated caspase-1 expression in synovium of CIA (n = 7) and the healthy group (n = 5). Image J was used to analyze the western blotting pictures by gray value and p20/pro casp-1 ratio was used to define caspase-1 activation. B. Activated caspase-1 expression in myocardial tissues of CIA (n = 7) and the healthy group (n = 5). C. MIAT relative expression in LPS, LPS/ATP and LPS/ATP +Inhibitor group of J774A.1.Data of cells are collected from three independent experiments. ANOVA was used for statistical analysis among groups. Student’s t test was performed for two-group samples, *P < 0.05
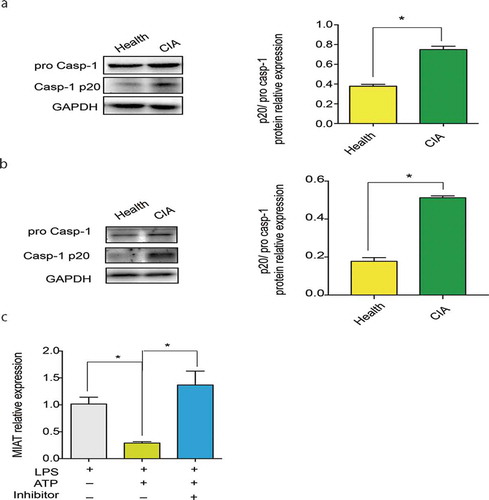
In vitro, we used 100 ng/mL LPS and 5 mM ATP to induce NLRP3 inflammasome activation in J774A.1 [Citation29]. Then, we applied a specific P2X7 receptor inhibitor, A-804,598 to block ATP when J774A.1 was stimulated with LPS/ATP. We found that MIAT had high expressions in both of LPS group and LPS/ATP group. But MIAT had lower expression in LPS/ATP group than in LPS group when displayed higher expression in LPS/ATP +inhibitor group than in LPS/ATP group ( C). These results showed that ATP-induced NLRP3 inflammasome activation might inhibit MIAT expression.
3. Discussion
In our study, we firstly submitted that MIAT showed an abnormal expression by mining the data from GEO database. The result seemed to show that the increase of MIAT expression was relative to the degrees of inflammation in CIA. Subsequently, we established a CIA mice model, on the one hand, that demonstrated that arthritis can drive cardiac macrophage infiltration increased, on the other hand, confirmed that the abnormal increase of MIAT expression in the knee joints synovium and heart muscles of arthritis mice.
Macrophage inflammation bears an important responsibility for arthritis and atherosclerotic CVD [Citation30–32]. At the above studies, we used J774A.1, a murine macrophage cell line, to investigate the potential role of MIAT in macrophage inflammation. We built an inflammation model through the stimulation with 100 ng/mL LPS [Citation29]. As was expected, macrophage inflammation might regulate MIAT expression positively. Interestingly, our data also showed that pro-inflammatory cytokines IL-1β, TNF-ɑ might increase expressions with MIAT knocked down. These results suggested that MIAT might have a suppressed influence on inflammation. However, MIAT was a possibly positive inducer for IL-6. Based on previous research, IL-6 induces macrophage to differentiate into M2 for inhibiting inflammation [Citation33,Citation34]. Although we trended to believe that MIAT in macrophage perhaps played a big role in inflammatory inhibition, our limitation was that lacking the data in vivo for further verification, especially for the role of MIAT in IL-6.
Besides, NLRP3 inflammasome activation in macrophage is also considered as a favorite cytokines-mediated pathway to the pathogenesis of RA and CVD [Citation35–37]. In our study, LPS/ATP was used to establish a NLRP3 inflammasome activation model [Citation29]. Our data showed that MIAT expression in J774A.1 was suppressed by LPS/ATP than by LPS. Actually, NLRP3 inflammasome activation usually induces macrophage death, which is called pyroptosis [Citation38]. An alternative explanation is reduced MIAT expression due to cell death. However, no significant difference of GAPDH expression (the endogenous control of groups) denied this explanation. Subsequently, we controlled ATP-related cell receptor P2X7 receptor [Citation39] inhibited by A-804,598 to block ATP-induced NLRP3 inflammasome pathway. Notably, the results demonstrated that ATP-induced pathway might regulate MIAT expression down. It seemed to opposite to the data in vivo, showing NLRP3 inflammasome activation appeared with high MIAT expression at the same time. The data in vivo was from synthetically different phenotypes of inflammation in a complex animal model. In our opinion, NLRP3 pathway is one of a plenty of inflammatory phenotypes existing in CIA model. Other ways we did not find might the key to upregulate MIAT expression in mice. We would explore them further in future. Moreover, IL-1β level in cell supernatant in our study are also used to verify that NLRP3 inflammasome activation seemed to be a stronger, more responsive pathway for inflammation. Therefore, our perspective tends that the more violent inflammation in macrophage may weaken anti-inflammatory effect via making MIAT expression falling down.
