ABSTRACT
Circular RNAs (circRNAs) are considered as a new regulatory factor in growth, metastasis and therapeutic resistance of human cancers. But the clinical significance and underlying mechanism of circular RNA ITCH (circ-ITCH) in gastric cancer (GC) remain unknown. In the present study, we found that circ-ITCH was down-regulated in GC cell lines, GC tissues and their serum-derived exosomes. The level of circ-ITCH was related to invasion depth. Functional assays showed that circ-ITCH overexpression inhibited the proliferation, migration, invasion and epithelial mesenchymal transition (EMT) of GC cells, whereas circ-ITCH knockdown appeared an opposite effect. Bioinformatic analysis and luciferase reporter assay confirmed that circ-ITCH acted as miR-199a-5p sponge and increased the level of Klotho. The expression level of miR-199-5p was up-regulated in GC tissues and negatively correlated with that of circ-ITCH. MiR-199a-5p mimics reversed the effects on inhibiting metastasis induced by circ-ITCH overexpression and decreased the level of Klotho in GC cells. Our findings indicate that circ-ITCH suppresses metastasis of GC by acting as the sponge of miR-199a-5p and increasing Klotho expression, which serves as a potential biomarker and targets for the diagnosis and therapy of GC.
Abbreviations: CircRNAs: circular RNAs; GC: gastric cancer; circ-ITCH: circular RNA Itchy E3 ubiquitin protein ligase; ceRNA: competitive endogenous RNA; EMT: Epithelial–mesenchymal transition; siRNA: Small interfering RNA; TEM: transmission electron microscope; NTA: nanoparticle tracking analysis.
Introduction
Gastric cancer (GC), the fifth most frequently diagnosed cancer and the third leading cause of cancer death around the world [Citation1], threatens human health, especially in China [Citation2]. Due to the absence of specific symptoms and diagnostic indicators, most GC patients are at an advanced stage and the overall 5-years survival rate is very low. Moreover, because of the frequent occurrence of metastasis, the prognosis of gastric cancer is not well [Citation3,Citation4]. Therefore, it is urgent to reveal the molecular mechanisms and search for specific diagnostic marker and therapeutic target of GC.
Circular RNAs (circRNAs) comprise a large class of non-coding RNAs (ncRNAs) known for their covalently closed loop structures with neither 5' to 3' polarity nor polyadenylated tail. CircRNAs, which are characterized by abundance, conservative and related to ALU sequences [Citation5], are widely present in a variety of mammalian cells. Increasing evidence indicates that the alteration in circRNAs expression is common in various cancer. The SCD-circRNA 2 regulated by the RNA-binding protein RBM3 is crucial that high expression of SCD-circRNA 2 promotes cell proliferation in hepatocellular carcinoma [Citation6]. A series of circRNAs have been reported about their roles in the diagnosis and treatment of GC. For example, circNRIP1 sponges miR-149-5p to regulate miR-149-5p-AKT1/mTOR axis and eventually acts as a tumor promotor in GC [Citation7]. Exosomal circSHKBP1 is a promising circulating biomarker via regulating the miR-582-3p/HUR/VEGF axis and suppressing HSP90 degradation in GC [Citation8]. A clinical study proves that circPVT1, one of circRNAs identified among differential expression profile, is a new prognostic marker in GC [Citation9]. Now, the specific and functional circRNAs are gradually emerging in GC.
Present study focuses on Circular RNA Itchy E3 ubiquitin protein ligase (circ-ITCH) which generates from 6 to 13 exons of ITCH gene in GC. Accumulating evidences indicate that circ-ITCH is universally down-regulated in several cancers and plays an important role in the way of miRNA sponge. Circ-ITCH acts as the sponge of miR-17/miR-224 to regulate p21 and PTEN expression in bladder cancer [Citation10]. Circ-ITCH is a tumor suppressor by increasing HOXB13 expression via sponging miR-17-5p in prostate cancer [Citation11]. In ovarian cancer, circ-ITCH sponges miR-106a to increase CDH1 expression and suppresses proliferation, invasion, and glycolysis of ovarian cancer cells [Citation12]. However, the specific mechanism by which circ-ITCH plays a role in the progression of GC is unclear.
