ABSTRACT
To date, proposed therapies and antiviral drugs have been failed to cure coronavirus disease 2019 (COVID-19) patients. However, at least two drug companies have applied for emergency use authorization with the United States Food and Drug Administration for their coronavirus vaccine candidates and several other vaccines are in various stages of development to determine safety and efficacy. Recently, some studies have shown the role of different human and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) microRNAs (miRNAs) in the pathophysiology of COVID-19. miRNAs are non-coding single-stranded RNAs, which are involved in several physiological and pathological conditions, such as cell proliferation, differentiation, and metabolism. They act as negative regulators of protein synthesis through binding to the 3′ untranslated region (3′ UTR) of the complementary target mRNA, leading to mRNA degradation or inhibition. The databases of Google Scholar, Scopus, PubMed, and Web of Science were searched for literature regarding the importance of miRNAs in the SARS-CoV-2 life cycle, pathogenesis, and genomic mutations. Furthermore, promising miRNAs as a biomarker or antiviral agent in COVID-19 therapy are reviewed.
1. Introduction
The global pandemic of coronavirus disease 2019 (COVID-19) caused by a highly contagious and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged on 31 December 2019, in Wuhan city, Hubei province, China [Citation1]. Subsequently, infection by the virus has led to high morbidity and mortality rates [Citation2]. SARS-CoV-2 is a positive-sense, single-stranded enveloped ribonucleic acid (+ssRNA) virus that belongs to the genus Betacoronavirus and the family Coronaviridae [Citation3]. COVID-19 infection presents with a myriad of complications from symptoms of the common cold (such as fever, chills, coughing, fatigue, headache, sore throat, or unusual symptoms (loss of smell or taste), breathing problems to gastrointestinal symptoms including nausea, vomiting, diarrhea, and abdominal pain. The condition can progress to life-threatening issues such as severe pneumonia and acute respiratory distress syndrome (ARDS), ultimately terminating in death [Citation4,Citation5]. Although numerous medicines have claimed to treat the infection, no remedy or vaccine for treatment/prevention of COVID-19 has been approved in the United States or the European Union [Citation6–8]. However, Pfizer and BioNTech are the first companies to file for an emergency use authorization with the United States Food and Drug Administration for their coronavirus vaccine candidate. It is anticipated that several other companies are close to filing for similar authorization in the United States and elsewhere.
It has been demonstrated that microRNAs (miRNAs) play a crucial role in the host antiviral responses and the viral pathogenesis in cases of the herpes virus, polyomavirus, retroviruses, pestivirus, and hepacivirus, as well as coronavirus [Citation9,Citation10]. miRNAs are non-coding single-stranded RNAs of 6–8 nm long with 19–28 nucleotides that regulate gene expression and protein synthesis at the translational level. As a negative regulator of mRNA, gene silencing is accomplished by miRNAs through direct binding to the 3′ untranslated region (3′ UTR) of the target mRNA and induction of mRNA degradation or translation suppression. It also has been reported that miRNAs can interact with other regions of the mRNA sequence such as 5′ UTR, gene promoters, and coding sequences [Citation11]. Furthermore, miRNAs have been shown to activate gene expression and transcription by targeting enhancer regions under certain conditions [Citation12]. miRNAs participate in many physiological processes and pathophysiological stages such as proliferation, metabolism, differentiation, and apoptosis, and miRNAs can modulate innate and adaptive immunity by affecting protein levels. miRNAs can be encoded by the genome of eukaryotic organisms and some viruses [Citation13,Citation14]. Moreover, host miRNAs expression changes during viral infections, involving various signaling pathways, modulation of host-virus interactions, regulation of viral infectivity, and transmission and activation of antiviral immune responses. On the other hand, the viral genome can express its miRNAs and can even hijack human miRNAs to the repertoire of the infected cells [Citation15,Citation16].
miRNAs are known to play a role in the corroboration or contravention of other coronaviruses pathogenesis, such as SARS-CoV and the middle east respiratory syndrome coronavirus (MERS-CoV) that caused epidemic outbreaks in 2003 and 2012, respectively [Citation17,Citation18]. Since the outbreak of COVID-19, the significance of miRNAs in the pathophysiology of COVID-19 has been postulated. Understanding the role of miRNAs during SARS-CoV-2 infection may lead to the discovery of novel mechanisms that could help with the diagnose of the disease, combat the virus pathogenesis, and potentially blocking the virus RNA entry, replication, and invasion. We searched the scientific databases in Google Scholar, Scopus, PubMed, and Web of Science to obtain relevant studies published in this context through November 2020. Based on the retrieved articles, the role of human and viral miRNAs in different stages of the SARS-CoV-2 life cycle and pathogenesis and genomic mutations are reviewed. miRNAs induction as a promising biomarker or antiviral agents in COVID-19 is suggested.
2. Virus and human miRNAs in SARS-CoV-2 life cycle
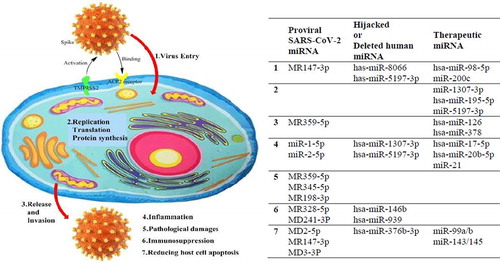
The SARS-CoV-2 life cycle includes attachment and entry, replication/translation/protein synthesis, and release and invasion of the virus. Transmembrane serine protease 2 (TMPRSS2) activates the spike protein subunits of SARS-CoV-2 to bind to the host cell angiotensin-converting enzyme 2 (ACE2) surface receptor [Citation19]. Then, membrane fusion and virus endocytosis is initiated. The RNA genome of the virus is released following proteolysis and uncoating, using the host cell replication and protein translation machinery to make structural proteins such as spikes (S), envelopes (E), membranes (M), nucleocapsides (N), and nonstructural and accessory proteins. After packaging and viral assembly, SARS-CoV-2 is released by exocytosis to invade and infect other cells [Citation20,Citation21]. The SARS-CoV-2 genome consists of 11 open reading frames (ORF) such as ORF1ab, encoding 16 nonstructural proteins, followed by encoding structure proteins, including S, E, M, and N proteins, as well as six accessory proteins, including 3a, 6, 7a, 7b, 8, and 10 [Citation22].
