ABSTRACT
MicroRNA plays an important regulatory role in the development of all organisms, including hair follicle development. In order to improve domestic cashmere yield, the role of miRNA in hair follicle cycle has become a research hotspot. However, the molecular and cellular mechanisms by which miRNA-203 regulates hair follicle development are not completely understood. In this study, we found that the relevant target genes of miRNA-203 (DDOST and NAE1) were less expressed in telogen by qPCR and Immunoblotting analysis, contrary to the expression mode of miRNA-203. The Dual-Luciferase reporter assay was used to verify the correlation between miRNA-203 and its target gene expression. The results showed that miRNA-203 specifically binds to the 3 ‘UTR of DDOST and NAE1, and the expression of miRNA-203 significantly down-regulates the expression of DDOST and NAE1 mRNA and protein. Therefore, this study demonstrates that miRNA-203 may regulate hair follicle development in Cashmere goats by targeting DDOST and NAE1.
Introduction
miRNAs are non-coding RNAs of about 20–25 nucleotides that affect gene expression by binding to target messenger mRNA through complementary seed regions [Citation1]. Recent studies have shown that miRNA is involved in physiological processes such as epidermal and hair follicle development, pigmentation and hair follicle cycle development. According to the study, [Citation2] found that some miRNA expressions were different in alpaca skin with different hair colors using Illumina sequencing technology. miRNA has been shown to be involved in the synthesis of melanin [Citation3]. In our previous studies, it has been shown that miR-202 negatively regulates the expression of target genes wnt5a, kit and tcf7 in the skin tissues of C57BL/6 black mice and BALB/c mice [Citation4].
[Citation5] an intracellular regulatory network has been constructed, indicating that the miRNAs studied are directly or indirectly involved in several signal transduction pathways through their target genes. [Citation6] it has been proved that miR-22 is significantly up-regulated in the middle hair growth period and in the peak resting period, directly inhibiting the regulators of hair cycle. In our previous studies, it has been proved that miR-let7a is differentially expressed in the follicular cycle of Liaoning Cashmere goats [Citation7]. [Citation8] have compared the miRNA transcriptome in goats and sheep by Solexa sequencing, and identified the differences in miRNA and found that miRNA-203 was highly expressed in the sheep library. Thus, miRNA-203 may regulate hair follicle growth, but no further studies have shown this.
Results
Differential expression of miRNA-203 and target genes in telogen and anagen
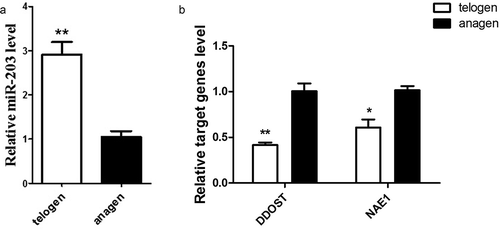
qPCR was used to detect the expression patterns of miRNA-203 and target genes in telogen and anagen. The expression of miRNA-203 in anagen of hair follicle was found to be decreased (, p < 0.01). Nevertheless, the expression of DDOST and NAE1 in anagen was higher (, p < 0.01). Correlation analysis showed that there was a significant negative correlation between miRNA-203 and target genes (DDOST and NAE1) expression.
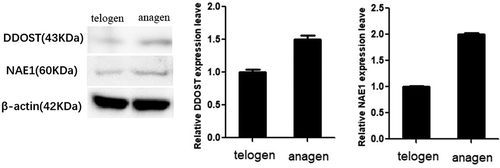
Figure 1. mRNA expression of miR-203 and target gene DDOST were detected in telogen and anagen skin tissue of Cashmere goats. In this figure, detection of miR-203 in telogen and anagen(Figure 1a);detection of DDOST and NAE1 mRNA in telogen and anagen(Figure 1b). The results showed that the expression level of miR-203 was reversed to the expression level of DDOST and NAE1. All data in this figure represent the mean ±SEM of three independent experiments, **P < 0.01
The molecular weights of DDOST and NAE1 were 43 kDa and 60kDa, respectively, and the expressions of DDOST and NAE1 were consistent with the results of qPCR analysis, which were higher in anagen than in telogen (, p < 0.05).
miRNA-203 down-regulates DDOST and NAE1 genes expression
DNA sequencing results showed that the vector was triumphantly constructed (). Dual-Luciferase reporter system showed that the WT expression of the target gene reduced than in the mutant and blank group (, p < 0.01).