Previous studies indicate that circRNAs can act as competitive endogenous RNA (ceRNA) to sponge several miRNAs and regulate gene expression [Citation13,Citation14]. The dysregulation of miRNA is an important factor of the occurrence and progression of cancers [Citation15,Citation16]. MiR-199a-5p, a member of the microRNA (miR)-199a gene, has been considered to involve in multiple types of malignancies. Previous studies indicate that the expression of miR-199a-5p shows tissue heterogeneity in different tumors. MiR-199a-5p is down-regulated in triple-negative breast cancer, targeting PIK3CD to alter EMT-related genes CDH1, ZEB1, and TWIST expression [Citation17]. MiR-199a-5p plays a tumor suppression role in NSCLC via targeting MAP3K11 [Citation18]. However, miR-199a-5p can directly target both PIAS3 and p27 to promote osteosarcoma growth and it is up-regulated in gastric cancer [Citation19,Citation20]. Nevertheless, mechanism of interactions between circRNAs and miR-199a-5p in GC is unknow.
This study aims to clarify the role of circ-ITCH in GC and the underlying mechanism by which circ-ITCH regulates miR-199a-5p. We find that circ-ITCH, which may be novel biomarker and target for the diagnosis and therapy of GC, inhibits metastasis of GC by acting as the sponge of miR-199a-5p and increasing Klotho expression.
Materials and methods
Clinical specimens
A total of 61 pairs of GC patient tissues, paired adjacent tissues (5 cm away from the tumor edge), 33 serum samples of preoperative GC patients and healthy donors were collected from Department of General Surgery, the Affiliated People’s Hospital of Jiangsu University between March 2017 and August 2018. They were classified according to the WHO criteria and staged according to the tumor-node-metastasis (TNM). All GC tissue samples were stored at −80°C after being collected. The serum samples were centrifuged at 3000 g for 10 min and were stored at −80°C until RNA extraction. Informed consents were obtained from each participant. And the study was approved by the Medical Ethics Committee of Jiangsu University ((IRB protocol number: 2,020,161).
Cell culture
Human GC cell lines HGC-27, AGS and MKN-45 were purchased from Runyan Biotechnology Company (Nanjing, China). Human GC cell MGC-803 and HEK-293 T cell were obtain from Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Human normal gastric mucosa epithelial cell line GES-1 was purchased from Gefan Biological Technology (Shanghai, China). All the gastric cancer cell lines had identification certificates in supplementary materials. HGC-27, MKN-45, GES-1 cells were cultured in RPMI 1640 (Bioind, Israel)). MGC-803 and HEK-293 T cells were cultured in high glucose-DMEM (Bioind, Israel) and AGS was cultured in DMEM/F-12 (Bioind, Israel). All cell lines were maintained in DMEM supplemented with 10% fetal bovine serum (Bovogen, Australia) in a humidified incubator with 5% CO2 at 37°C.
Lentiviral and siRNAs transfection
The circ-ITCH overexpression and the control lentivectors were purchased from Hanbio Biotechnology (Shanghai, China). MGC-803 and MKN-45 cells were seeded in 12-wells plate (8 × 10⁴cells per well) for 24 h. Then the appropriate MOI lentiviral particle was then added to the medium and incubated for 24 h. Stably transfected clones were selected by culturing in the appropriate concentration of puromycin for two weeks. Small interference RNAs (siRNAs) of circ-ITCH, miR-199a-5p mimics/inhibitor and their negative controls were obtained from GenePharma (Suzhou, China). HGC-27 and MGC-803 cells were seeded in 6-wells plate (2 × 10⁵ cells per well). After 24 h, siRNAs, mimics or inhibitor were transfected into the cells by using Lipofectamine™ 2000 (Life Technologies, Shanghai, China) in serum-free medium. Cells were changed to complete medium at 6 h after transfection and cultured for another 36 h. The sequences for siRNAs and mimics/inhibitor were listed in .