Viral miRNAs encoded by the SARS-CoV-2 genome can target several host genes. In one study, 3377 human target genes were predicted as unique targets of 170 mature miRNAs from SARS-CoV-2. Additionally, many human miRNAs were suggested as potential targets for viral genes involved in the SARS-CoV-2 life cycle, such as the genome of S, M, N, and E proteins and ORF1ab, ORF3a, ORF8, ORF7a, ORF10, and ORF6. The S glycoprotein and ORF1ab gene are known to be targeted by 67 and 369 different human miRNAs, respectively. Furthermore, 10 human miRNAs have been identified to possess binding sites across the SARS-CoV-2 genome. These host cellular miRNAs, by targeting the SARS-CoV-2 genome, produce antiviral effects [Citation23–25]. In another study, miRNAs in human lung epithelium were predicted to have potential binding sites in the SARS-CoV-2 genome; 128 human miRNAs had a very low level of expression in lung epithelia, and six of 128 miRNAs exhibited differential expression upon in vitro infection with SARS-CoV-2. It was hypothesized that the endogenous or therapeutic upregulation of these miRNAs would effectively boost the protective capacity of the respiratory epithelial cells against COVID-19. The important virus and human miRNAs involved in different stages of the SARS-CoV-2 life cycle are reviewed in the following sections.
2.1. Attachment and entry
The virus-encoded MR147-3p has been shown to induce TMPRSS2 gene expression in the gut [Citation26]. TMPRSS2 is involved in the activation of the S protein of the virus binding to ACE2 on human cells [Citation27]. The gastrointestinal symptoms of COVID-19 might be mediated by virus-derived MR147-3p which facilitates effective evasion of the virus into gut cells. has-miR-8066 and has-miR-5197-3p are associated with mucin-type O-glycan biosynthesis, which is important in SARS-CoV-2 S protein synthesis. has-miR-3934-3p is involved in the biosynthesis of heparin sulfate/heparin proteoglycans, which can provide binding sites for SARS-CoV-2 [Citation28].
In addition, host miRNAs might block attachment and entry of SARS-CoV-2 because several miRNA families were found to target ACE2 and TMPRSS2 genes in different tissues. In an in vitro study, hsa-miR-98-5p was demonstrated to directly target the 3’ UTR of TMPRSS2 gene transcription, both in human lung microvascular endothelial cells (HMVEC-L) and human umbilical vein endothelial cells (HUVEC) [Citation29]. The hsa-let-7e/hsa-mir-125a and hsa-mir-141/hsa-miR-200 miRNA families inhibit ACE2/TMPRSS2 gene transcription. To be more exact, hsa-miR-125a-5p together with miRNAs from the miR-200 family targets 3′ UTR of ACE2 mRNA while hsa-let-7e-5p targets 3′ UTR of TMPRSS2 [Citation30]. hsa-miR-4661-3p binds to the 3′ UTR of the S gene and downregulates its expression [Citation26]. hsa-miR-23b and hsa-miR-98, by cleaving VP1 and interferon (IFN)-β genes, target the S protein of SARS-CoV-2 [Citation31]. hsa-miR-510-3p, hsa-miR-624-5p, and hsa-miR-497-5p have been reported to target RNA S glycoprotein of SARS-CoV-2 sequence [Citation32]. In addition, hsa-miR-622, hsa-miR-761, miR-A3r, hsa-miR-15b-5p, miR-A2r, and has-miR-196a-5p have been shown to target the S protein gene [Citation33]. miR-338-3p, miR-4661-3p, miR-4761-5p, hsa-miR-4464, hsa-miR-1234-3p, hsa-miR-7107-5p, and hsa-miR-885-5p have been shown to bind to the receptor-binding region of the S gene [Citation25]. The CLEC4M gene is also related to the expression of S glycoprotein [Citation34], and it was reported that CLEC4M could be suppressed by hsa-miR-4462 and hsa-miR-5187-5p. Similarly, the ACE2 receptor was suppressed by targeting its 3’ UTR gene by hsa-miR-23b-5p and hsa-miR-769-5p [Citation35]. 31 miRNAs and 29 miRNAs can downregulate hypothalamic ACE2 and TMPRSS2 gene expression, respectively, suggesting that they might be useful in treating SARS-CoV-2 neuroinvasion, by blocking virus entry [Citation36]. In another study, miR-200 c suppressed ACE2 mRNA and protein expression in rat primary cardiomyocytes and human-induced pluripotent stem cell-derived cardiomyocytes, suggesting a promising strategy to treat SARS-CoV-2 cardiovascular complications [Citation37].
Acute renal injury and nephropathy are additional challenging complications in COVID-19 patients [Citation38]. Several miRNAs are upregulated in nephropathies, such as miR-18 or miR-125b, and both are reported to act as an ACE2 gene enhancer. Therefore, they may contribute to ACE2 related kidney injuries following SARS-CoV-2 infection. AntimiR-18 and antimiR-125b have also been reported to be efficient against nephropathy associated with COVID-19 [Citation39].