Figure 3. Luciferase reporter assay. In this figure, luciferase reporter vectors of DDOST were constructed, and it has been assayed by sequencing. The sequencing results confirmed that luciferase reporter vectors were constructed successfully. mut, mutant; WT, wild type
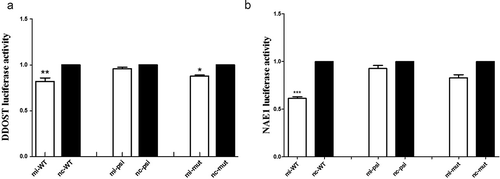
Figure 4. The targets of miR-203. MiR-203 binding site on the target genes 3ʹ UTR is shown, together with its homology across species. Target points were detected by Dual-Luciferase Reporter gene assay. In the experiment, transfection was divided into three groups, including WT group, MT group, and psicheck-2 group (Psi). Mimics (mi) and NC were, respectively, transfected with WT and mut. Detection of expression of DDOST and NAE1 in three groups by Dual-Luciferase Reporter gene detection system. NC, negative control.(Figure 4a-b) All data in this figure represent the mean ±SEM of three independent experiments, *p < 0.05, **P < 0.01
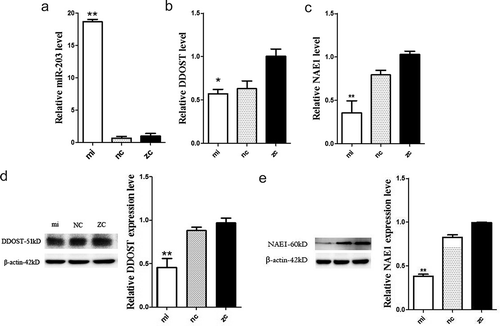
The RNA extracted from the transfected cells was amplified by qPCR, and it was found that miRNA-203 was expressed in the miRNA-203 mimics group higher than the negative group (-a, p < 0.01). However, the expression of DDOST and NAE1 in the miRNA-203 mimics and NC groups was opposite to that of miRNA-203 (, p < 0.01). Interestingly, the expression of miRNA-203, DDOST and NAE1 in the miRNA-203 mimics and NC groups was lower than in the blank group. The results of protein expression experiments showed that DDOST and NAE1 expressions were significantly reduced in the miRNA-203 mimics group (, p < 0.01).
Figure 5. Over-expression of miR-203 in HEK-293 T cells. The relative expressions of miR-203 in miR-203 over-expression and negative control were detected by qPCR(Fig5a);the relative expressions of DDOST in miR-203 over-expression and negative control were detected by qPCR(Fig5b-c). The protein expressions of DDOST and NAE1 in miR-203 over-expression and negative control were detected by western blot. mi, NC, and ZC, respectively, represent the cells transfected with miR-203, miR-203 negative control and the blank group(Fig5d-e). All data in this figure represent the mean ±SEM of three independent experiments, **P < 0.01
Discussion
In recent years, accumulating research data have shown that miRNAs are effective regulators in the pathophysiological processes of the body [Citation9–14]. A large number of studies have shown the expression and function of miRNA in the skin of mice, alpacas and other animals [Citation2,Citation4,Citation15–17]. It has been reported that Wnt3a can promote the formation of melanogen in mouse melanocytes, while miR-27-3p regulates the formation of melanogen by inhibiting the expression of Wnt3a [Citation3]. [Citation5] studied the number of miRNAs selectively expressed in the hair follicle cycle of Liaoning Cashmere goats. It provides a basis for further understanding of the role of miRNA in hair follicle circulation. [Citation18] have shown that miR-339-5p induces HFSC differentiation by targeting DLX5 to negatively regulate loureirin A. Meanwhile, our previous studies have also shown that miRNA plays an important role in hair follicle development regulation of Cashmere goats and Fine wool sheep [Citation8]. [Citation7] have provided new information on the regulation of miR-let7a and its related target genes in hair follicle.
miRNA-203, a skin-specific miRNA, involved in keratinocyte [Citation19]. Warshauer et al. (2015) showed that RBM28 participates in the growth of HF by regulating the activity of miRNA-203 and p63. miRNA-203 regulates epidermal keratinocyte differentiation [Citation20]. The miRNA transcriptome of goat and sheep skin was compared using Solexa sequencing to understand the development of HF; the expression of miRNA-203 was significantly higher in the sheep library [Citation8]. [Citation21] have shown that the expression levels of miRNAs such as miRNA-203, miRNA-205, miRNA-96 occured three transitions in one hair follicle cycle. These studies suggest that miRNA-203 is involved in hair follicle development.
There have been many reports on DDOST and NAE1 as potential targets for the action of pathological drugs [Citation22,Citation23]. Used MTFP technology and random cloning methods to obtain the characteristics of target genes regulated by miRNA-203, including DDOST and NAE1, but no further studies. At the same time, we used the online biological software miRBase to predict the target genes of miRNA-203, and determined DDOST and NAE1 as the research objects. In this study, we have confirmed that DDOST and NAE1 are involved in hair follicle development under the regulation of miRNA-203, and target genes (DDOST and NAE1) is negatively correlated with the expression of miRNA-203.