Table 1. Sequences of siRNAs
Cell counting and cell colony formation assay
The transfected cells were seeded in 24-well plates (1 × 10⁴per well) and were counted for 6 days. The medium was changed every 2 days. Colony formation assay was used to incubate the transfected cells seeded in 6-well plates (1 × 103 per well) for 7–14 days. The medium was changed every 3 days. And then the cells were fixed with 4% paraformaldehyde and stained with crystal violet. The number of clones were photographed and counted under a microscope. All experiments were repeated for triplicate.
Cell counting Kit-8 (CCK8) assay
For CCK8 assay, the treated cells were plated in 96-well plates (1 × 103 cells/well). Cell proliferation was detected every 24 h for 5 days. 100 µl medium containing 10% CCK-8 solution (Vazyme, Nanjing, China) was added to each well and incubated for 2 h at 37°C. The absorbance of each well at 450 nm was measured by microplate spectrophotometer. Each group was measured at least three times.
Cell migration and invasion assay
For migration assay, 600 μL medium containing 10% FBS was added into the lower chamber of transwell with 8 μm pore size (Corning, MA, USA), while the transfected cells were seeded into the upper chamber in 200 μL serum-free medium. As for invasion assay, the diluted Matrigel (BD Biosciences, NY, USA) was firstly added to the upper chamber and incubated for 30 min at 37°C. The other steps were consistent with the migration assay. After 24–48 h, the cells on the upper surface were removed with a cotton swab and the cells on the lower surface were fixed in 4% paraformaldehyde for 30 min followed by staining with crystal violet for 15 min. Finally, the cells were photographed and counted under an inverted microscope. All experiments were repeated for triplicate and the results should be the mean of all experiments.
Isolation and identification of serum exosomes
The exosome quick extraction solution (SBI, CA, USA) was added to serum samples at 1:5 ratio and the mixture was stored at 4°C for at least 12 h. Then the supernatant was discarded after centrifuged at 1500 × g for 30 min. The exosome-rich precipitation was dissolved with phosphate buffered saline (PBS) and stored at −80°C. The morphology of exosomes was shown by transmission electron microscope (TEM). The particle size of exosomes was measured by nanoparticle tracking analysis (NTA).
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was isolated from GC tissues or cell lines using Trizol reagent (Invitrogen, MA, USA) according to the manufacturer’s procedures. Total RNA in serum exosomes was separated by utilizing miRNeasy serum/plasma kit according to the manufacturer’s instructions (Qiagen, CA, USA). The reverse transcription (RT) and quantitative real time polymerase chain reaction were performed with the corresponding kits (Vazyme, Nanjing, China). The target genes were normalized to β-actin to obtain the relative expression level. The expression of genes was evaluated by the 2−ΔΔCt method. All the sequences of the primers were listed in .
Table 2. The primer information of qRT-PCR
Western blot
Proteins of cells were obtained by Radio-Immunoprecipitation Assay (RIPA) extraction reagent supplemented with 1% protease inhibitors (Roche, CA, USA). An equal amount of extracted protein was evaluated by 12% SDS-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to 0.45 μm PVDF membranes (Millipore, MA, USA) followed by blocking with 5% nonfat milk for 1 h. Afterward, the appropriate primary antibodies against PCNA, Bcl2, Bax, Vimentin, Slug, Twist, MMP9, E-cadherin (E-cad), N-cadherin(N-cad), Cyclin D1, Caspase3, Caspase9 and Klotho (CST, MA, USA) were diluted and conducted with the membranes at 4°C overnight. After incubated with HRP-conjugated goat anti-rabbit/mouse IgG secondary antibody (Invitrogen, CA, USA) for 1 h at room temperature, the protein bands were visualized by using chemiluminescence (Millipore, Carrigtwohill, Ireland). β-actin (CST, MA, USA) was used as loading control.
Dual-luciferase reporter assay
The circ-ITCH and Klotho 3'UTR binding sites of miR-199a-5p were predicted by StarBase (http://starbase.sysu.edu.cn/). The luciferase reporter vector containing wild type (WT) or mutant type (MUT) of circ-ITCH were purchased from GenePharma (Suzhou, China). The HEK-293 T cell was co-transfected with miR-199a-5p mimics and two different luciferase reporter vectors for 24 h. The luciferase activity was detected using the Dual Luciferase Assay Kit (Promega, MA, USA) according to the manufacturer’s instructions. This experiment was performed for at least three times.