In conclusion, we firstly observed significant high expression of MIAT in joints synovium and myocardial tissues of CIA mice and macrophage inflammation. Although its high expression may inhibit inflammation in our research, the high expression of MIAT in the heart can also stimulate heart injury and leads to cardiovascular disease through other pathways [Citation27,Citation40], which perhaps is a bridge of RA and CVD. It needs further study for the CVD-related mechanism of MIAT in macrophage. This is a novel finding for macrophage inflammation, which might guide a new research direction for CV complications of RA.
Author contributions
All authors have equal contributions to this study.
Fund
This study was funded by the National Natural Science Foundation of China (Grant no. 81671603 and Grant no. 81201060), the Shandong Provincial Natural Science Foundation (Grant no. ZR2016HM56) and the Scientific Research Award Fund of Outstanding Young Scientist in Shandong Province (Grant no. 2006BS03005).
Disclosure statement
The authors have no potential financial conflicts of interest.
Additional information
Funding
References
- Scott DL, Wolfe F, Huizinga TW. Rheumatoid arthritis. Lancet. 2010;376(9746):1094–1108.
- Gerlag DM, Raza K, van Baarsen LG, et al. EULAR recommendations for terminology and research in individuals at risk of rheumatoid arthritis: report from the study group for risk factors for rheumatoid arthritis. Ann Rheum Dis. 2012;71(5):638–641.
- Nomeir AM, Turner R, Watts E, et al. Cardiac involvement in rheumatoid arthritis. Ann Intern Med. 1973;79(6):800–806.
- Liao KP. Cardiovascular disease in patients with rheumatoid arthritis. Trends Cardiovasc Med. 2017;27(2):136–140.
- Mavrogeni S, Dimitroulas T, Sfikakis PP, et al. Heart involvement in rheumatoid arthritis: multimodality imaging and the emerging role of cardiac magnetic resonance. Semin Arthritis Rheum. 2013;43(3):314–324.
- England BR, Thiele GM, Anderson DR, et al. Increased cardiovascular risk in rheumatoid arthritis: mechanisms and implications. BMJ. 2018;361:k1036.
- Khan F. Assessment of endothelial function as a marker of cardiovascular risk in patients with rheumatoid arthritis. Int J Rheum Dis. 2010;13(3):189–195.
- Lazúrová I, Tomáš Ľ. Cardiac impairment in rheumatoid arthritis and influence of Anti-TNFα treatment. Clin Rev Allergy Immunol. 2017;52(3):323–332.
- Siouti E, Andreakos EJBP. The many facets of macrophages in rheumatoid arthritis. Biochem Pharmacol. 2019;165:152–169.
- Udalova IA, Mantovani A, Feldmann M. Macrophage heterogeneity in the context of rheumatoid arthritis. Nat Rev Rheumatol. 2016;12(8):472–485.
- Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140(6):821–832.
- Jianhui L, Rosenblatt-Velin N, Loukili N, et al. Endotoxin impairs cardiac hemodynamics by affecting loading conditions but not by reducing cardiac inotropism. Am J Physiol Heart Circ Physiol. 2010;299(2):H492–501.
- House LM, Morris RT, Barnes TM, et al. Tissue inflammation and nitric oxide-mediated alterations in cardiovascular function are major determinants of endotoxin-induced insulin resistance. Cardiovasc Diabetol. 2015;14(1):56.
- Swanson KV, Deng M, Ting JP. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat Rev Immunol. 2019;19(8):477–489.
- Rada B, Park JJ, Sil P, et al. NLRP3 inflammasome activation and interleukin-1β release in macrophages require calcium but are independent of calcium-activated NADPH oxidases. Inflammation Res. 2014;63(10):821–830.
- He Y, Hara H, Núñez G. Mechanism and regulation of NLRP3 inflammasome activation. Trends Biochem Sci. 2016;41(12):1012–1021.
- Shen HH, Yang YX, Meng X, et al. NLRP3: A promising therapeutic target for autoimmune diseases. Autoimmun Rev. 2018;17(7):694–702.
