ABSTRACT
This study designs to investigate the role and potential mechanism of lncNRA HOTTIP in OS progression in vitro and in vivo. HOTTIP, PTBP1, and KHSRP expression levels were tested through qRT-PCR and western blot in OS tissues or cell lines. Cell proliferation was examined via CCK-8 and colony formation. Cell cycle and apoptosis were analyzed via flow cytometry analysis. The invasive and migratory abilities of OS cells were evaluated by transwell and wound-healing assays. The localization of HOTTIP in OS cells was determined by subcellular fractionation assay. RNA pull down and RNA immunoprecipitation were allowed to assess the interaction between HOTTIP and PTBP1. Xenograft tumor growth assay was employed to test the role of HOTTIP and KHSRP in OS progression. Our data demonstrated HOTTIP was upregulated in OS tissues. HOTTIP knockdown resulted in a suppression of OS cell proliferation, invasion and migration, as well as a promotion of OS cell apoptosis, while HOTTIP overexpression exhibited opposite effects. In mechanism, PTBP1 and KHSRP highly expressed in OS and HOTTIP was identified to interact with PTBP1 to promote KHSRP expression. Meanwhile, we found that overexpression of KHSRP or PTBP1, individually, can partially remove the repression of HOTTIP suppression for OS cell progression. Moreover, xenograft tumor growth assay revealed that HOTTIP knockdown significantly inhibited tumor growth, and this inhibitory effect was abolished by KHSRP overexpression. Collectively, these findings confirmed that HOTTIP facilitates OS cell proliferation, invasion and migration by binding to PTBP1 to promote KHSRP level.
Abbreviation: LncRNA: long noncoding RNA; HOTTIP: HOXA distal transcript antisense RNA; KHSRP: KH-Type Splicing Regulatory Protein; qRT-PCR: quantitative real-time PCR; OS: osteosarcoma; OST: osteosarcoma tissues; ANT: adjacent normal tissue.
KEYWORDS:
Introduction
Osteosarcoma (OS), as a common malignant bone tumor, has become the primary cause of tumor-related death in children and adolescents [Citation1,Citation2]. Due to its invasive and metastatic properties, the morbidity and mortality of OS were both higher than other primary bone tumors [Citation3]. Clinical data showed the five-year survival rate of the OS patients with distant metastasis is even less than 30% [Citation4]. Although some treatments, such as chemotherapy, radiotherapy and surgery, have been adopted to increase the survival rate of the OS patients, the prognosis remains poor due to the multiple and complex mechanisms [Citation5]. Therefore, seeking for several new therapeutic targets of OS and finding out innovative therapies are highly expected. Nowadays, with the exploration of the research development at the molecular level, the related molecular mechanism of OS has attracted lots of attention.
With the rapid development of next-generation sequencing, more and more RNA molecules have been identified to be correlated with the pathogenesis of various human diseases. Recently, many studies indicated that long non-coding RNAs (lncRNAs), more than 200 nucleotides, may be related to the modulation of the development of OS [Citation6,Citation7]. Among which, lncRNA Homeobox A transcript at the distal tip (HOTTIP), originated from HOTTIP gene that located at 5ʹ end of the HOXA gene cluster, was originally identified in distal human fibroblasts such as those from the foreskin, foot or hand [Citation8,Citation9]. It has been reported to be upregulated in the OS tissues and its upregulation was closely related with the poor prognosis of the patients [Citation10]. Furthermore, HOTTIP could promote OS cell migration and invasion [Citation11]; however, the mechanisms in OS progression are still unclear.
KH-type splicing regulatory protein (KHSRP) is a multifunctional molecule that has a positive role in cell proliferation, invasion, and migration in cancers [Citation12,Citation13]. Also, KHSRP was identified to be increased in primary cell culture of OS, and its silencing significantly suppressed OS cell proliferation and migration, indicating that KHSRP is an oncogene of OS [Citation14]. On the other hand, Gou et al. found that lncRNA AB074169 bound KHSRP and decreased its expression in papillary thyroid carcinoma [Citation15]. In addition, KHSRP was found to be modulated by HOTTIP in OS cells. Nevertheless, how lncRNA HOTTIP regulates the level of KHSRP in OS needs to be further explored.
Polypyrimidine tract-binding protein 1 (PTBP1), known as an RNA binding protein, regulates nearly all processes of mRNA like alternative splicing, mRNA stability. Previous study has revealed that lncRNA could increase gene mRNA stability via interacting with PTBP1 [Citation16]. Moreover, PTBP1 was reported to accelerate tumor cell growth, migration, and invasion in breast cancer [Citation17], bladder cancer [Citation18], clear-cell renal cell carcinoma [Citation19], and so on. Prediction based on starBase database (http://starbase.sysu.edu.cn/), HOTTIP and KHSRP may potentially bind to PTBP1. However, little is known regarding the function of PTBP1 in OS.