2.2. Viral genomic replication and translation and protein synthesis
It is well known that viral genomic replication, translation, and protein synthesis require the transcription and translation machinery of the host cells [Citation40]. SARS-CoV-2 miRNAs, therefore, should play a role somewhere in this machinery. Viral miRNAs, by suppression of some mRNA deadenylases such as subunits of the CCR4-NOT transcription complex (CNOT4, CNOT10, and CNOT6L), protect the SARS-CoV-2 mRNA from turnover and degradation in human cells. Viral miRNAs also prevent RNA polymerase II attachment to promoters of host genes at the initiation step by targeting transcriptional regulators involved in both basal transcription machinery elements including basal transcription factors (TAF4, TAF5, and TAF7L), site-specific transcription factors (MAFG, TCF4, TFDP2, TRPS1, BRF1, and SOX11), and components of the human mediator complex (MED1, MED9, MED12L, and MED19) [Citation24].
Contrarily, many known human miRNAs have been shown to target and inhibit viral genes involved in replication, translation, and protein synthesis. miR-1307-3p and miR-3613-5p were predicted to prevent virus replication by targeting 3’ UTR of the virus genome [Citation41]. Some human miRNAs target ORF1ab which encodes enzymes for virus replication and translation [Citation26]. ORF1ab and ORF3a have been reported to be targeted by hsa-miR-203b-3p which suppresses viral replication in the case of influenza A [Citation24]. hsa-let-7a and hsa-miR-101 have been shown to target nonstructural protein synthesis in SARS-CoV-2, by cleaving IFN-β and ATP5B genes, respectively [Citation31]. hsa-miR-497-5p, hsa-miR-21-3p, and hsa-miR-195-5p are widely expressed in respiratory epithelial cells, have a high target score against the SARS-CoV-2 genome, and are predicted to be effective against the virus through nucleotide deletion in the coding regions of the viral ss-RNA [Citation32].
Complimentary miRNAs (cc-miRNA) have been utilized to target viral genes and inhibit their posttranscriptional expression [Citation42]. The cc-miRNAs (modified to 25–27 nucleotides), namely, cc-miR1c, cc-miR2c, cc-miR3c, cc-miR4c, cc-miR5c, cc-miR6c, and cc-miR7c, are formed on the basis of ID02510.3p-miRNA, ID00448.3p-miRNA, miRNA 3154, miRNA 7114–5p, miRNA 5197–3p, ID02750.3p-miRNA, and ID01851. 5p-miRNA has shown a strong binding with the SARS-CoV-2 viral genome. In this study, human miRNAs including miR-1273a, miR-1273d, miR-1272, miR-1292-5p, miR-3143, miR-1226-5p, and miR-7161-3p were reported to interact with the RNA of SARS-CoV-2 as part of the RNA-induced silencing complex (RISC) [Citation43]. In another study, miR-5197-3p interacted effectively and without competition with the mRNA genome of SARS-CoV-2. miR-5197-3p did not target human coding genes, which implies the absence of side effects of this cc-miRNA on the translation of human mRNAs [Citation44]. In another study, 12 cc-miRNAs were reported to target the SARS-CoV-2 genome [Citation33]. These cc-miRNAs had previously been reported to efficiently inhibit HIV (Human Immunodeficiency Virus)-1 replication, without any reported off-target effects [Citation45]. Together, cc-miRNAs might be efficient agents against SARS-CoV-2 infection.
2.3. Release and invasion
Targeting and upregulation of human genes by viral miRNAs may support SARS-CoV-2 release and escape from human cells and their invasion into other tissues. MR359-5p via inducing the overexpression and activity of MYH9 (non-muscle myosin heavy chain 9) facilitates virus invasion, trafficking within the cell, and release [Citation26]. MYH9 has been reported to bridge between the attachment of virus particles to cell surface receptors and subsequent the uncoating event required for genomic release into the host cell [Citation46]. Integrin β-5 gene expression was also enhanced by MR369-5p [Citation26]. Integrin β-5 is related to cell skeleton-related functions such as actin filament-associated processes and actin cytoskeleton organization. The actin cytoskeleton is critical for viral replication at different stages of the virus life cycle, and viruses can subvert the force-generating and macromolecular scaffolding properties of the actin cytoskeleton to propel viral surfing, internalization, and migration within the cell [Citation47,Citation48].
Human miRNAs might prevent SARS-CoV-2 release and invasion. The N glycoprotein is an important antigen of SARS-CoV-2 which participates in the genomic package and virus particle release [Citation49]. ORF9 encodes N protein [Citation50]. A mix of antisense miRNAs targets 3’ UTR, 5’ UTR, and ORF9 regions hypothesized to inhibit virus translation process and particle assembly [Citation51]. hsa-miR-126 and hsa-miR-378 inhibit virus N protein synthesis via translation suppression and cleavage of the IFN-β gene, respectively [Citation31].
3. The role of miRNAs in pathogenicity of SARS-CoV-2
SARS-CoV-2 proliferation and invasion in human cells especially the lungs more often than not will result in inflammation, pathological damage, and disruption of cellular signaling pathways and metabolism. Furthermore, human antiviral defensive mechanisms i.e. the immune system and apoptotic activities are suppressed by the virus, increasing viral susceptibility [Citation52,Citation53]. The role of miRNAs in inducing inflammation and pathological damages, immunosuppression, and reducing host cell apoptosis is review below.
3.1. Inflammation and pathological damages
Activation of the immune system and inflammation are observed following proliferation and invasion of SARS-CoV-2 in human cells; these events cause tissue damage and organ dysfunction, especially in the respiratory tract, the gastrointestinal tract, brain, liver, and kidneys [Citation54,Citation55].
The work of Khan et al. (2020) showed that SARS-CoV-2 miRNAs participate in these complications. For example, brain development, heart development, and insulin signaling pathway were predicted to be targeted by the virus miRNAs. Patients with underlying disorders like cardiovascular diseases, diabetes, or breathing complications are more susceptible to SARS-CoV-2 infection, which suggests an epigenetic modulatory role of virus miRNAs and even targeting host miRNAs [Citation23]. Host miRNAs targeted by the virus are regulated vital functions such as kidney and heart development, neuronal processes, metabolic processes, fatty acid metabolism, insulin resistance, glucagon signaling pathway, and peroxisome proliferator-activated (PPAR) signaling. In COVID-19 patients, dysregulation of these processes follows inhibition of human miRNAs, overcomplicating the condition especially in individuals with comorbidity [Citation23].
