ABSTRACT
Accumulating evidence has reported the role of microRNA (miR) in retinoblastoma (RB). Therefore, the objective was to discuss how miR-362-3p exerted its function in RB cell progression via regulating ubiquitin-specific protease 2 (USP22) and lysine-specific histone demethylase 1 (LSD1). MiR-362-3p, USP22 and LSD1 expression in RB cells and tissues were tested. The biological functions of RB cells were detected via over-expressing miR-362-3p and down-regulating USP22. The target relationship of USP22 and miR-362-3p as well as the interaction of USP22 and LSD1 in RB was verified. Down-regulated miR-362-3p and up-regulated USP22 and LSD1 were demonstrated in RB tissues and cells. Restoring miR-362-3p and depleting USP22 attenuated invasion, proliferation and migration, and facilitated apoptosis of RB cells. USP22 was a target gene of miR-362-3p. USP22 deubiquitinated LSD1 in RB. It is revealed that miR-362-3p targets USP22 and then restrains invasion, proliferation and migration while promotes apoptosis of RB via reducing LSD1 modified by deubiquitination.
Introduction
As the major eye malignancy in infancy and childhood, retinoblastoma (RB) is resulted from RB1 gene mutations [Citation1]. The incidence of RB accounts for about 3% of all malignant tumors in children, about 1 in 20,000 live births globally [Citation2]. Clinically, RB is manifested with conjunctival edema and congestion, corneal edema, increased intraocular pressure, vitreous strabismus and opacity [Citation3]. The treatment of RB has recently been developed to include precision diagnostic as well as therapeutic tools containing optical coherence tomography, molecular diagnosis, intra-arterial chemotherapy and intravitreal chemotherapy, leading to raised eye salvage and vision potential [Citation4]. Regardless of the advancements in RB treatment in the last few years, the survival rate of patients with RB is much less improved, which is basically limited by early diagnosis [Citation5]. Therein, the urgency to clarify an accurate therapeutic target to treat RB registers a priority.
MicroRNAs (miRNAs) are a kind of small non-coding RNAs comprised of 22–25 nucleotides, and are frequently involved in regulating pathophysiologic mechanisms via repressing target gene expression [Citation6]. A previous review has concluded the dysregulated miRNAs in RB, such as miR-145, miR-181b and miR-498, and further manifested the functional role of miRNAs in biological functions of RB cells [Citation7]. Demonstrated by a prior study, miR-362-3p is examined to down-regulate in cervical cancer and miR-362-3p can disturb malignant cell migration and invasion [Citation8]. With respect to hepatocellular carcinoma, overexpressing miR-362-3p can retard proliferation, invasion and migration of cancer cells [Citation9]. Studies have mentioned the potentials of miR-362-3p overexpression in controlling cancers, which may enlighten its possible role in RB. Ubiquitin specific protease 22 (USP22) is a subunit of the human Spt-Ada-Gcn5-Acetyltransferase complex and an ubiquitin hydrolase that catalyzes the removal of monoubiquitination from histone H2B [Citation10]. A study has demonstrated that knockdown of USP22 promotes human RB cell aging and apoptosis [Citation11]. Also, another study has presented that depleting USP22 reduces the levels of phosphorylated RB protein [Citation12]. Abnormally expressed lysine-specific histone demethylase 1 (LSD1) is generally connected with inferior clinical outcomes in solid malignancies [Citation13]. According to Charles et al., it is revealed the cell cycle correlation of the RB protein Rb and the histone demethylase LSD1 with the Epstein-Barr virus latency promoter Cp [Citation14]. Therefore, it is hypothesized that miR-362-3p may be adopted as a novel biomarker in RB. In addition, we aim to clarify how miR-362-3p disturbs RB cell progression via regulating USP22 and LSD1.
Materials and methods
Ethics statement
The study was endorsed by the Institutional Review Board of The First Affiliated Hospital of Zhengzhou University. Ethical agreements were gained from the donors by written informed consent. All animal experiments were in compliance with the Guide for the Care and Use of Laboratory Animal by International Committees. The protocol was ratified by the Institutional Animal Care Use Committee of The First Affiliated Hospital of Zhengzhou University.