To our understanding, lncRNAs can bind to PTBP1 to enhance mRNA stability and regulate various biological processes. Here, we hypothesize a regulation mechanism that HOTTIP facilitates cell proliferation, invasion, and migration through binding to PTBP1 to promote KHSRP level, aiming to better understand the function mechanism of HOTTIP in OS progression.
Materials and methods
Clinical specimen collection
Twenty human OS tissues (OST) and the matched normal ones (ANT, located >3 cm distal margin of the tumor) were removed from OS patients (the clinical characteristics in ) who were underwent surgery at Hunan Cancer Hospital (Changsha, China). All specimens were stored at −80°C for subsequent experiments, and the informed consent had been signed by all patients. The application of tissues was approved by the Medical Ethics committees of Hunan Cancer Hospital.
Table 1. The clinicopathological features of patients with osteosarcoma
Cell culture
Saos-2 (Catalog No: TCHu114), MG63 (Catalog No: TCHu124), U2OS (Catalog No: TCHu 88), HOS (Catalog No: TCHu1167) and hFOB 1.19 (Catalog No: GNHu14) cell lines were all obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China), and maintained in a humidified atmosphere (37°C, 5% CO2) using DMEM medium (Gibco, CA, USA) added with 10% (v/v) fetal bovine serum (Gibco, CA, USA) and 100 U/mL penicillin-streptomycin (Invitrogen, CA, USA). OS cells were cultured at 37°C in a humidified 5% CO2 atmosphere, while hFOB 1.19 cells were placed under the condition of 5% CO2 and 34°C.
Cell transfection
To silence the expression of HOTTIP and PTBP1, negative control (si-NC), si-HOTTIP and si-PTBP1 were purchased from Genepharma (Shanghai, China). To enhance the expression of HOTTIP, PTBP1, and KHSRP, HOTTIP, PTBP1, and KHSRP cDNA was cloned into the multiple cloning site of the pcDNA3.1 vector termed pcDNA3.1-HOTTIP/PTBP1/KHSRP, and negative control was pcDNA3.1-NC (RiboBio, Guangzhou, China). Cell transfection was achieved using Lipofectamine 3000 (Invitrogen, USA) following the manufacturer’s protocols at probably 80% cell confluence.
Total RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA of OS tissues and cells were prepared using TRIzol® reagent (Invitrogen) and reverse transcripted into cDNA. qRT-PCR processes were done with SYBR Green qPCR Master Mix using a 7500 Thermocycler (Applied Biosystems, USA). The relative expression levels of genes were normalized to GAPDH through a 2−ΔΔCt method. The primer sequences of genes are listed in .
Table 2. The primer sequences of genes used in qRT-PCR
CCK-8 assay
Cell viability was examined using a cell counting kit-8 (CCK-8; Dojindo, Japan). In brief, 100 µL cell suspension (1 × 105 cells/mL) was seeded in the 96-well plates before transfection. After the time point of 24, 48, 72 h, CCK-8 solution (10 µL) was added and incubated for additional 4 h at room temperature. The absorbance was determined by a microplate reader at 450 nm.
Colony formation assay
To evaluate the ability of colony formation, treated OS cells were seeded into 6-well plates and transfected with plasmids. After two weeks of culture, colonies were fixed with 10% methanol, and then stained with 1% crystal violet for 5 minutes. Subsequently, the colonies were imaged and counted by an optical microscope.
Cell cycle analysis
Treated OS cells were stained with propidium iodide (Sigma, USA) for 10 min in dark after fixation in 75% ethanol. After washed three times with PBS, cell cycle was tested via flow cytometry (FACSC alibur, BD, USA).
Cell apoptosis assay
For the determination of cell apoptosis, the collected OS cells were seeded into 6-well plates. Then, the cell samples were fixed with prechilled 70% alcohol (room temperature, 1 h). After washed with PBS, the cells were stained with 10 μL propidium iodide (PI) buffer and 5 μL Annexin V-fluorescein isothiocyanate (FITC) for 15 min (protected from light). Apoptotic rate was detected using a flow cytometry (Beckman Coulter, USA) and analyzed with FlowJo software (LLC, USA).
Cell invasion assay
Cell invasion assay was carried out in a Matrigel coated transwell chamber (Corning, NY, USA). The upper chamber was filled with 200 μL of FBS-free DMEM medium contained 1 × 105 cells. DMEM containing 10% FBS was then added into the lower chamber. After incubation, the non-invading cells were removed, and the invaded cells were fixed with 4% paraformaldehyde and stained with 1% crystal violet solution for 30 min. After washed, the invasive cells were observed by an optical microscope and counted in five randomly selected fields.
Cell migration assay
Cell migration ability was investigated by a wound‐healing assay. Briefly, the transfected cells were plated into the 6‐well plates at a concentration of 5 × 104 cells per well and grown to nearly 100% confluence. Then, a straight scratch was made in the cell monolayer through a sterile plastic micropipette tip. Subsequently, the cells were cultured at 37°C in the cell incubator. Images were taken at 0 and 24 h, respectively, using an optical microscope. The wound areas were analyzed by Image J software and calculated using the formula (S0 h – S24 h)/S0 h × 100%.