SARS-CoV-2 miRNAs have been shown to inhibit ribosomal translation of some key human proteins, via hybridizing their mRNAs [Citation56]. These proteins include hemoglobin subunit beta and gamma-globin 2, IFN 1, and olfactory receptor proteins. Marked perturbation of oxygen distribution in vital organs, immune response, and dysosmia in COVID-19 patients have been suggested to result from virus miRNA-like inhibition [Citation56]. Some host miRNAs alteration, mediated by SARS-CoV-2, were strongly associated with comorbidities. hsa-miR-3611 was associated with high severity of sensitivity toward viral respiratory infections in opioid addicts. hsa-miR-1307-3p, hsa-miR-8066, hsa-miR-5197-3p, and hsa-miR-3691-3p were shown to be associated with transforming growth factor (TGF)-β pathways, inflammatory response, cytokine-cytokine receptor interaction, and oxidative stress, all hallmarks of pulmonary damage [Citation28]. In addition, hsa-miR-1468-5p was reported to mediate similar damage in cardiac tissues [Citation28].
The results of one study showed that target genes of six SARS-CoV-2 miRNAs, including miR-1-5p, miR-2-5p, miR-3-5p, miR-4-5p, miR-5-5p, and miR-6-5p, were directly/indirectly involved in the upregulation of the immune response and the chemokine signaling pathway. It was concluded that SARS-CoV-2 miRNAs are critical in cytokine storm events of COVID-19 [Citation57].
Computational identification of SARS-CoV-2 miRNAs in different tissues was reported by Liu et al. (2020). In the lung, SARS-CoV-2-encoded miRNAs were suggested to increase inflammation by inducing the chemokine signaling pathway. The putative enhancer region of arrestin beta-2 and CXC chemokine ligand 16 (CXCL16), two pro-inflammatory cytokines, may be targets of MR147-5p. MR66-3p was indicated to target the transcription enhancer of TNF-α, an important cytokine in the inflammatory response, in the spleen [Citation26].
Virus-encoded MR345-5p was reported to bind to the 5’ UTR of the TGF-β receptor 3 (TGFBR3) gene and enhance its expression [Citation26]. TGFBR3 function leads to the activation of the innate and adaptive immunity system by controlling regulatory T-cells and promoting Th1 differentiation [Citation58]. The virus-encoded miRNAs were also predicted to contribute to the pathological damage. MD2-5p, MR359-5p, and MR345-5p may contribute to liver dysfunction by targeting the FOXO3, ADIPOQ, and ADIPOR1 genes involved in the biological processes of cellular response to peptide hormone stimulus and regulation of fatty acid metabolic processes [Citation59,Citation60]. In the liver, MR198-3p acts on the enhancer region of adenosine deaminases acting on the RNA (ADARs) gene. ADARs mainly regulate genes involved in the function of actin filament severing and regulation of cellular protein metabolic processes [Citation61] resulting in the activation of FOXO3, ADIPOQ, ADIPOR1, and ADAR genes by the virus miRNAs, suggesting a possible mechanism of liver damages observed in COVID-19 patients [Citation26].
On the other hand, some of the host miRNAs such as hsa-miR-17-5p, hsa-miR-20b-5p, and hsa-miR-323a-5p have a preventive role against the inflammatory cascade and reduce acute lung injury and other organ damages seen in SARS-CoV-2 infection [Citation23]. The NF-ĸB p65 (transcription factor p65) subunit is a protein encoded by the RELA gene and mediates immune responses, inflammation, and apoptosis [Citation62]. hsa-miR-516b-3p, hsa-miR-3529-3p, and hsa-miR-6749-3p by targeting 3’ UTR of the RELA could suppress its expression and alleviate COVID-19 [Citation35]. Application of specific antagomiRs against miRNAs involved in the inflammatory process including miR-21, miR-125b, miR-199a, miR-211, miR-138, miR-211, miR-146a, and miR-146b have been proposed to attenuate the cytokine storm and to decrease acute lung damage in COVID-19 patients [Citation63].
3.2. Immunosuppressive activity
Virus miRNAs can evade components of the immune system, resulting in the attenuated or abnormal activity of the immune system mechanisms. As a result of SARS-CoV-2’s escape from the immune response, the virus prolongs its latency period inside human cells and therefore, less transition in lytic viral replication occurs [Citation64].
SARS-CoV-2 miRNAs were found to downregulate some immune-signaling pathways, such as IFN-I signaling and autophagy [Citation23]. Virus-encoded MR328-5p has been shown to activate the retinoid X receptor α (RXRα) gene. RXRα over-expression attenuates host immunity response by suppressing the IFN1 response [Citation26]. Other genes targeted by SARS-CoV-2 miRNAs are STAT1 and STAT5B. The STAT family transcription factors are signal transducers activated by cytokine-induced stimuli and regulate the IFN response. So, targeting the STAT family by viral miRNAs could lead to deregulation of the IFN response [Citation24]. SARS-CoV-2 matures miRNA MD241-3P targets and suppresses the morphogenetic protein receptor type 2 (BMPR2) gene in bone. Since BMPR2 is involved in the TGF-β signaling pathway and plays a role in pulmonary vascular homeostasis, its inhibition reduces the antiviral response of the body and might induce respiratory lung disease [Citation65].
SARS-CoV-2 has been reported to hijack some human miRNAs involved in the suppression of the immune system and therefore, the host miRNAs are employed as proviral factors that contribute to SARS-CoV-2 pathogenesis. For example, the genes of hsa-miR-146b and hsa-miR-939, two key modulators of immunity responses, were reported to be hijacked by the SARS-CoV-2 genome [Citation26]. Highly expressed and functionally crucial miRNAs in immune cells titrated by the virus resulted in post-transcriptional dysregulation of genes of the immunity system and modulation of the propagation of SARS-CoV-2 infection [Citation66].