Study subjects
From January 2016 to January 2019, RB tissues (n = 46) and normal retina tissues (n = 11) were gathered from the ophthalmology department of The First Affiliated Hospital of Zhengzhou University. Patients who received radiotherapy or chemotherapy prior to surgical resection were excluded. The normal retina tissues were gathered from 11 patients who experienced ruptured globes in The First Affiliated Hospital of Zhengzhou University.
Cell culture
Human retinal pigment epithelium cell line ARPE-19 and human RB cell lines WERI-Rb1, Y79 were available from the American Type Culture Collection (VA, USA). The RB cell lines were fostered in RPMI-1640 or Dulbecco’s modified Eagle medium (DMEM) while ARPE-19 cells in DMEM/F-12 (Hyclone, Logan, UT, USA) (all containing 10% fetal bovine serum (FBS), 100 U/mL penicillin and 100 mg/mL streptomycin, 5% CO2). All cell culture medium were bought from Gibco (Thermo Fisher, California, USA) [Citation15].
Cell transfection
Transfection with mimic negative control (NC), miR-362-3p mimic, inhibitor NC, miR-362-3p inhibitor, short hairpin RNA (sh)-NC, sh-USP22, overexpression (oe)-NC or oe-USP22 (all from GenePharma Co., Ltd., Suzhou, Jiangsu, China) was performed on WERI-Rb1 and Y79 cells. The transfection was performed in strictly accordance with LipofectamineTM 2000 reagent (Invitrogen, California, USA). After transfection, cells were incubated with 5% CO2 at 37°C for 48 h for the follow-up experiments.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
The total RNA was extracted from RB tissues and cell lines by Trizol (Ambion, Life Technologies, Austin, TX, USA). The reverse transcription of RNA was implemented by PrimeScript RT kit (Promega, Madison, WI, USA). USP22, LSD1 and miR-362-3p levels were determined by SYBR Premix ExTaq and Mir-X miRNA First Strand Synthesis Kit (Takara, Dalian, China). Glyceraldehyde phosphate dehydrogenase (GAPDH) and U6 were utilized as the standard controls. The relative expression was tested by 2−ΔΔCt method. The primer sequences were showed in .
Table 1. Primer sequence
Western blot assay
Lysed in radio-immunoprecipitation assay lysis buffer, the proteins from cells and tissues were transferred to polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA) after isolated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The membranes were incubated with primary antibodies USP22 (ab195289) and LSD1 (ab17721) (Abcam, Cambridge, MA, USA), and GAPDH (sc-53,646) (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Then, the membrane was probed with the secondary antibody. The protein band was developed by chemiluminescence reagent, with GAPDH as the internal control [Citation16].
Cell counting kit (CCK)-8 assay
RB cell proliferation was verified by CCK-8 reagent (Dojindo Laboratories, Kumamoto, Japan). The cells were added into the 96-well plate (1 × 104 cells/well). Each well was added with CCK-8 reagent (10 μL) at 24 h, 48 h and 72 h, separately, and hatched for 4 h. Optical density values (450 nm) were measured by a microplate reader (Bio-Rad Laboratories, Hercules, CA, USA) [Citation17].
Transwell assay
Cell invasion assay was tested by transwell chamber whose well size was 8 μM (BD Biosciences, Bedford, MA, USA). Added into the apical chamber pre-coated with 50 μL Matrigel (BD Biosciences), cells were fostered with serum-free medium. The basolateral chamber was added with RPMI-1640 medium supplemented with 20% FBS. At 24 h post incubation, the invasive cells were fastened and then dyed with 1% crystal violet staining solution. A microscope was used to reckon the migrated and invaded cells (Olympus, Tokyo, Japan) [Citation18].
Scratch test
Seeded in the 6-well plate with 5 × 105 cells in each well, cells were grown to cover the bottom on the next day. Scratches were marked by a 200-μL pipette vertically along the edge of the ruler with three parallel marks for each well. Cells were added into serum-free medium to eliminate the effects of cell proliferation and pictured under the microscope after 24 h.
Flow cytometry
Cell apoptosis was detected by a FACScan flow cytometer with reference to the fluorescein isothiocyanate (FITC) Annexin V Apoptosis Detection kit (BD Biosciences). The cell apoptosis rate was analyzed by Cell Quest software (BD Biosciences) [Citation17].