Western blot
Total proteins were harvested from cells lysed on ice for 1 h and the proteins concentrations were then measured. Proteins (30 μg) were isolated with SDS-PAGE electrophoresi and then transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, Beford, MA, USA). After blocked with 5% nonfat milk, the membrane was incubated with the primary antibodies (PTBP1, 1:2000; β-catenin, 1:1000; cyclin D1, 1:5000; c-myc, 1:1000; GAPDH, 1:10000; Abcam, USA) followed by secondary antibodies (1:2000, Abcam, USA). The bands were visualized by enhanced chemiluminescence kit.
Subcellular fractionation assay
The cytoplasmic and nuclear RNAs were isolated using NE-PER Reagent (Thermo Scientific), followed by qRT-PCR examination. The percentage of RNA levels were normalized to U1 and GAPDH, respectively.
RNA pulldown assay
First, the biotinylated lncRNA HOTTIP RNAs and antisense transcripts were transcribed in vitro. Then, biotin-labeled RNAs were incubated with RNase-free DNase I (Invitrogen), and Sephadex G-50 Quick Spin Columns (Sigma, USA) were used for purification. Subsequently, cell lysates and obtained RNAs were co-incubated at 4°C for two hours. After 1 h treatment with Dynabeads™ MyOne™ Streptavidin T1 (Invitrogen), the proteins bound to HOTTIP were detected by western blot assay.
RNA immunoprecipitation (RIP) assay
For RIP assay (using Magna RIP Kit from Millipore), cells were treated with lysis solution for 0.5 h at 4°C. Then, obtained cell extracts were incubated overnight with RIP buffer containing magnetic beads conjugated with anti-Ago2 (Millipore) or negative control immunoglobulin G (IgG; Millipore) at 4°C. After incubation and washing, the precipitated RNAs were determined by qRT-PCR analysis.
RNA stability measurement
Cells were seeded into 6-well plates and cultured for 24 h, followed by incubation with 5 μg/mL actinomycin D (Sigma) for 0, 2, 4 h. After treatment, total RNAs were isolated and performed KHSRP mRNA level assessment by qRT-PCR analysis.
Xenograft tumor growth
Animals (nude mice, eight-week old, male) were supplied by SLAC Laboratory Animal Co., Ltd (Shanghai, China). Animal manipulations were performed following the NIH guidelines, and the protocols were approved by the Animal Care and Use Committee of our hospital. U2OS cells (2 × 105 cells in 100 μL) stably carrying si-HOTTIP or pcDNA3.1-KHSRP were subcutaneously deliver into the back of nude mice. Tumor size was examined every six days from the injection day to day 30 after injection. Tumor volume was measured by following formula: V = 0.5 (long diameter) × (short diameter)2. Animals were sacrificed on day 30 and tumors were removed and weighed.
Statistics
All experiments were replicated at least three times and all data were presented as mean ± SD. The Student’s t-test (for two groups) or One-way ANOVA (for multiple groups) were performed in the data analysis using GraphPad Prism 6.0 software. p value less than 0.05 was considered statistically significant.
Results
HOTTIP is strongly expressed in the OS tissues and cells
We firstly determined the expression pattern of HOTTIP in the OS tissues and cells. As results indicated that the level of HOTTIP was obviously increased in the tissues compared with adjacent normal ones ()). Moreover, the HOTTIP expression level in the OS cells were also remarkably higher than that in the hFOB 1.19 cells ()). Among these cell lines, the expression levels of HOTTIP were much higher in U2OS and Saos-2 cells that were chosen for further experiments. These results indicated that HOTTIP is highly expressed in the OS tissues and cells, and may play an important role in OS progression.
Figure 1. HOTTIP is strongly expressed in the OS tissues and cells. (a) qRT-PCR analysis of the expression level of HOTTIP in the Osteosarcoma tissues (OST) and adjacent normal tissue (ANT). (b) The relative HOTTIP expression in the OS cells (Saos-2, MG63, U2OS, HOS) and the normal osteoblast hFOB1.19 cells. All data were represented by 3 independent experiments. *P < 0.05; **P < 0.01
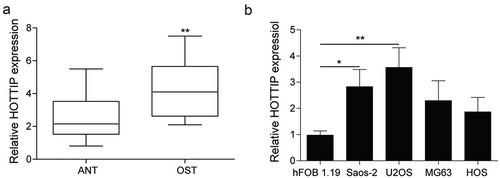
Downregulation of HOTTIP induces apoptosis and inhibits proliferation, invasion and migration in OS cells
For the investigation of the effect of HOTTIP on the OS development, we silenced the HOTTIP and observed the change of cell proliferation, invasion, and migration. As indicated in ), the HOTTIP level was obviously downregulated after the si-HOTTIP transfection. Downregulation of HOTTIP significantly decreased OS cell viability, clone number and promoted apoptosis, cell number of G0/G1 phase compared with cells transfected with si-NC (–e)). In addition, the biological function of HOTTIP on cell invasion and migration in OS cells were determined by transwell invasion assay and wound healing assay, respectively. HOTTIP silence in OS cells results in an apparent downregulation in the invaded cell number ()) and cell migration ability ()).