The genes of several host miRNAs were also induced during COVID-19. Some significant immune pathways and proinflammatory cytokines such as IFN-γ signaling, TGF-β signaling, interleukin (IL) signaling, IGF1 (insulin-like growth factor 1) signaling, tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) signaling, and toll-like receptors (TLRs) were shown to be downregulated by these human miRNAs, leading to host’s immune suppression [Citation23]. In another study, 22 human miRNAs were reported to be titrated by the SARS-CoV-2 genome, leading to altered and dysregulated post-transcriptional events in the infected cells, including CD8+T, CD4+T, natural killer (NK), CD14, and mast cells [Citation67]. Noncoding SARS-CoV-2 RNAs by acting as host miRNA sponges may dysregulate and repress human miRNA levels, including miR-376a-3p, miR-99b-5p, miR-10a-5p, miR-376a-3p, miR-548av-5p, and miR-99b-5p which are involved in immune responses [Citation68].
3.3. Reducing host cell apoptosis
Host cell apoptosis, a protective mechanism in cells infected with viruses, plays a critical role in normal physiological processes and virus pathogenesis [Citation69]. Although apoptotic defense is one mechanism to curtail virus spread, SARS-CoV-2 has been shown to employ sophisticated molecular strategies such as miRNAs to subvert and neutralize host cell apoptosis [Citation70]. For instance, two virus-encoded miRNAs, MD2-5p and MR147-3p, have been hypothesized to repress the expression of pro-apoptotic elements and cation transport regulator-like protein 1 (CHAC1) and RAD9A genes, respectively [Citation26]. SARS-CoV-2 altered host miRNA MD3-3P has been shown to target and inhibit the tumor suppressor p53 gene. p53 is a primary inducer of apoptosis during viral infection and plays a role in the activation of innate immunity [Citation65]. SARS-CoV-2 by sponging and depletion of the human miR-376b-3p could potentially enhance the mammalian target of rapamycin (mTOR) activity to reduce the autophagy pathway [Citation68].
On the other hand, some miRNAs are speculated to efficiently combat SARS-CoV-2 by reducing host cell apoptosis and invoke a defensive response against the virus. mTOR is an anti-apoptosis agent that mediates cell proliferation whereas p53 is involved in cell apoptosis. mTOR inactivation and p53 activation could impede virus replication and proliferation. Interestingly, p53-dependent miRNAs, including miR-101, miR-100, miR-99a/b, miR-7, miR-107, let-7, and miR-199, bind to the 3′ UTR of mTOR mRNA and inactivates it. Furthermore, miR-107, miR-7, miR-429, miR-200, miR-15/16, miR-223, miR-143/145, and miR-17 inactivate the downstream target of mTOR [Citation71].
An illustration of SARS-CoV-2 life cycle and pathogenesis with some proviral, hijacked and therapeutic miRNAs is presented in .
4. Discussion
miRNAs are implicated in numerous physiological and pathological processes. For example, in preclinical studies, it was shown that miRNAs modulate apoptosis during myocardial ischemia/reperfusion injury and/or regulate endoplasmic reticulum stress in myocardial ischemia and cardiac remodeling [Citation72,Citation73]. Several interventional clinical trials -phase 1 and 2- on miRNAs are ongoing to examine the efficacy of miRNAs in the treatment of polycystic kidney disease, lung and liver cancer, lymphoma, hepatitis C virus, wound healing, heart failure, mesothelioma, and keloid and scar tissue formation [Citation74,Citation75].
The following two reasons may help explain the clinical importance of understanding the role of human and viral miRNAs in COVID-19 pathogenicity: First, miRNAs might be promising biomarkers in COVID-19 diagnosis. Importantly, a recent study reported that COVID-19 severity and mortality in aged patients may have resulted from defects in and lower abundance of human miRNAs [Citation76]. The normal function of miRNAs in the human body is required for the maintenance of cellular homeostasis. Host miRNAs targeting SARS-CoV-2 regulate critical biological processes and signaling pathways such as biosynthesis, immune response, circadian rhythm, and cardiomyopathy among several cellular processes. Therefore, miRNAs levels should be considered potential biomarkers of diagnostic value in COVID-19 patients which may help in the prediction of different stages of the disease. The differential expression of human miRNAs could establish the presence of infection and potentially its severity, especially in patients with comorbidities [Citation77]. For instance, in COVID-19 patients with diabetes, increased risk of heart failure was shown to be due to reduced cardioprotective miRNA 133a [Citation78]. In another study, differential miRNA expression in peripheral blood samples was observed in ten COVID-19 patients and four healthy controls [Citation79]. It also was reported that miR-6501-5p, miR-16-2-3p, and miR-618 were expressed to a greater degree in COVID-19 patients than in healthy subjects while miR-144-3p, miR-627-5p, and miR-183-5p were less expressed in COVID-19 patients. This study suggested that differential miRNA expression during SARS-CoV-2 infection might contribute to virus replication and regulation of the immune response. It is possible that miR-618 could be both a promising biomarker of and a therapeutic agent for COVID-19 [Citation79].
Second, miRNAs can be regulated artificially to treat COVID-19. Here, an important question remains to be answered: Would such regulated human and viral miRNAs significantly affect the disease pathophysiology?