Dual luciferase reporter gene assay
The wild type (WT) and mutant type (MUT) reporter plasmids fragment of USP22 (WT-USP22, MUT-USP22) compounded by GenePharma were bound to miR-362-3p and inserted into psiCHECK-2 vectors (Invitrogen). Co-transfection with WT or MUT reporter and miRNA mimic or miRNA NC was implemented on WERI-Rb1 and Y79 cells for luciferase activity assay. At 48 h post transfection, the luciferase activity was tested by a Dual Luciferase Reporter Assay kit (Promega) [Citation18].
Immunoprecipitation
WERI-Rb1 cells (about 1 × 107) in nuclear lysis buffer were equally distributed and incubated with anti-USP22 antibody (ab4812) or anti-immunoglobulin G (IgG) antibody (ab109489; both from Abcam). At 6 h post incubation, non-labeled protein A + G beads (Santa Cruz Biotechnology) were supplemented to the samples, which was followed by centrifugation at 8000 g. The beads were cleaned with 1 mL ice-cold lysis buffer, the sample buffer was boiled for 2 min and the immunoprecipitated proteins were released from the beads [Citation16].
Statistical analysis
All data were analyzed by SPSS 22.0 software (IBM Corp. Armonk, NY, USA). The measurement data were represented as mean ± standard deviation. The differences between groups were conducted by t-test, while those among multiple groups were by one-way analysis of variance (ANOVA), and Tukey’s post hoc test was used after ANOVA. Statistical significance was set at P < 0.05.
Results
Down-regulated miR-362-3p and miR-362-5p and up-regulated USP22 and LSD1 are found in RB tissues and cells
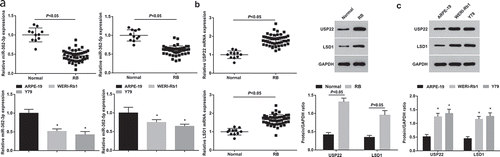
MiR-362-3p, miR-362-5p, USP22 and LSD1 expression in human RB tissues and normal retina tissues, WERI-Rb1, Y79 and ARPE-19 were tested. It was demonstrated that miR-362-3p and miR-362-5p levels were depressed while USP22 and LSD1 levels were heightened in the RB tissues and cells (). The differential expression of miR-362-3p was more significant, so we selected miR-362-3p for subsequent experiments.
Figure 1. MiR-362-3p and miR-362-5p expression is decreased while USP22 and LSD1 expression is increased in RB tissues and cells. (a), Expression of miR-362-3p and miR-362-5p in RB tissues and cell lines tested by RT-qPCR. (b), Expression of USP22 and LSD1 in RB tissues tested by RT-qPCR. (c), Expression of USP22 and LSD1 in RB cell lines WERI-Rb1 and Y79 tested by western blot assay. * P < 0.05 vs. ARPE-19 cells. + P < 0.05 vs. normal group. Normal (n = 11), RB (n = 46), WERI-Rb1, Y79, ARPE-19 (N = 3). Measurement data were depicted as mean ± standard deviation, comparison between two groups was conducted by t test, comparisons among multiple groups were assessed by one-way ANOVA followed with Tukey’s post hoc test
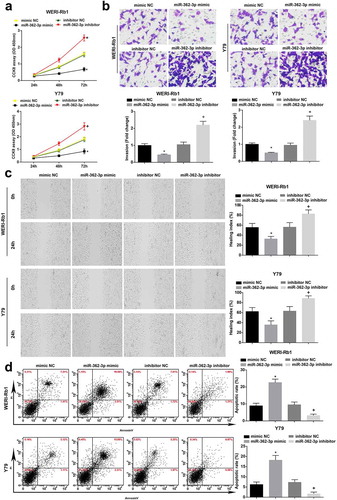
Up-regulating miR-362-3p restrains RB cell progression
CCK-8 assay, Transwell assay, scratch test and flow cytometry were adopted to test the biological functions of RB cells. miR-362-3p overexpression depressed RB cell progression while miR-362-3p inhibition functioned oppositely ().