Statistical analysis
Statistical analyses were performed using the SPSS 22.0 software (Chicao, IL, USA) and GraphPad Prism 5.0 (LaJolla, CA, USA). The results of various experiments were performed as the mean ± SD. Differences between experimental groups were assessed by the Student’s t-test or one-way analysis of variance (ANOVA). The associations between circ-ITCH and the clinicopathological features were evaluated by the Pearson χ2 test. The receiver operating characteristic (ROC) curve was used to assess the diagnostic value of circ-ITCH. P value < 0.05 was considered statistically significant.
Results
Circ-ITCH is down-regulated in GC cell lines, tissues and serum-derived exosomes of GC patients
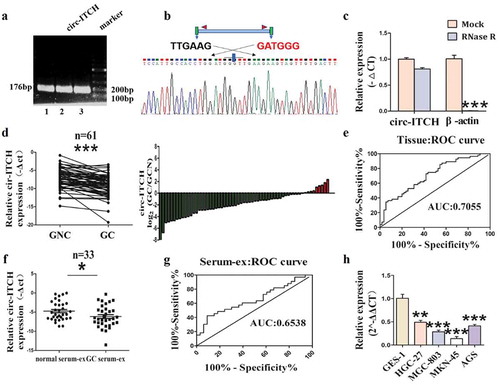
To explore whether circ-ITCH involved in GC, we designed specific PCR primers for circ-ITCH utilizing Primer Premier 5. The results of agarose gel electrophoresis and sanger sequencing showed that the primer design was successful (). RNase R assay confirmed that the level of circ-ITCH showed no distinct change in HGC-27 cells under RNase R treatment whereas the expression of β-actin was remarkably reduced ()). Then we examined the expression of circ-ITCH in 61 paired of GC tissues and matched adjacent normal tissues utilizing qRT-PCR. The expression of circ-ITCH was significantly decreased in tumor tissues ()). The area under the ROC curve was 70.55% ()). Furthermore, combined with clinicopathological parameters, the level of circ-ITCH was related to invasion depth (). The morphology and particle size of exosomes were shown by TEM and NTA (Figure S1(A, B)). Their average particle size of serum-derived exosomes was about 100 nm. Circ-ITCH was undetectable in serum. But interestingly, it was detectable in serum exosomes, suggesting that circ-ITCH could be enriched in exosomes relative to serum. And as expected, circ-ITCH expression level in GC serum-derived exosomes was also down-regulated, compared to serum-derived exosomes of healthy donors ()). But the area under the ROC curve was 65.38% ()), lower than that of tissues. In addition, circ-ITCH expression level in each GC cell line was uniformly down-regulated ()). Given the above results, circ-ITCH was consistently down-regulated in GC tissues, cell lines and GC serum-derived exosomes, which may be a molecular biomarker in the diagnosis of GC.