A growing body of evidence suggests that genomic SARS-CoV-2 mutations influence miRNA expression, thereby influencing the pathophysiology of diseases. Mutant type of SARS-CoV-2 might induce a lower immune response and a weaker cytokine storm. In other words, mutations in the region overlapped with viral miRNAs involved in inflammation enhancement (i.e. MR288-5p and MD202-5p) could disrupt their activity. Also, hsa-miR-939-5p and hsa-miR-146b-3p (two inflammatory modulators) were reported not capable of binding to the mutant SARS-CoV-2 genome. On the other hand, the wild-type genome by attracting different batches of host miRNAs might change the actin filament fragmentation and epithelial differential regulation to enhance the virus invasion [Citation26]. The target sequence of miRNA 197–5p lost by the Nsp3 synonymous C3037U conserved mutation has been identified [Citation25]. miRNA 197–5p was reported to be upregulated in cardiovascular patients and miRNA 197–5p has been suggested as a prognostic and risk factor of such cardiac events. miRNA 197–5p also mediates defense against SARS-CoV-2. Thus, a higher COVID-19 mortality rate and susceptibility in patients with cardiovascular disease might be related to the loss of the miRNA 197–5p binding site across the SARS-CoV-2 genome [Citation25]. A mutation in the S glycoprotein (A930V (24,351 C > T)) was found in the Indian SARS-CoV-2 strain. It was the target location of hsa-miR-27b-3p, which is responsible for reducing the SARS-CoV-2 entry receptor by downregulating ACE2. Therefore, the mutant variant could cause a more severe condition due to the absence of the hsa-miR-27b-3p sequence [Citation31]. An amino acid substation of D614G at the S glycoprotein changed its miRNA sequence from hsa-miR-4793-5p to hsa-miR-3620-3p and increased the target score due to the nucleotide substitution 1841A > G at the SARS-CoV-2 RNA [Citation32]. The artificial regulation of human and viral miRNAs could also change COVID-19 pathophysiology.
A major concern about developing miRNA-based therapies is designing efficient and stable delivery systems for miRNAs and/or their antagonists. Viral and non-viral vectors, including exosomes and bacteriophages, liposomes, modified micelles, polymers, conjugates, nanoparticles, intravenous injection, intranasal, and even oral delivery systems, have been applied in this context [Citation80,Citation81]. The administration of the plant MIR2911 in a honeysuckle decoction (HD) significantly blocked virus replication and accelerated the negative conversion in patients with moderate COVID-19. However, HD-MIR2911 failed to be absorbed from the gastrointestinal tract in individuals with the SID1 transmembrane family member 1 polymorphism, but it proposes a promising insight considering a potential clinical application of miRNA against COVID-19 [Citation82,Citation83].
In conclusion, human and viral miRNAs play a determinant role in every stage of the SARS-CoV-2 life cycle, pathogenesis, and genomic mutations. Among them, some miRNAs have been suggested as potential biomarkers or antiviral agents, or for prediction and potential treatment of COVID-19. However, comprehensive preclinical and clinical studies on suitable drug delivery systems should be conducted to develop effective miRNAs-based therapeutics against COVID-19.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733.
- Matta S, Chopra K, Arora V. Morbidity and mortality trends of Covid 19 in top 10 countries. Indian J Tubercul. 2020;67(4):167–172.
- Of the International CSG. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536.
- Yang Y, Xiao Z, Ye K, et al. SARS-CoV-2: characteristics and current advances in research. Virol J. 2020;17:1–17.
- Reddm WD, Zhou JC, Hathorn KE, et al. Prevalence and characteristics of gastrointestinal symptoms in patients with severe acute respiratory syndrome Coronavirus 2 infection in the United States: A multicenter cohort study. Gastroenterology. 2020;159(2):765–767.
- Costanzo M, De Giglio MAR, Roviello GN. SARS-CoV-2: recent reports on antiviral therapies based on lopinavir/ritonavir, darunavir/ umifenovir,hydroxychloroquine, remdesivir, favipiravir and other drugs for the treatment of the new coronavirus. Current medicinal chemistry; 2020.
- McKee DL, Sternberg A, Stange U, et al. Candidate drugs against SARS-CoV-2 and COVID-19. Pharmacol Res. 2020;157:104859.
- Dong Y, Dai T, Wei Y, et al. A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduct Target Ther. 2020;5:1–14.
- Bernier A, Sagan SM. The diverse roles of microRNAs at the host–virus interface. Viruses. 2018;10:440.
- Leon-Icaza SA, Zeng M, Rosas-Taraco AG. microRNAs in viral acute respiratory infections: immune regulation, biomarkers, therapy, and vaccines. ExRNA. 2019;1(1):1–7.
- Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141(4):1202–1207.
- O’Brien J, Hayder H, Zayed Y, et al. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne). 2018;9:402.
- Shi Y, Jin Y. MicroRNA in cell differentiation and development. Sci China Ser C Life Sci. 2009;52:205–211.
- Rodrigues D, Monteiro VVS, Navegantes-Lima KC, et al. MicroRNAs in cell cycle progression and proliferation: molecular mechanisms and pathways. Non-Cod RNA Invest. 2018;2:28.
- Bruscella P, Bottini S, Baudesson C, et al. Viruses and miRNAs: more friends than foes. Front Microbiol. 2017;8:824.
- Grundhoff A, Sullivan CS. Virus-encoded microRNAs. Virology. 2011;411:325–343.
- Mallick B, Ghosh Z, Chakrabarti J. MicroRNome analysis unravels the molecular basis of SARS infection in bronchoalveolar stem cells. PloS One. 2009;4:e7837.
- Hasan MM, Akter R, Ullah M, et al. A computational approach for predicting role of human microRNAs in MERS-CoV genome. Adv Bioinformatics. 2014;2014.
- Shang J, Wan Y, Luo C, et al. Cell entry mechanisms of SARS-CoV-2. Proceedings of the National Academy of Sciences; Saint Paul, MN; 2020;117:11727–11734.
- V’kovski P, Kratzel A, Steiner S, et al. Coronavirus biology and replication: implications for SARS-CoV-2. Nature Rev Microbiol. 2020;1–16.
- Poduri R, Joshi G, Jagadeesh G. Drugs targeting various stages of the SARS-CoV-2 life cycle: exploring promising drugs for the treatment of Covid-19. Cell Signal. 2020;74:109721.
- Kim D, Lee J-Y, Yang J-S, et al. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181(4):914–921.