Figure 2. Up-regulating miR-362-3p restrains cell progression in RB. (a), Comparison of proliferative capacity of WERI-Rb1 and Y79 cells by CCK-8 assay. (b), Comparison of invasive ability of WRI-Rb1 and Y79 cells by Transwell assay. (c), Comparison of migration ability of WERI-Rb1 and Y79 cells by scratch test. (d), Analysis of apoptosis rate of WRI-Rb1 and Y79 cells by flow cytometry. * P < 0.05 vs. mimic NC group. + P < 0.05 vs. inhibitor NC group. N = 3. Measurement data were depicted as mean ± standard deviation, comparisons among multiple groups were assessed by one-way ANOVA followed with Tukey’s post hoc test
USP22 is targeted by miR-362-3p
The target relationship between miR-362-3p and USP22 was forecasted by bioinformatics website (http://www.targetscan.org/vert_72/). A special binding site existed between USP22 and miR-362-3p. In addition, dual luciferase reporter gene assay suggested that miR-362-3p mimic decreased the luciferase activity of USP22 3’UTR Wt reporter vector ()).
Figure 3. USP22 is targeted by miR-362-3p. (a), The binding site of miR-362-3p on USP22 3’UTR and the luciferase activity of WERI-Rb1 and Y79 cells. (b), miR-362-3p and USP22 expression in WERI-Rb1 cells by RT-qPCR and Western blot assay. (c), miR-362-3p and USP22 expression in Y79 cells by RT-qPCR and Western blot assay. * P < 0.05 vs. mimic NC group. + P < 0.05 vs. inhibitor NC group. N = 3. Measurement data were depicted as mean ± standard deviation, comparison between two groups was conducted by t test, comparisons among multiple groups were assessed by one-way ANOVA followed with Tukey’s post hoc test
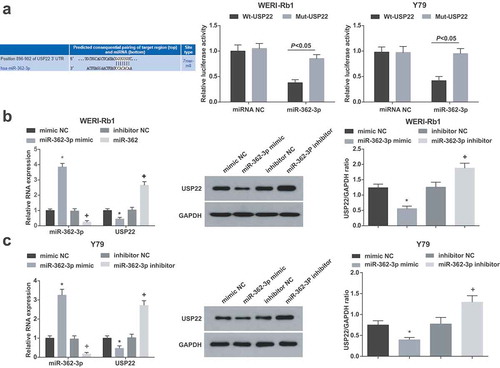
Detected by western blot assay and RT-qPCR, it was manifested that in WERI-Rb1 and Y79 cells (), miR-362-3p overexpression raised miR-362-3p and decreased USP22 expression. Suppression of miR-362-3p reduced miR-362-3p and elevated USP22 expression levels.
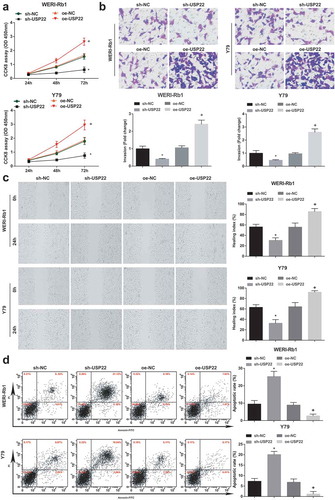
Down-regulating USP22 attenuates RB cell progression
The biological functions of WERI-Rb1 and Y79 cells were tested by CCK-8 assay, Transwell assay, scratch test and flow cytometry (). It was revealed that down-regulating USP22 blocked RB cell progression while up-regulating USP22 worked in an opposite way.
Figure 4. Down-regulating USP22 inhibits cell progression in RB. (a), Comparison of the proliferative capacity of WERI-Rb1 and Y79 cells by CCK-8 assay. (b), Comparison of the invasive ability of WRI-Rb1 and Y79 cells by Transwell assay. (c), Comparison of migration ability of WERI-Rb1 and Y79 cells by scratch test. (d), Analysis of apoptosis rate of WRI-Rb1 and Y79 cells by flow cytometry. * P < 0.05 vs. sh-NC group. + P < 0.05 vs. oe-NC group. N = 3. Measurement data were depicted as mean ± standard deviation, comparisons among multiple groups were assessed by one-way ANOVA followed with Tukey’s post hoc test
USP22 deubiquitinates LSD1 for protein stabilization in RB
To examine the interaction between endogenous USP22 and LSD1 in RB cells, endogenous LSD1 was immunoprecipitated by USP22 antibody or IgG-conjugated agarose beads in WERI-Rb1 cells. The experimental results displayed that endogenous LSD1 was detected in the USP22 antibody group (), lane 2) but not detected in the IgG group (), lane 1).