Table 3. Correlation between circ-ITCH expression and clinical pathological characteristic in GC (n = 61)
Figure 1. Circ-ITCH was down-regulated in GC tissues, GC cell lines and GC serum-derived exosomes. (a) The results of agarose gel electrophoresis. (b) Sanger sequencing results from PCR products of circ-ITCH. (c) The level of circ-ITCH in HGC-27 cells under Rnase R treatment compared with the level of β-actin. (d) The expression of circ-ITCH in 61 paired of GC tissues and matched adjacent normal tissues. (e) The area under the ROC curve of the GC tissues (sensitivity%:52.71, specificity%:74.55). (f) The expression of circ-ITCH in 33 paired of GC patient serum-derived exosomes. (g) The area under the ROC curve of the serum-derived exosomes (sensitivity%:42.42, specificity%:90.91). (h) Circ-ITCH expression level in each GC cell line was also uniformly downregulated. *P < 0.05, **P < 0.01, ***P < 0.001
Circ-ITCH overexpression inhibits GC cell proliferation, migration and invasion
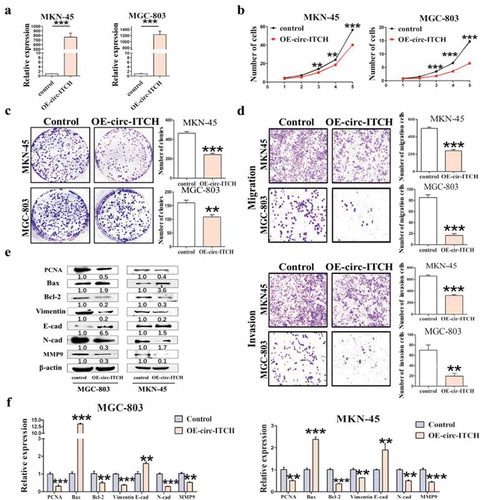
To further investigate the biological function of circ-ITCH, we overexpressed circ-ITCH in MGC-803 and MKN-45 cells via lentivirus transfection. Both the results of fluorescence (Figure S2(A)) and qRT-PCR ()) showed that its overexpression was successful. Stably transfected clones were used for subsequent cell functional assays. The cell counting and colony formation assays revealed that circ-ITCH overexpression inhibited the proliferation of MKN-45 and MGC-803 cells ()). Compared to GC cells treated with empty vector, we found that the number of migrated and invaded cells obviously decreased in overexpressing circ-ITCH GC cells using migration and invasion assays ()). The expression of PCNA and Bcl-2 were decreased after circ-ITCH overexpression in both GC cells. Meanwhile, the level of Bax was increased. Indicators of metastasis Such as N-cad, Vimentin and MMP9 were decreased uniformly, whereas E-cad was increased ()), which was consistent with qRT-PCR ()). Taken together, circ-ITCH overexpression significantly inhibited the capacity of GC cell proliferation, migration and invasion.
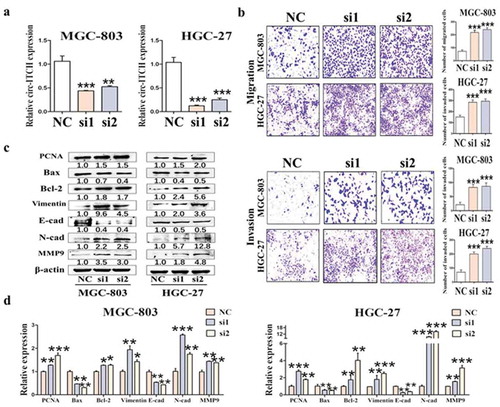
Figure 2. Circ-ITCH overexpression inhibited GC cell proliferation, migration and invasion. (a) Circ-ITCH overexpression efficiency was assessed by qRT-PCR. (b) The results of cell counting assay after circ-ITCH overexpression in MKN-45 and MGC-803 cells. (c) The number of cell clones after circ-ITCH overexpression in MKN-45 and MGC-803 cells. (d) Circ-ITCH overexpression inhibited the migration and invasion of MKN-45 and MGC-803 cells. (e) The expression of proliferation and metastasis-related proteins in GC cells after circ-ITCH overexpression. (f) The results of qRT-PCR matching all the protein indicators. *P < 0.05, **P < 0.01, ***P < 0.001. (a) The knockdown efficiency of circ-ITCH after circ-ITCH siRNAs transfection in MGC-803 and HGC-27 cells. (b) The number of migrated and invaded cells increased in MGC-803 and HGC-27 cells after circ-ITCH knockdown compared with control cells. (c) The expression of proliferation and metastasis-related proteins in GC cells after circ-ITCH knockdown. (d) The results of mRNA level matching all the protein indicators. *P < 0.05, **P < 0.01, ***P < 0.001
Circ-ITCH knockdown promotes GC cell proliferation, migration and invasion
Next, we knocked down circ-ITCH in MGC-803 and HGC-27 cells by siRNAs transfection. The results confirmed that the knockdown efficiency of circ-ITCH was more than 50% ()). As expected, contrary to the overexpression results, circ-ITCH knockdown promoted the progression of GC, especially in terms of metastasis. Cell transwell assay appeared that the reduction of circ-ITCH promoted the migration and invasion of GC cells ()). Circ-ITCH knockdown also facilitated the proliferation of GC cells (Figure S2(B, C)). Meanwhile, we also measured the protein and mRNA levels of related indicators. The expression levels of PCNA and Bcl-2 were increased and the expression level of Bax was decreased. Silencing of circ-ITCH also promoted the expression of EMT-related N-cad, Vimentin, MMP9, while inhibiting the expression of E-cad ()). Collectively, combined with overexpression and knockdown of circ-ITCH, we demonstrated that circ-ITCH acted as a potential tumor suppressor in GC.