Figure 2. Downregulation of HOTTIP induces apoptosis and inhibites proliferation, invasion and migration in OS cells. (a) qRT-PCR analysis of the HOTTIP expression level in U2OS and Saos-2 cells after HOTTIP silencing. (b) CCK-8 assay was used to evaluate the effect of HOTTIP knockdown on U2OS and Saos-2 cells viability. (c) Cell cycle of U2OS and Saos-2 cells treated with si-NC or si-HOTTIP was analyzed using flow cytometry analysis. (d) Colony formation assay analyzed the function of si-HOTTIP on cell colony formation capacity. (e) Cell apoptosis in OS cells was measured with Annexin V-PI staining followed by flow cytometry. (f) Invasion of OS cells was determined by transwell assay. (g) Wound healing assay was performed to assess the migratory capacity of U2OS and Saos-2 cells. All data were represented by 3 independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001
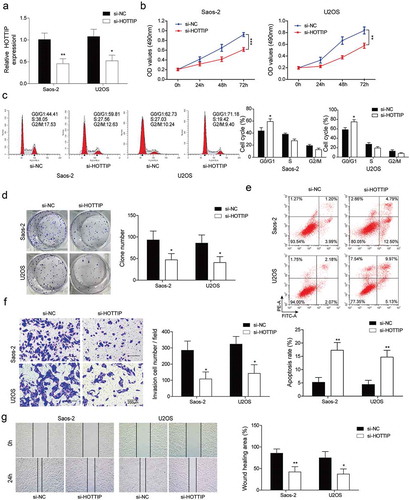
Overexpression of HOTTIP promotes proliferation, invasion and migration in OS cells
Next, we examined the influences of HOTTIP overexpression on OS progression in vitro. After transfection of pcDNA3.1-HOTTIP, the expression of HOTTIP was dramatically increased in Saos-2 and U2OS cells ()). Overexpression of HOTTIP in Saos-2 and U2OS cells obviously increased the cell viability ()), decreased the cell number of G0/G1 phase ()), and increased colony number ()). These findings suggested that HOTTIP overexpression facilitated OS cell proliferation in vitro. Moreover, we found that HOTTIP overexpression significantly enhanced the invasive and migratory abilities of Saos-2 and U2OS cells ()). These results provided evidence that HOTTIP positively regulates OS development and promotes cell proliferation, invasion, and migration.
Figure 3. Overexpression of HOTTIP promotes proliferation, invasion and migration in OS cells. (a) qRT-PCR analysis of the HOTTIP expression level in U2OS and Saos-2 cells after HOTTIP overexpression. (b) CCK-8 assay was used to evaluate the effect of HOTTIP overexpression on U2OS and Saos-2 cells viability. (c) Flow cytometry was adopted to detect cell cycle of U2OS and Saos-2 cells after HOTTIP overexpression. (d) Colony formation assay was employed to test the influences of HOTTIP overexpression on cell proliferation. (e, f) Transwell and wound-healing assays were performed to test the invasive and migratory abilities of OS cells after HOTTIP overexpression. All data were represented by 3 independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001
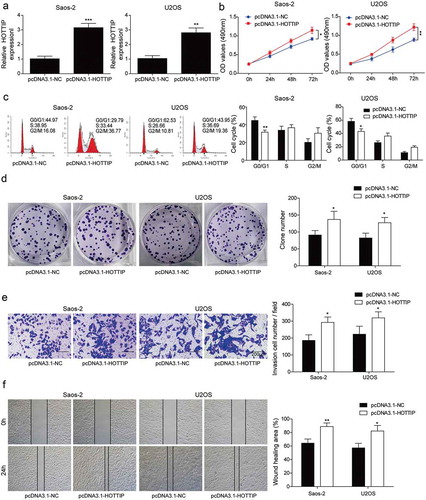
HOTTIP interacts with PTBP1 and increases its expression
In previous study, lncRNA could increasing its target gene via interacting with PTBP1 [Citation16]. And we predicted the binding between PTBP1 and HOTTIP through starBase and used the RNA pull down and RIP assays to validate the predicted correlation. First, we found that lncRNA HOTTIP was located in both cytoplasm and nucleus in ). Taken together, results of RNA pull down and RIP suggested that HOTTIP specifically interacted with PTBP1 ()). Furthermore, the expression levels of PTBP1 (mRNA and protein) were significantly down-regulated by si-HOTTIP while up-regulated by pcDNA3.1-HOTTIP in OS cells (–g)). These results demonstrated that HOTTIP might bind to PTBP1 to regulate the target gene expression.