Figure 5. USP22 deubiquitinates LSD1 for protein stabilization in RB. (a), Immunoprecipitation of USP22 in WERI-Rb1 cells with USP22 antibody. (b), USP22 and LSD1 expression in WERI-Rb1 cells by RT-qPCR. (c), USP22 and LSD1 expression in Y79 cells by RT-qPCR.* P < 0.05 vs. sh-NC group. + P < 0.05 vs. oe-NC group. N = 3. Measurement data were depicted as mean ± standard deviation, comparisons among multiple groups were assessed by one-way ANOVA followed with Tukey’s post hoc test
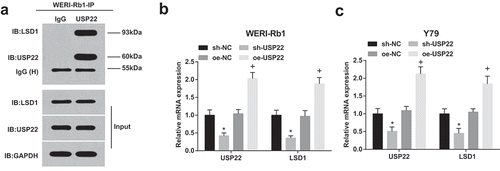
USP22 and LSD1 expression in WERI-Rb1 and Y79 cells were tested by RT-qPCR ()). It was suggested that USP22 inhibition reduced USP22 and LSD1 expression while overexpression of USP22 heightened USP22 and LSD1 expression.
Discussion
RB is the commonest intraocular malignancy in children, and the therapeutic strategies include concurrent chemotherapy, radiotherapy, local therapies and intra-arterial chemotherapy [Citation19]. Intriguingly, it is manifested that miRNAs act as therapeutic biomarkers and potential diagnostic agents in RB [Citation20]. Zhou et al. have made it clear that USP22 is crucial in cell proliferation/aging/apoptosis of RB [Citation11]. The current study was designed to decipher how miR-362-3p functioned in RB cells via regulating USP22 and LSD1.
Tested by this study, down-regulated miR-362-3p and up-regulated USP22 and LSD1 were present in RB tissues and cells. Some studies have manifested that miR-362-3p expression is down-regulated in non-small cell lung cancer [Citation21] and in glioma [Citation22], but there was no study pivoting on miR-362-3p expression in RB. It is displayed that USP22 is markedly heightened in gastric cancer (GC) and prostate cancer tissue samples and cell lines relative to that in adjacent noncancerous tissue samples [Citation23,Citation24]. However, no study have implied the overexpressed USPP in RB. Several articles have reported that LSD1 expression is elevated in colorectal cancer and breast cancer in comparison to that in corresponding adjacent non-tumor tissue [Citation25,Citation26]. Also, no study has shown the overexpression of LSD1 in RB. It was verified in our study that USP22 was targeted by miR-362-3p and USP22 deubiquitinated LSD1 for protein stabilization in RB. There were no studies revealing the target relationship between miR-362-3p and USP22 in RB, but some studies have shown the target relationship of miRs and USP22 in other diseases. A study has purported that miR-30e-5p suppresses nasopharyngeal carcinoma cell proliferation and metastasis via targeting USP22 [Citation27]. Another study has reported that USP22 is directly targeted by miR-29 c in pancreatic cancer [Citation28]. However, the target relationship between miR-362-3p and USP22 as well as the interaction between USP22 and LSD1 needs further study.
The biological functions of RB cells were validated in our study by multiple assays and the results presented that restoring miR-362-3p and depleting USP22 restrained the proliferation, migration and invasion and facilitated the apoptosis of RB cells. It has been depicted previously that depletion of USP22 dramatically limits cell proliferation potency and raises cell apoptosis or DNA damage [Citation11]. It is reported that the down-regulation of USP22 markedly restrains migration, survival, proliferation and invasion of GC cells relative to that in the controls [Citation23]. A study has verified that the highly expressed USP22 stimulates colony formation ability and cell growth of breast cancer [Citation29]. Another study has reported that lowly expressed USP22 represses proliferation, invasion, migration of nasopharyngeal carcinoma cells [Citation27]. It is displayed that up-regulation of miR-362-3p restrains cell proliferation, migration and invasion, and stimulates cell apoptosis in renal cancer [Citation30]. It is suggested that overexpressed miR-362-3p is capable of attenuating cell aggressive behaviors in hepatocellular carcinoma [Citation9]. It is confirmed that restoration of miR-362 obviously inhibits cell migration, proliferation and invasion in cervical cancer [Citation31]. Nevertheless, the interplay of miR-362-3p and USP22 in RB needs further study. A study has reported that other treatments, such as down-regulation of PlncRNA-1, silencing ANRIL or up-regulating ATM impedes cell migration, proliferation and invasion of RB [Citation32,Citation33].