Taken together, this study demonstrates that miRNA-203 plays a regulatory role in hair follicle development of Cashmere goats by targeting target genes (DDOST and NAE1). This finding is helpful for further studies on the role of miRNA in improving the quality and yield of villi.
Materials and methods
Animal experiments were conducted in accordance with the regulations on the administration of experimental animals and under the guidance of the animal protection and research committee of Jilin University.
Animal and sample collection
Cashmere goats come from the Baicheng Breeding Center of Jilin Province. Approximately 1cm2 skin samples of anagen and telogen were harvested from adult Cashmere goats in September and March, and frozen in liquid nitrogen for analysis.
RNA preparation, qPCR analysis
Total RNA was extracted from samples using Trizol (Invitrogen, Rockville, MD) according to manufacturer’s instructions. RNA integrity was checked on Nanodrop 2000 (Thermo, China). Primer sequences were designed and synthesized by Comate Bioscience co., LTD. (changchun, China), as shown in Table 1. MiRNA expression analysis; the First Strand was synthesized using Revert AidTM First Strand cDNA Synthesis Kit (Takara, Dalian, China); all experimental procedures were performed at low temperature. In order to determine the expression of miRNA-203 and its target genes (DDOST and NAE1) in anagen and telogen, qPCR was performed using the FastStart Essential DNA Green Master (Roche). The relative content of miRNA was normalized to U6.
Western blot analysis
Protein was extracted using RIPA total protein extraction lysis buffer (Bioworld, USA), and the BCA protein quantitative kit (Beyotime, Shanghai) was used to detect the protein concentration. One hundred and fifty micrograms of protein extract per sample was separated at 10% SDS-PAGE, and the separated protein was transferred to PVDF (Bio-Rad Laboratories, Inc.). Membranes with 1:1000 DDOST antibody (from rabbit, absin, China); 1:600 NAE1 antibody (human, abdominal muscle, China) and 1:2000 β-actin antibody (rabbit, abdomen China) were incubated, 4°C for the night. After washing three times with TBST, secondary anti-horseradish peroxidase (HRP)-conjugated goat anti-rabbit-IgG was incubated at 1:5000 (Bioworld, America) for 1 h at 37°C. Western blot was analyzed using ECL’s advanced visual western blot detection equipment (Invitrogen).
Cell culture, transfection
HEK-293 T cells were obtained from the animal basic pathological anatomical laboratory of the college of veterinary medicine, and cultured in DMEM medium (Thermo, Grand Island, NY) containing 10% heat-inactivated fetal bovine serum ([FBS], MRC, Jiangsu, China) and 1% penicillin-streptomycin (HyClone, Beijing, China) in an incubator containing 5% CO2. On the day before transfection, 5•105 cell densities were inoculated on 6-well plates and co-cultured with DMEM containing 10% FBS. DMEM (3 ml) was mixed with Lipofectamine 3000 (20 µl) and miRNA-203 mimics/NC (11 µl 20 mol), incubated at room temperature for 15–20 min, and the complex was transfected into cells for 24 h. The expression differences of miRNA-203, DDOST and NAE1 were detected from the mRNA and protein expression levels.
Dual-luciferase reporter system
We used Targetscan and miRBase for computational analysis to predict the target sites of miRNA-203 binding to DDOST and NAE1 3ʹUTR (-a). The 3ʹUTR region of DDOST and NAE1 was amplified with a pair of WT and mut primers. Using Xho I and Not I (Takara, Dalian) endonucerase as the cleavage sites, the amplification sequence was cloned into the downstream PSI-check2 vector to prepare DDOST-3ʹUTR-WT, DDOST-3ʹUTR-mut, NAE1-3ʹUTR-WT and NAE1-3ʹUTR-mut reporter genes. After the DNA sequence was confirmed by Comate Bioscience Limited (Changchun, China), vectors of reporter gene system were constructed (Table 2). Before transfection, 2•105 cell densities were inoculated on 96-well plates until confluence reached 70% or higher. DMEM (including FBS, 100 µl) and Lipofectamine 3000 (1 µl) and miRNA-203 mimics/NC (0.9 μl, 20 µmol), pmiR-DDOST/NAE1-REPORT-WT-vector (0.33 µg), pmiR-DDOST/NAE1-REPORT-mut-vector (0.33 µg) and pmiR-RB-REPORT-psi-vector (0.33 µg) were mixed and incubated for 15–20 min. After co-transfection of miRNA-203 mimics/NC and DDOST/NAE1 vectors into HEK-293 T cells, luciferase activity was determined by the Dual-Luciferase reporter gene assay (Promega). All reporter gene assays were performed in triplicate.
Statistical analysis
A difference with p<0.05 was considered significant.