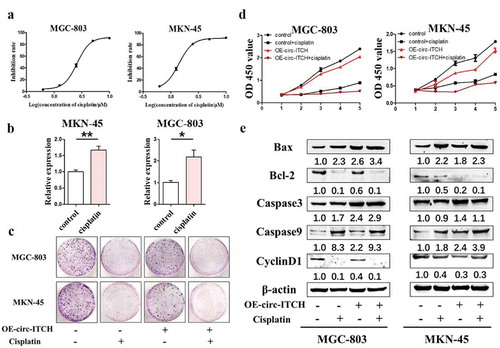
Circ-ITCH has partial effect on anticancer chemotherapy
We also explored whether circ-ITCH had effect on chemotherapy. We used the broad-spectrum chemotherapy drug cisplatin for control. We determined the IC50 value of cisplatin treatment in MKN-45 and MGC-803 by CCK8 assay ()) and treated cells according to the IC50 value subsequently. Then we detected the level of circ-ITCH after treating with cisplatin, the results of qRT-PCR showed that the expression of circ-ITCH was up-regulated in MKN-45 and MGC-803 cells ()). These results suggested that circ-ITCH may play a role in anticancer chemotherapy. Next, we compared the effects of cisplatin treatment and circ-ITCH overexpression on GC cells. For colony formation assay, the number of colonies was decreased in MKN-45 and MGC-803 cells, especially after treating with cisplatin ()). Similar results were obtained in the CCK8 assay after cisplatin treatment or circ-ITCH overexpression and the proliferation of the GC cells was most inhibited under the combination ()). Moreover, we detected several apoptosis-related indicators such as Bax, Bcl-2, Caspase3 and Caspase9 and Cyclin proteins by western blot. The result showed that the expression of Bax, Caspase3 and Caspase9 were increased, the expression of Bcl-2 and Cyclin D1 were decreased after circ-ITCH overexpression or cisplatin treatment ()). Actually, some indicators showed better trends after combination. These results proved that circ-ITCH has partial effect on anticancer chemotherapy.
Figure 4. Circ-ITCH has partial effect on anticancer chemotherapy. (a) The IC50 value of cisplatin was determined by CCK8 experiment (MKN-45:1.5μΜ, MGC-803:2.6μΜ). (b) The expression level of circ-ITCH was up-regulated in MKN-45 and MGC-803 cells after treating with cisplatin. (c) Cisplatin treatment or overexpression of circ-ITCH could inhibit the formation of colonies in GC cells, but cisplatin had a stronger effect. (d) Cisplatin treatment or overexpression of circ-ITCH could inhibit the proliferation of GC cells, but cisplatin was stronger. (e) The changes of apoptosis-related indicators and Cyclin D1 after cisplatin treatment or overexpression of circ-ITCH
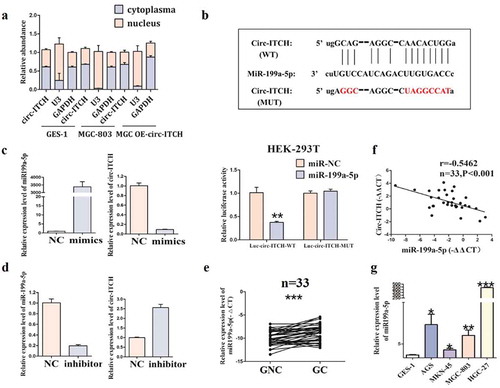
Circ-ITCH acts as the sponge of mir-199a-5p in GC
Emerging evidence indicates that the function of circRNA is usually related to its subcellular localization. To explore the specific mechanism of circ-ITCH function in GC, we detected the subcellular localization of circ-ITCH in GC cells. The results showed that circ-ITCH was mainly localized in the cytoplasm, and the localization did not change after its overexpression ()). Subsequently, we found that circ-ITCH possessed a binding site with miR-199a-5p through bioinformatics analysis. miR-199a-5p overexpression decreased the luciferase activity of Luc-circ-ITCH wild type (WT) plasmids, whereas the significant difference was not found in the luciferase activity of Luc-circ-ITCH mutant type (MUT) plasmids ()). To further confirm, mimics and inhibitor of miR-199a-5p were transfected with GC cells, respectively. As expected, miR-199a-5p mimics and miR-199a-5p inhibitor had opposite effects on the expression level of circ-ITCH ()). Moreover, we also detected the expression of miR-199a-5p in GC tissues and cell lines. The results showed that miR-199a-5p was up-regulated in both GC tissues and cell lines ()). And there was a negative correlation between the expression level of circ-ITCH and that of miR-199a-5p in GC tissues by using the Pearson χ2 test analysis ()). Our results demonstrated that circ-ITCH suppressed the progression of GC, especially in terms of metastasis, by acting as the sponge of miR-199a-5p.