To briefly conclude, our study suggests that miR-362-3p targets USP22 and then represses proliferation, invasion and migration as well as promotes apoptosis of RB via reducing LSD1 modified by deubiquitination. It has widened our insight into novel agents for RB treatment. However, the promoter methylation of miR-362-3p needs further exploration to decode the upstream mechanism of miR-362-3p in RBa. Additionally, the correlation of the downstream gene expression profiles of USP22 and LSD1 in RB cells by microarray analysis shall be further studied in the future.
Acknowledgments
We would like to acknowledge the reviewers for their helpful comments on this paper.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Yang L, Zhang L, Lu L, et al. lncRNA UCA1 increases proliferation and multidrug resistance of retinoblastoma cells through downregulating miR-513a-5p. DNA Cell Biol. 2020;39(1):69–77.
- Deng L, Li J, Lu S, et al. Crocin inhibits proliferation and induces apoptosis through suppressing MYCN expression in retinoblastoma. J Biochem Mol Toxicol. 2019;33(5):e22292.
- Yu F, Pang G, Zhao G. ANRIL acts as onco-lncRNA by regulation of microRNA-24/c-Myc, MEK/ERK and Wnt/beta-catenin pathway in retinoblastoma. Int J Biol Macromol. 2019;128:583–592.
- Soliman SE, VandenHoven C, Mackeen LD, et al. Vision and visual potential for perifoveal retinoblastoma after optical coherence tomographic-guided sequential laser photocoagulation. Br J Ophthalmol. 2019;103(6):753–760.
- Yang M, Wei W. Long non-coding RNAs in retinoblastoma. Pathol Res Pract. 2019;215(8):152435.
- Zhang W, Ren Y, Li J. Application of miR-193a/WT1/PODXL axis to estimate risk and prognosis of idiopathic membranous nephropathy. Ren Fail. 2019;41(1):704–717.
- Delsin LEA, Salomao KB, Pezuk JA, et al. Expression profiles and prognostic value of miRNAs in retinoblastoma. J Cancer Res Clin Oncol. 2019;145(1):1–10.
- Yang S, Zhang X, Sun Y, et al. MicroRNA-362-3p Inhibits Migration and Invasion via Targeting BCAP31 in Cervical Cancer. Front Mol Biosci. 2020;7:107.
- Li Z, Hu Y, Zeng Q, et al. Circular RNA MYLK promotes hepatocellular carcinoma progression by increasing Rab23 expression by sponging miR-362-3p. Cancer Cell Int. 2019;19(1):211.
- Zhang K, Yang L, Wang J, et al. Ubiquitin-specific protease 22 is critical to in vivo angiogenesis, growth and metastasis of non-small cell lung cancer. Cell Commun Signal. 2019;17(1):167.
- Zhou D, Liu P, Sun D-W, et al. USP22 down-regulation facilitates human retinoblastoma cell aging and apoptosis via inhibiting TERT/P53 pathway. Eur Rev Med Pharmacol Sci. 2017;21(12):2785–2792.
- Dou Y, LIN J, SHU H, et al. Role of ubiquitin-specific peptidase 22 in carcinogenesis of human pharyngeal squamous cell carcinoma. Mol Med Rep. 2014;10(6):2973–2978.
- Cusan M, Cai SF, Mohammad HP, et al. LSD1 inhibition exerts its antileukemic effect by recommissioning PU.1- and C/EBPalpha-dependent enhancers in AML. Blood. 2018;131(15):1730–1742.
- Chau CM, Deng Z, Kang H, et al. Cell cycle association of the retinoblastoma protein Rb and the histone demethylase LSD1 with the Epstein-Barr virus latency promoter Cp. J Virol. 2008;82(7):3428–3437.
- Yang Y, Wu N, Wu Y, et al. Artesunate induces mitochondria-mediated apoptosis of human retinoblastoma cells by upregulating Kruppel-like factor 6. Cell Death Dis. 2019;10(11):862.