Figure 4. HOTTIP interacts with PTBP1 and increases its expression. (a) qRT-PCR detection of the percentage of HOTTIP, GAPDH and U1 in the cytoplasm and nuclear fractions of U2SO and Saos-2 cells. (b) RNA pull down analysis on the binding of HOTTIP to PTBP1. (c) RIP assay was used to detected the interaction of HOTTIP with PTBP1 in U2SO and Saos-2 cells. (d, e) The mRNA and protein levels of PTBP1 were detected by qRT-PCR and western blot in OS cells treated with pcDNA3.1-HOTTIP. All data were represented by 3 independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001
PTBP1 restoration attenuates the effects of HOTTIP knockdown on proliferation, migration, and invasion in OS cells
We then investigated whether PTBP1 involves in the functional effects of HOTTIP on OS cell. By transfecting Saos-2 and U2OS cells with si-HOTTIP and pcDNA3.1-PTBP1, we found that the repressive effects of si-HOTTIP on cell viability, proliferation, invasion, and migration were abolished by the co-transfection of si-HOTTIP and pcDNA3.1-PTBP1 ()). These results indicated that PTBP1 restoration attenuates the effects of HOTTIP knockdown on proliferation, migration, and invasion in OS cells.
Figure 5. PTBP1 restoration attenuates the effects of HOTTIP knockdown on proliferation, survival, migration, and invasion in OS cells. Saos-2 and U2OS cells co-transfected with siHOTTIP and pcDNA3.1-PTBP1 were subjected for (a) CCK-8 assay, (b) flow cytometry analysis, (c) colony formation assay, (d) transwell analysis and (e) wound-healing assay. All data were represented by 3 independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001
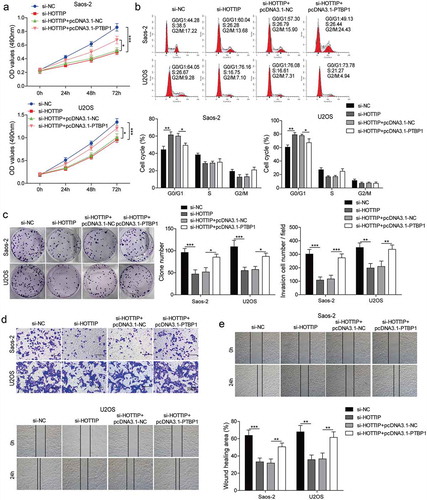
HOTTIP modulates the expression of KHSRP through binding to PTBP1
In order to further investigate the mechanism of HOTTIP in regulating OS development, we also predicted the binding between PTBP1 and KHSRP by starBase. As can be seen in ), the expression levels of PTBP1 and KHSRP were higher in OS tissues. We then found a positive correlation between the expression level of PTBP1 and KHSRP or HOTTIP ()). Moreover, both mRNA and protein levels of KHSRP were markedly decreased by suppression of PTBP1 and increased by overexpression of PTBP1 in Saos-2 and U2OS cells (–g)). Then, we assessed the effect of PTBP1 silencing and PTBP1 overexpression on the stability of KHSRP mRNA by qRT-PCR. Knockdown of PTBP1 in OS cells could decrease the mRNA stability of KHSRP while PTBP1 up-regulation increased the mRNA stability of KHSRP when treated with actinomycin D ()). And we demonstrated that PTBP1 overexpression partly reversed the downregulation of KHSRP induced by HOTTIP suppression in OS cell lines ()). Collectively, HOTTIP promotes the expression of KHSRP via binding to PTBP1.
Figure 6. HOTTIP modulates the expression of KHSRP through binding to PTBP1. (a, b) qRT-PCR assay analyzed the expression levels of PTBP1 and KHSRP. (c, d) Correlation analysis between the expression level of PTBP1 and KHSRP or HOTTIP in clinical samples. (e, f) Expression of PTBP1 and KHSRP mRNAs of Saos-2 and USOS cells were examined by qRT-PCR after transfection of pcDNA3.1 or pcDNA3.1-PTBP1. (g) Expression of PTBP1 and KHSRP proteins of Saos-2 and USOS cells were tested by western blot after transfection of pcDNA3.1 or pcDNA3.1-PTBP1. (h, i) The levels of KHSRP were detected via qRT-PCR in si-PTBP1/pcDNA3.1-PTBP1 transfected OS cells treated with actinomycin D at 0, 2, 4 h. 18s rRNA was used as negative control. (g, h) The expression levels of KHSRP were detected via qRT-PCR and western blot in cells transfected with si-HOTTIP and pcDNA3.1-PTBP1. All data were represented by 3 independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001
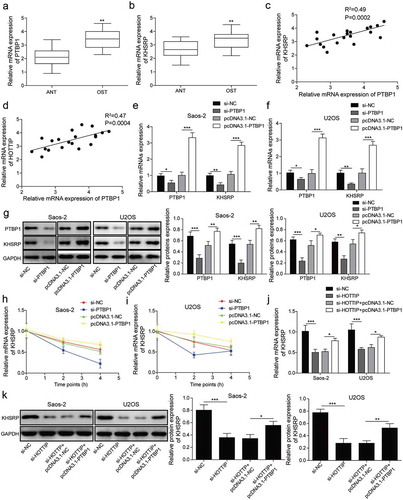
KHSRP involves in HOTTIP facilitated OS progression in vitro
Lastly, the rescue assays were performed to further validate whether HOTTIP promoted OS cell proliferation, invasion, and migration by upregulating KHSRP. From the results of CCK-8, colony formation and flow cytometry assays, the HOTTIP downregulation significantly inhibited the cell viability, arrested cell cycle, attenuated colony-formation ability and promoted apoptosis, however, KHSRP overexpression could relieve these effects of HOTTIP suppression ()). Meanwhile, upregulation of KHSRP abolished the repression of si-HOTTIP in cell invasion and migration ()). Additionally, we found that HOTTIP knockdown significantly decreased the protein expression of β-catenin, cyclin D1 and c-myc, while KHSRP overexpression abolished this trend ()). These results further confirmed the hypothesis that HOTTIP facilitates OS progression via Wnt/β-catenin signaling pathway by interacting with PTBP1 to increase KHSRP level.