- Mathews RJ, Robinson JI, Battellino M, et al. Evidence of NLRP3-inflammasome activation in rheumatoid arthritis (RA); genetic variants within the NLRP3-inflammasome complex in relation to susceptibility to RA and response to anti-TNF treatment. Ann Rheum Dis. 2014;73(6):1202–1210.
- Liu D, Zeng X, Li X, et al. Role of NLRP3 inflammasome in the pathogenesis of cardiovascular diseases. Basic Res Cardiol. 2018;113(1):5.
- Sun Q, Hao Q, Prasanth KV. Nuclear long noncoding RNAs: key regulators of gene expression. Trends Genet. 2018;34(2):142–157.
- Boon RA, Jaé N, Holdt L, et al. Long Noncoding RNAs: from Clinical Genetics to Therapeutic Targets? J Am Coll Cardiol. 2016;67(10):1214–1226.
- Xu F, Jin L, Jin Y, et al. Long noncoding RNAs in autoimmune diseases. J Biomed Mater Res A. 2019;107(2):468–475.
- Li Z, Li X, Jiang C, et al. Long non-coding RNAs in rheumatoid arthritis. Cell Prolif. 2018;51(1). DOI:10.1111/cpr.12404
- Yuan M, Wang S, Yu L, et al. Long noncoding RNA profiling revealed differentially expressed lncRNAs associated with disease activity in PBMCs from patients with rheumatoid arthritis. PloS One. 2017;12(11):e0186795.
- Sun C, Huang L, Li Z, et al. Long non-coding RNA MIAT in development and disease: a new player in an old game. J Biomed Sci. 2018;25(1):23.
- Qu X, Du Y, Shu Y, et al. MIAT is a pro-fibrotic long non-coding RNA governing cardiac fibrosis in post-infarct myocardium. Sci Rep. 2017;7(1):42657.
- Yu B, Wang S. Angio-LncRs: lncRNAs that regulate angiogenesis and vascular disease. Theranostics. 2018;8(13):3654–3675.
- Augello A, Tasso R, Negrini SM, et al. Cell therapy using allogeneic bone marrow mesenchymal stem cells prevents tissue damage in collagen-induced arthritis. Arthritis Rheumatism. 2007;56(4):1175–1186.
- Hirano S, Zhou Q, Furuyama A, et al. Differential regulation of IL-1β and IL-6 release in murine macrophages. Inflammation. 2017;40(6):1933–1943.
- Tu J, Hong W, Guo Y, et al. Ontogeny of synovial macrophages and the roles of synovial macrophages from different origins in arthritis. Front Immunol. 2019;10:1146.
- Kurowska-Stolarska M, Alivernini S. Synovial tissue macrophages: friend or foe? RMD Open. 2017;3(2):e000527.
- Nicolás-Ávila JA, Hidalgo A, Ballesteros I. Specialized functions of resident macrophages in brain and heart. J Leukoc Biol. 2018;104(4):743–756.
- Fu XL, Duan W, Su CY, et al. Interleukin 6 induces M2 macrophage differentiation by STAT3 activation that correlates with gastric cancer progression. Cancer Immunol Immunother. 2017;66(12):1597–1608.
- Dubey A, Izakelian L, Ayaub EA, et al. Separate roles of IL-6 and oncostatin M in mouse macrophage polarization in vitro and in vivo. Immunol Cell Biol. 2018;96(3):257–272.
- Zhang Y, Zheng Y, Li H. NLRP3 inflammasome plays an important role in the pathogenesis of collagen-induced arthritis. Mediators Inflamm. 2016;(2016):9656270.
- Zhao C, Gu Y, Zeng X, et al. NLRP3 inflammasome regulates th17 differentiation in rheumatoid arthritis. Clin Immunol. 2018;197:154–160.
- An N, Gao Y, Si Z, et al. Regulatory mechanisms of the NLRP3 inflammasome, a novel immune-inflammatory marker in cardiovascular diseases. Front Immunol. 2019;10:1592.
- Lima Jr. H, Jacobson LS, Goldberg MF, et al. Role of lysosome rupture in controlling NLRp3 signaling and necrotic cell death. Cell Cycle. 2013;12(12):1868–1878.
- Yin J, Wang Y, Hu H, et al. P2X receptor inhibition attenuated sympathetic nerve sprouting after myocardial infarction via the NLRP3/IL-1β pathway. J Cell Mol Med. 2017;21(11):2695–2710.
- Liao J, He Q, Li M, et al. LncRNA MIAT: myocardial infarction associated and more. Gene. 2016;578(2):158–161.