- Qiu GZ, Mao X-Y, Ma Y, et al. Ubiquitin-specific protease 22 acts as an oncoprotein to maintain glioma malignancy through deubiquitinating B cell-specific Moloney murine leukemia virus integration site 1 for stabilization. Cancer Sci. 2018;109(7):2199–2210.
- Xu C, Hu C, Wang Y, et al. Long noncoding RNA SNHG16 promotes human retinoblastoma progression via sponging miR-140-5p. Biomed Pharmacother. 2019;117:109153.
- Yang L, Zhang L, Lu L, et al. <p>Long Noncoding RNA SNHG16 Sponges miR-182-5p And miR-128-3p To Promote Retinoblastoma Cell Migration And Invasion By Targeting LASP1. Onco Targets Ther. 2019;12:8653–8662.
- Buaboonnam J, Narkbunnam N, Vathana N, et al. Outcomes of pediatric retinoblastoma treated with ICEV regimen: A single-center study. Pediatr Hematol Oncol. 2019;36(2):73–81.
- Golabchi K, Soleimani-Jelodar R, Aghadoost N, et al. MicroRNAs in retinoblastoma: potential diagnostic and therapeutic biomarkers. J Cell Physiol. 2018;233(4):3016–3023.
- Jiang Z, Yin J, Peng G, et al. Circ_0074027 contributes to the progression of non-small cell lung cancer via microRNA-362-3p/clathrin heavy chain axis. Anticancer Drugs. 2021;32(1):1–10.
- Xu G, Fang P, Chen K, et al. MicroRNA-362-3p Targets PAX3 to Inhibit the Development of Glioma through Mediating Wnt/beta-Catenin Pathway. Neuroimmunomodulation. 2019;26(3):119–128.
- Liu H, Liu N, Zhao Y, et al. Oncogenic USP22 supports gastric cancer growth and metastasis by activating c-Myc/NAMPT/SIRT1-dependent FOXO1 and YAP signaling. Aging (Albany NY). 2019;11(21):9643–9660.
- McCann JJ, Vasilevskaya IA, Poudel Neupane N, et al. USP22 Functions as an Oncogenic Driver in Prostate Cancer by Regulating Cell Proliferation and DNA Repair. Cancer Res. 2020;80(3):430–443.
- Zhang H-S, Liu H-Y, Zhou Z, et al. TSPAN8 promotes colorectal cancer cell growth and migration in LSD1-dependent manner. Life Sci. 2020;241:117114.
- Feng J, Xu G, Liu J, et al. Phosphorylation of LSD1 at Ser112 is crucial for its function in induction of EMT and metastasis in breast cancer. Breast Cancer Res Treat. 2016;159(3):443–456.
- Ma YX, Zhang H, Li X-H, et al. MiR-30e-5p inhibits proliferation and metastasis of nasopharyngeal carcinoma cells by target-ing USP22. Eur Rev Med Pharmacol Sci. 2018;22(19):6342–6349.
- Huang L, Hu C, Cao H, et al. MicroRNA-29c Increases the Chemosensitivity of Pancreatic Cancer Cells by Inhibiting USP22 Mediated Autophagy. Cell Physiol Biochem. 2018;47(2):747–758.
- Kim D, Hong A, Park HI, et al. Deubiquitinating enzyme USP22 positively regulates c-Myc stability and tumorigenic activity in mammalian and breast cancer cells. J Cell Physiol. 2017;232(12):3664–3676.
- Zou X, ZHONG J, LI J, et al. miR-362-3p targets nemo-like kinase and functions as a tumor suppressor in renal cancer cells. Mol Med Rep. 2016;13(1):994–1002.
- Shi C, Zhang Z. MicroRNA-362 is downregulated in cervical cancer and inhibits cell proliferation, migration and invasion by directly targeting SIX1. Oncol Rep. 2017;37(1):501–509.
- Wang S, Liu J, Yang Y, et al. PlncRNA-1 is overexpressed in retinoblastoma and regulates retinoblastoma cell proliferation and motility through modulating CBR3. IUBMB Life. 2018;70(10):969–975.
- Yang Y, Peng XW. The silencing of long non-coding RNA ANRIL suppresses invasion, and promotes apoptosis of retinoblastoma cells through the ATM-E2F1 signaling pathway. Biosci Rep. 2018;38(6). DOI:10.1042/BSR20180558