Figure 7. KHSRP involves in HOTTIP facilitated OS progression in vitro. (a) Cell viability of OS cells transfected with si-HOTTIP and pcDNA3.1-KHSRP was detected using CCK-8. (b) Cell cycle of OS cells treated with si-HOTTIP and pcDNA3.1-KHSRP was analyzed by flow cytometry analysis. (c) Colony formation assay was used to estimate colony-formation ability of cells transfected as described in (A). (d) Flow cytometry was allowed to test the effects of HOTTIP/KHSRP axis on cell apoptosis. (e-f) Cell invasion and migration were measured by transwell and wound healing assays. (g) Protein expression of β-catenin, cyclin D1 and c-myc were examined by western blot in OS cells treated with si-HOTTIP and pcDNA3.1-KHSRP. All data were represented by 3 independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001
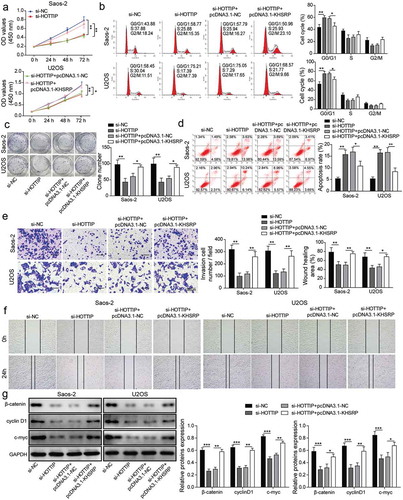
KHSRP involves in HOTTIP facilitated OS progression in vivo
We then investigate whether KHSRP involves in HOTTIP facilitated OS progression in vivo. By injecting U2OS cells carrying si-HOTTIP or pcDNA3.1-KHSRP into nude mice, we found that tumors originated from si-HOPPIP treated U2OS cells were obviously smaller compared to the one from si-NC group, while tumors originated from si-HOPPIP and pcDNA3.1-KHSRP treated U2OS cells were restored to si-NC level (–c)). Moreover, we found the expression of KHSRP in the tumors of si-HOPPIP treated group was significantly downregulated, while it was restored to normal level in the tumors of si-HOPPIP and pcDNA3.1-KHSRP treated group ()). Therese results indicated that KHSRP overexpression reversed the inhibitory effects of HOTTIP knockdown on OS tumor growth.
Figure 8. KHSRP overexpression reverses the inhibitory effects of si-HOTTIP on OS tumor growth in vivo. (a) Representative of OS tumors derived from U2OS cells treated with si-HOTTIP plus pcDNA3.1-KHSRP. (b, c) Tumor weight and volume were examined. (d) Protein expression of KHSRP was examined in tumors derived from U2OS cells treated with si-HOTTIP plus pcDNA3.1-KHSRP. All data were represented by 3 independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001
Discussion
Currently, researchers have highlighted the important roles of lncRNAs in the initiation and progression of human tumors by acting as tumor oncogenes or suppressors through multiple mechanisms [Citation20,Citation21]. As reported, increasing lncRNAs have been validated to participate in the progression of OS [Citation6,Citation22]. Fei et al. showed that lncRNA ROR promoted OS progression by targeting miR-206 [Citation23]. Additionally, lncRNA ANRIL was associated with a poor prognosis of OS and promotes tumorigenesis via PI3K/Akt pathway [Citation24]. These results implied that lncRNAs may be potential diagnostic and therapeutic targets of OS. Among these lncRNAs, HOTTIP was found to be increased and correlated with poor prognosis in OS patients by Li et al. in 2015 [Citation10]. Furthermore, HOTTIP overexpression could enhance the chemo-resistance of OS cells to cisplatin [Citation25]. HOTTIP may serve as one of the potential biomarkers and therapeutic targets of OS. In the present study, we demonstrated HOTTIP was upregulated in OS, and silence of the HOTTIP decreased the cell proliferation, migration, invasion, and increased the cell apoptosis, while HOTTIP overexpression exhibited opposite effects. Taken together, HOTTIP positively regulated OS progression, which provided a potential target for treating OS. However, the regulatory mechanisms of HOTTIP in OS need further to be explored.