- Khan M-A-A-K, Sany MRU, Islam MS, et al. Epigenetic regulator miRNA pattern differences among SARS-CoV, SARS-CoV-2 and SARS-CoV-2 world-wide isolates delineated the mystery behind the epic pathogenicity and distinct clinical characteristics of pandemic COVID-19. bioRxiv. 2020;11:765.
- Demirci MDS, Adan A. Computational analysis of microRNA-mediated interactions in SARS-CoV-2 infection. PeerJ. 2020;8:e9369.
- Rad AH, McLellan AD. Implications of SARS-CoV-2 mutations for genomic RNA structure and host microRNA targeting. bioRxiv. 2020;21(13):4807.
- Liu Z, Wang J, Xu Y, et al. Implications of the virus-encoded miRNA and host miRNA in the pathogenicity of SARS-CoV-2. arXiv. 2020;arXiv preprint arXiv:200404874. 2020.
- Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280.
- Arisan ED, Dart A, Grant GH, et al. The prediction of miRNAs in SARS-CoV-2 genomes: hsa-miR databases Identify 7 key miRs linked to host responses and virus pathogenicity-related KEGG pathways significant for comorbidities. Viruses. 2020;12:614.
- Matarese A, Gambardella J, Sardu C, et al. miR-98 regulates TMPRSS2 expression in human endothelial cells: key implications for COVID-19. Biomedicines. 2020;8(11):462.
- Nersisyan S, Shkurnikov M, Turchinovich A, et al. Integrative analysis of miRNA and mRNA sequencing data reveals potential regulatory mechanisms of ACE2 and TMPRSS2. Plos One. 2020;15:e0235987.
- Sardar R, Satish D, Birla S, et al. Comparative analyses of SAR-CoV2 genomes from different geographical locations and other coronavirus family genomes reveals unique features potentially consequential to host-virus interaction and pathogenesis. bioRxiv. 2020.
- Haddad H, Al-Zyoud W. miRNA target prediction might explain the reduced transmission of SARS-CoV-2 in Jordan, Middle East. Noncoding RNA Res. 2020;5:135–143.
- Sardar R, Satish D, Birla S, et al. Integrative analyses of SARS-CoV-2 genomes from different geographical locations reveal unique features potentially consequential to host-virus interaction, pathogenesis and clues for novel therapies. Heliyon. 2020;6:e04658.
- Tang H, Lu X, Qie S, et al. Thoughts on detecting tissue distribution of potential COVID-19 receptors. Future Virol. 2020;15:489–496.
- Bahrami A, Bakherad M Comparative genomics identifies key genes and miRNAs that may be used as a strategy to control and treatment of COVID-19; 2020.
- Mukhopadhyay D, Mussa BM. Identification of novel hypothalamic microRNAs as promising therapeutics for SARS-CoV-2 by regulating ACE2 and TMPRSS2 expression: an in silico analysis. Brain Sci. 2020;10:666.
- Lu D, Chatterjee S, Xiao K, et al. MicroRNAs targeting the SARS-CoV-2 entry receptor ACE2 in cardiomyocytes. J Mol Cell Cardiol. 2020;148:46–49.
- Shao M, Li X, Liu F, et al. Acute kidney injury is associated with severe infection and fatality in patients with COVID-19: a systematic review and meta-analysis of 40 studies and 25,278 patients. Pharmacol Res. 2020;;161:105107.
- Widiasta A, Sribudiani Y, Nugrahapraja H, et al. Potential role of ACE2-related microRNAs in COVID-19-associated nephropathy. Noncoding RNA Res. 2020;5:153–166.
- Walsh D, Mathews MB, Mohr I. Tinkering with translation: protein synthesis in virus-infected cells. Cold Spring Harb Perspect Biol. 2013;5:a012351.
- Chen L, Zhong L Genomics functional analysis and drug screening of SARS-CoV-2. Genes & Diseases; 2020.
- Cullen BR. Viruses and microRNAs: rISCy interactions with serious consequences. Genes Dev. 2011;25:1881–1894.
- Rakhmetullina A, Ivashchenko A, Akimniyazova A, et al. The miRNA complexes against coronaviruses COVID-19, SARS-CoV, And MERS-CoV; 2020.
- Ivashchenko A, Rakhmetullina A, Aisina D How miRNAs can protect humans from coronaviruses COVID-19, SARS-CoV, and MERS-CoV; 2020.
- Zhang T, Cheng T, Wei L, et al. Efficient inhibition of HIV-1 replication by an artificial polycistronic miRNA construct. Virol J. 2012;9:118.
- Gao J, Xiao S, Xiao Y, et al. MYH9 is an essential factor for porcine reproductive and respiratory syndrome virus infection. Sci Rep. 2016;6:1–13.
- Orozco-García E, Trujillo-Correa A, Gallego-Gómez JC Cell biology of virus infection. The Role of Cytoskeletal Dynamics Integrity in the Effectiveness of Dengue Virus Infection. Cell Biology-New Insights: IntechOpen; 2016.
- Spear M, Wu Y. Viral exploitation of actin: force-generation and scaffolding functions in viral infection. Virol Sin. 2014;29:139–147.
- Cong Y, Ulasli M, Schepers H, et al. Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J Virol. 2020;94(4).
- Yoshimoto FK. The proteins of severe acute respiratory syndrome coronavirus-2 (SARS CoV-2 or n-COV19), the cause of COVID-19. Protein J. 2020;39(3):198-216.
- El-Nabi SH, Elhiti M, El-Sheekh M. A new approach for COVID-19 treatment by micro-RNA. Med Hypotheses. 2020;143:110203.
- Martines RB, Ritter JM, Matkovic E, et al. Pathology and pathogenesis of SARS-CoV-2 associated with fatal coronavirus disease, United States. Emerg Infect Dis. 2020;26:2005.