Acknowledgments
This study was funded by the Liaoning Cashmere Goat Breeding Center. And this work was funded by the Natural Science Foundation of Jilin Province. (20170101156JC)
Disclosure statement
The author states that there is no conflict of interest.
Additional information
Funding
References
- Hochfeld LM, Anhalt T, Reinbold CS, et al. Expression profiling and bioinformatic analyses suggest new target genes and pathways for human hair follicle related microRNAs. BMC Dermatol. 2017;17:3.
- Tian X, Jiang J, Fan R, et al. Identification and characterization of microRNAs in white and brown alpaca skin. BMC Genomics. 2012;13:555.
- Zhao Y, Wang P, Meng J, et al. MicroRNA-27a-3p inhibits melanogenesis in mouse skin melanocytes by targeting Wnt3a. Int J Mol Sci. 2015 May;16:10921–10933.
- Qu L, Li J, Zhao Z, et al. Differential expression of miR-202 and validation of predicted target genes in the skin tissue of C57BL/6 black mice and BALB/c white mice. DNA Cell Biol. 2017;36:443–450.
- Bai WL, Dang YL, Yin RH, et al. Differential expression of microRNAs and their regulatory networks in skin tissue of liaoning cashmere goat during hair follicle cycles. Anim Biotechnol. 2016;27:104–112.
- Yuan S, Li F, Meng Q, et al. Post-transcriptional regulation of keratinocyte progenitor cell expansion, differentiation and hair follicle regression by miR-22. PLos Genet. 2015;11:e1005253.
- Ma T, Li J, Jiang Q, et al. Differential expression of miR-let7a in hair follicle cycle of Liaoning cashmere goats and identification of its targets. Funct Integr Genomics. 2018;18:701–707.
- Li JP, Qu HE, Jiang HZ, et al. Transcriptome-wide comparative analysis of microRNA profiles in the telogen skins of liaoning cashmere goats (capra hircus) and fine-wool sheep (ovis aries) by solexa deep sequencing. DNA Cell Biol. 2015;35:696–705.
- Wang P, Zhao Y, Fan R, et al. MicroRNA-21a-5p functions on the regulation of melanogenesis by targeting Sox5 in mouse skin melanocytes. Int J Mol Sci. 2016;17:E959.
- Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110.
- Suo HB, Zhang KC, Zhao J. MiR-200a promotes cell invasion and migration of ovarian carcinoma by targeting PTEN. Eur Rev Med Pharmacol Sci. 2018;22:4080–4089.
- Wu N, Han Y, Liu H, et al. miR-5590-3p inhibited tumor growth in gastric cancer by targeting DDX5/AKT/m-TOR pathway. Biochem Biophys Res Commun. 2018;503:1491–1497.
- Tao Y, Ma C, Fan Q, et al. MicroRNA-1296 facilitates proliferation, migration and invasion of colorectal cancer cells by targeting SFPQ. J Cancer. 2018;9:2317–2326.
- Rong E, Zhang Z, Qiao S, et al. Functional characterization of a single nucleotide polymorphism in the 3ʹ untranslated region of sheep DLX3 gene. PLoS One. 2015;10:e0137135.
- Gao W, Sun W, Yin J, et al. Screening candidate microRNAs (miRNAs) in different lambskin hair follicles in Hu sheep. PLoS One. 2017;12:e0176532.
- Liu Z, Xiao H, Li H, et al. Identification of conserved and novel microRNAs in Cashmere goat skin by deep sequencing. PLoS One. 2012;7:e50001.
- Ji K, Zhang P, Zhang J, et al. MicroRNA 143-5p regulates alpaca melanocyte migration, proliferation and melanogenesis. Exp Dermatol. 2018;27:166–171.
- Li X, Wu Y, Xie F, et al. miR‑339‑5p negatively regulates loureirin A‑induced hair follicle stem cell differentiation by targeting DLX5. Mol Med Rep. 2018;18:1279–1286.
- Sonkoly E, Wei T, Janson PCJ, et al. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS One. 2007;2:e610.
- McKenna DJ, McDade SS, Patel D, et al. MiR-203 expression in keratinocytes is dependent on regulation of p53 levels by E6. J Virol. 2010;84:10644–10652.
- Fu S, Zhao H, Zheng Z, et al. Melatonin regulating the expression of miRNAs involved in hair follicle cycle of Cashmere goats skin. Yi Chuan. 2014;36:1235–1242.
- Santos-Bezerra DP, Machado-Lima A, Monteiro MB, et al. Dietary advanced glycated end-products and medicines influence the expression of SIRT1 and DDOST in peripheral mononuclear cells from long-term type 1 diabetes patients. Diab Vasc Dis Res. 2018;15:81–89.
- Jones MA, Ng BG, Bhide S, et al. DDOST mutations identified by whole-exome sequencing are implicated in congenital disorders of glycosylation. Am J Hum Genet. 2012;90:363–368.