With the rapid development of the biomedical field, researchers gradually discovered that the organism is a complex system, there may be the mutual regulation relationship between the various genes, thus forming a complex regulatory network. And they found some mutual regulations between mRNA and lncRNA [Citation26,Citation27]. The lncRNA can be a competitive endogenous RNA (ceRNA) to combine with the microRNA and further involved in the regulation of mRNA expression [Citation28]. HOTTIP was revealed to play a critical role in the tumorigenesis of various human tumors by acting as microRNA sponges to regulate corresponding oncogene or tumor suppressor gene [Citation29,Citation30]. LncRNA was also reported to bind to enhancer of zeste homolog 2 (EZH2), thereby repressing LATS2 expression in renal cell carcinoma [Citation27]. In addition, lncRNA could increasing its target gene level via interacting with RNA-binding proteins and further participate in disease process [Citation16,Citation31]. PTBP1 is one of the most investigated multifunctional RNA-binding proteins, controlling almost all steps of mRNA metabolism and processing. However, there are no reports on PTBP1 in OS. It is reported that lncRNA H19 could bind to PTBP1 to promote mRNA and protein expression levels of target gene in the development of fatty liver [Citation32]. In the present work, for the investigation of the regulatory mechanism of HOTTIP in OS, we investigated the mutual relationship between HOTTIP and PTBP1, PTBP1, and KHSRP. LncRNA HOTTIP regulates KHSRP expression by interacting with PTBP1. KHSRP is a tumor-promoting effector in various cancers. KHSRP could control colorectal cancer cell growth and modulates the tumor microenvironment [Citation13]. In this study, KHSRP was highly expressed in OS tissues, and a positive correlation between the expression level of PTBP1 and KHSRP or HOTTIP in clinical samples. Besides, PTBP1 overexpression not only increased the mRNA level of KHSRP but also abolish the downregulation of KHSRP induced by si-HOTTIP. Further, PTBP1 or KHSRP could partially reverse the function of HOTTIP inhibition in cell proliferation, apoptosis, migration, and invasion. As reported, Wnt/β-catenin signaling pathway was involved in the tumor cell proliferation, apoptosis, and metastasis [Citation25]. We also found HOTTIP knockdown significantly decreased the activation of Wnt/β-catenin pathway, while KHSRP overexpression abolished this effect. These results confirmed that HOTTIP facilitated OS progression via Wnt/β-catenin pathway by regulating PTBP1/KHSRP axis in vitro. Additionally, we further confirmed the inhibitory effects on OS tumor growth using in vivo xenograft tumor growth assay. Nevertheless, there are some limitations, like roles in multidrug resistance, etc., are also need to explore in the future.
In general, our study revealed the regulation mechanism of HOTTIP in the OS and figured out HOTTIP regulates KHSRP expression by interacting with PTBP1. This research contributed to a better understanding of the development mechanism of OS at the molecular level and provided a new therapeutic target for OS.
Ethics approval and consent to participate
The application of tissues was approved by the Medical Ethics committees of Hunan Cancer Hospital.
Acknowledgments
Thanks to the members of our laboratory for their contributions.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Ritter J, Bielack SS. Osteosarcoma. Ann Oncol. 2010;21(Suppl 7):vii320–5.
- Kager L, Tamamyan G, Bielack S. Novel insights and therapeutic interventions for pediatric osteosarcoma. Future Oncol. 2017;13:357–368.
- Wang WG, Wan C, Liao GJ. The efficacy of high-dose versus moderate-dose chemotherapy in treating osteosarcoma: a systematic review and meta-analysis. Int J Clin Exp Med. 2015;8:15967–15974.
- Zhang C, Hu J, Zhu K, et al. Survival, complications and functional outcomes of cemented megaprostheses for high-grade osteosarcoma around the knee. Int Orthop. 2018;42:927–938.
- Aljubran AH, Griffin A, Pintilie M, et al. Osteosarcoma in adolescents and adults: survival analysis with and without lung metastases. Ann Oncol. 2009;20:1136–1141.
- Li Z, Dou P, Liu T, et al. Application of long noncoding RNAs in osteosarcoma: biomarkers and therapeutic targets. Cell Physiol Biochem. 2017;42:1407–1419.
- Liu B, Zhao H, Zhang L, et al. Silencing of long-non-coding RNA ANCR suppresses the migration and invasion of osteosarcoma cells by activating the p38MAPK signalling pathway. BMC Cancer. 2019;19:1112.
- Fan Y, Yan T, Chai Y, et al. Long noncoding RNA HOTTIP as an independent prognostic marker in cancer. Clin Chim Acta. 2018;482:224–230.
- Wang F, Tang Z, Shao H, et al. Long noncoding RNA HOTTIP cooperates with CCCTC-binding factor to coordinate HOXA gene expression. Biochem Biophys Res Commun. 2018;500:852–859.