- Benvenuto D, Angeletti S, Giovanetti M, et al. Evolutionary analysis of SARS-CoV-2: how mutation of non-structural protein 6 (NSP6) could affect viral autophagy. J Infect. 2020;81(1):24-27.
- García LF. Immune response, inflammation, and the clinical spectrum of COVID-19. Front Immunol. 2020;11:1441.
- Prasanna PL, Abilash V. Coronaviruses pathogenesis, comorbidities and multi-organ damage–A review. Life Sci. 2020;255:117839.
- Demongeot J, Seligmann H. SARS-CoV-2 and miRNA-like inhibition power. Med Hypotheses. 2020;144:110245.
- Satyam R, Bhardwaj T, Goel S, et al. miRNAs in SARS-CoV 2: A spoke in the wheel of pathogenesis. Current pharmaceutical design; 2020.
- Raman C, Ren C, Boas S, et al. TCR signaling strength dependent regulation of T cell proliferation, survival and Th differentiation by TGF-βR3 (betaglycn). Am Assoc Immnol. 2017;7:6.
- Tikhanovich I, Cox J, Weinman SA. Forkhead box class O transcription factors in liver function and disease. J Gastroenterol Hepatol. 2013;28:125–131.
- Gamberi T, Magherini F, Modesti A, et al. Adiponectin signaling pathways in liver diseases. Biomedicines. 2018;6:52.
- Samuel CE. ADARs: viruses and innate immunity. Adenosine deaminases acting on RNA (ADARs) and A-to-I editing. Springer; 2011. p. 163–195.
- Hayden M, West A, Ghosh S. NF-κ B and the immune response. Oncogene. 2006;25:6758–6780.
- Gangemi S, Tonacci A AntagomiRs: A novel therapeutic strategy for challenging COVID-19 cytokine storm. Cytokine & growth factor reviews; 2020.
- Kumar S, Nyodu R, Maurya VK, et al. Host immune response and immunobiology of human SARS-CoV-2 infection. Coronavirus disease 2019 (COVID-19). Singapore: Springer; 2020. p. 43–53.
- Saini S, Saini A, Jyoti Thakur C, et al. Genome-wide computational prediction of miRNAs in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) revealed target genes involved in pulmonary vasculature and antiviral innate immunity. Mol Biol Res Commun. 2020;9:83–91.
- Srivastava R, Daulatabad SV, Srivastava M, et al. Role of SARS-CoV-2 in altering the RNA-binding protein and miRNA-directed post-transcriptional regulatory networks in humans. Int J Mol Sci. 2020;21:7090.
- Srivastava R, Daulatabad SV, Srivastava M, et al. SARS-CoV-2 contributes to altering the post-transcriptional regulatory networks across human tissues by sponging RNA binding proteins and micro-RNAs. BioRxiv. 2020.
- Bartoszewski R, Dabrowski M, Jakiela B, et al. SARS-CoV-2 may regulate cellular responses through depletion of specific host miRNAs. Am J Physiol Lung Cell Mol Physiol. 2020;319:L444–L55.
- Nainu F, Shiratsuchi A, Nakanishi Y. Induction of apoptosis and subsequent phagocytosis of virus-infected cells as an antiviral mechanism. Front Immunol. 2017;8:1220.
- Ivanisenko NV, Seyrek K, Kolchanov NA, et al. The role of death domain proteins in host response upon SARS-CoV-2 infection: modulation of programmed cell death and translational applications. Cell Death Discov. 2020;6:1–10.
- Ramaiah MJ. mTOR inhibition and p53 activation, microRNAs: the possible therapy against pandemic COVID-19. Gene Rep. 2020;20:100765.
- Makhdoumi P, Roohbakhsh A, Karimi G. MicroRNAs regulate mitochondrial apoptotic pathway in myocardial ischemia-reperfusion-injury. Biomed Pharmacother. 2016;84:1635–1644.
- Omidkhoda N, Hayes AW, Reiter RJ, et al. The role of MicroRNAs on endoplasmic reticulum stress in myocardial ischemia and cardiac hypertrophy. Pharmacol Res. 2019;150:104516.
- Hanna J, Hossain GS, Kocerha J. The potential for microRNA therapeutics and clinical research. Front Genet. 2019;10:478.
- Chakraborty C, Sharma AR, Sharma G. Therapeutic advances of miRNAs: A preclinical and clinical update. J Adv Res. 2020;28:127–138.
- Fulzele S, Sahay B, Yusufu I, et al. COVID-19 virulence in aged patients might be impacted by the host cellular microRNAs abundance/profile. Aging Dis. 2020;11:509.
- Guterres A, de Azeredo Lima CH, Miranda RL, et al. What is the potential function of microRNAs as biomarkers and therapeutic targets in COVID-19? Infection. Gene Evolut. 2020;85:104417.
- Mishra PK, Tandon R, Byrareddy SN. Diabetes and COVID-19 risk: an miRNA perspective. Am J Physiol Heart Circ Physiol. 2020;319:H604–H9.
- Li C, Hu X, Li L, et al. Differential microRNA expression in the peripheral blood from human patients with COVID‐19. J Clin Lab Anal. 2020;34(10):e23590.
- Baumann V, Winkler J. miRNA-based therapies: strategies and delivery platforms for oligonucleotide and non-oligonucleotide agents. Future Med Chem. 2014;6:1967–1984.
- Fu Y, Chen J, Huang Z. Recent progress in microRNA-based delivery systems for the treatment of human disease. ExRNA. 2019;1:1–14.
- Zhou L-K, Zhou Z, Jiang X-M, et al. Absorbed plant MIR2911 in honeysuckle decoction inhibits SARS-CoV-2 replication and accelerates the negative conversion of infected patients. Cell Discov. 2020;6:1–4.
- Zhou Z, Zhou Y, Jiang X-M, et al. Decreased HD-MIR2911 absorption in human subjects with the SIDT1 polymorphism fails to inhibit SARS-CoV-2 replication. Cell Discov. 2020;6:1–4.