- Li F, Cao L, Hang D, et al. Long non-coding RNA HOTTIP is up-regulated and associated with poor prognosis in patients with osteosarcoma. Int J Clin Exp Pathol. 2015;8:11414–11420.
- Tang Y, Ji F. lncRNA HOTTIP facilitates osteosarcoma cell migration, invasion and epithelial-mesenchymal transition by forming a positive feedback loop with c-Myc. Oncol Lett. 2019;18:1649–1656.
- Tong L, Luo Y, Wei T, et al. KH-type splicing regulatory protein (KHSRP) contributes to tumorigenesis by promoting miR-26a maturation in small cell lung cancer. Mol Cell Biochem. 2016;422:61–74.
- Caiazza F, Oficjalska K, Tosetto M, et al. KH-type splicing regulatory protein controls colorectal cancer cell growth and modulates the tumor microenvironment. Am J Pathol. 2019;189:1916–1932.
- Pruksakorn D, Teeyakasem P, Klangjorhor J, et al. Overexpression of KH-type splicing regulatory protein regulates proliferation, migration, and implantation ability of osteosarcoma. Int J Oncol. 2016;49:903–912.
- Guo Q, Gao L, Nie X, et al. Long noncoding RNA AB074169 inhibits cell proliferation via modulation of KHSRP-mediated CDKN1a expression in papillary thyroid carcinoma. Cancer Res. 2018;78:4163–4174.
- Li J, Yang Y, Fan J, et al. Long noncoding RNA ANCR inhibits the differentiation of mesenchymal stem cells toward definitive endoderm by facilitating the association of PTBP1 with ID2. Cell Death Dis. 2019;10:492.
- Wang X, Li Y, Fan Y, et al. PTBP1 promotes the growth of breast cancer cells through the PTEN/Akt pathway and autophagy. J Cell Physiol. 2018;233:8930–8939.
- Jiang D, Zhang Y, Yang L, et al. Long noncoding RNA HCG22 suppresses proliferation and metastasis of bladder cancer cells by regulation of PTBP1. J Cell Physiol. 2020;235:1711–1722.
- Jiang J, Chen X, Liu H, et al. Polypyrimidine tract-binding protein 1 promotes proliferation, migration and invasion in clear-cell renal cell carcinoma by regulating alternative splicing of PKM. Am J Cancer Res. 2017;7:245–259.
- Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–3981.
- Renganathan A, Felley-Bosco E. Long noncoding RNAs in cancer and therapeutic potential. Adv Exp Med Biol. 2017;1008:199–222.
- He R, Wu JX, Zhang Y, et al. LncRNA LINC00628 overexpression inhibits the growth and invasion through regulating PI3K/Akt signaling pathway in osteosarcoma. Eur Rev Med Pharmacol Sci. 2018;22:5857–5866.
- Fei D, Sui G, Lu Y, et al. The long non-coding RNA-ROR promotes osteosarcoma progression by targeting miR-206. J Cell Mol Med. 2019;23:1865–1872.
- Yu G, Liu G, Yuan D, et al. Long non-coding RNA ANRIL is associated with a poor prognosis of osteosarcoma and promotes tumorigenesis via PI3K/Akt pathway. J Bone Oncol. 2018;11:51–55.
- Li Z, Zhao L, Wang Q. Overexpression of long non-coding RNA HOTTIP increases chemoresistance of osteosarcoma cell by activating the Wnt/beta-catenin pathway. Am J Transl Res. 2016;8:2385–2393.
- Wu Q, Guo L, Jiang F, et al. Analysis of the miRNA-mRNA-lncRNA networks in ER+ and ER- breast cancer cell lines. J Cell Mol Med. 2015;19:2874–2887.
- Peng F, Shi X, Meng Y, et al. long non-coding RNA HOTTIP is upregulated in renal cell carcinoma and regulates cell growth and apoptosis by epigenetically silencing of LAST2. Biomed Pharmacother. 2018;105:1133–1140.
- Qi X, Zhang DH, Wu N, et al. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015;52:710–718.
- Tsang FH, Au SL, Wei L, et al. Long non-coding RNA HOTTIP is frequently up-regulated in hepatocellular carcinoma and is targeted by tumour suppressive miR-125b. Liver Int. 2015;35:1597–1606.
- Yuan Q, Liu Y, Fan Y, et al. LncRNA HOTTIP promotes papillary thyroid carcinoma cell proliferation, invasion and migration by regulating miR-637. Int J Biochem Cell Biol. 2018;98:1–9.
- Chen L, Chen Q, Kuang S, et al. USF1-induced upregulation of LINC01048 promotes cell proliferation and apoptosis in cutaneous squamous cell carcinoma by binding to TAF15 to transcriptionally activate YAP1. Cell Death Dis. 2019;10:296.
- Liu C, Yang Z, Wu J, et al. Long noncoding RNA H19 interacts with polypyrimidine tract-binding protein 1 to reprogram hepatic lipid homeostasis. Hepatology. 2018;67:1758–1783.