Figure 5. Circ-ITCH acted as a sponge of miR-199a-5p in GC. (a) The subcellular localization of circ-ITCH. (b) The potential binding site of miR-199a-5p in circ-ITCH was predicted by bioinformatic software () above), and the binding of circ-ITCH and miR-199a-5p was proved through dual-luciferase reporter assay in HEK-293 T cells () below). (c) The changes of circ-ITCH expression after miR-199a-5p mimics transfection. (d) The changes of circ-ITCH expression after miR-199a-5p inhibitor transfection. (e) The level of miR-199a-5p in 33 pairs of GC tissues and matched adjacent normal tissues. (f) In the GC tissues, the level of circ-ITCH and miR-199a-5p were negatively correlated by using Pearson correlation analysis. (g) The expression level of miR-199a-5p in various GC cells compared with GES-1 cells. *P < 0.05, **P < 0.01, ***P < 0.001
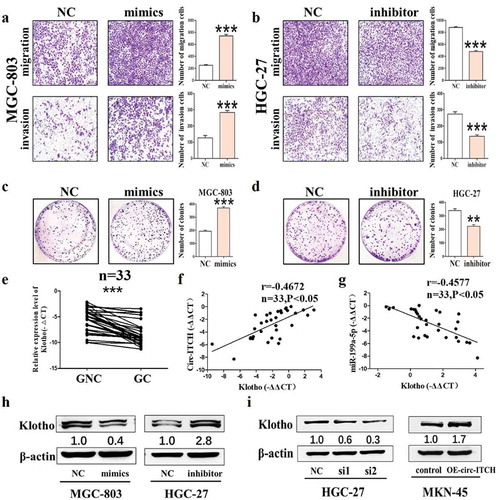
MiR-199a-5p promotes the metastasis of GC by targeting the Klotho
To further investigate the biological roles of miR-199a-5p in GC, the functional assays presented that the migration and invasion were enhanced and the number of cell clones was increased of MGC-803 cells after miR-199a-5p mimics were transfected into GC cells ()). MiR-199a-5p knockdown exerted the opposite effects ()). These results indicated that miR-199a-5p was a pro-cancer molecule in GC. Bioinformatics prediction indicate that there are two binding sites between Klotho 3'UTR and miR-199a-5p (Figure S2(D)). As expected, the expression level of Klotho was down-regulated in GC tissues, which was positively correlated with the expression of circ-ITCH and negatively correlated with the expression of miR-199a-5p. As shown in ), western blot results showed that miR-199a-5p overexpression reduced the level of Klotho in MGC-803 cells but the inhibitor of miR-199a-5p had an opposite effect. Moreover, we also detected the level of Klotho after circ-ITCH overexpression/knockdown in GC cells. The expression of Klotho was remarkably reduced in the circ-ITCH knockdown HGC-27 cells. Meanwhile, its level was increased in the circ-ITCH overexpression MKN-45 cells ()). These results suggested that circ-ITCH may play an important role in GC via circ-ITCH/miR-199a-5p/Klotho signaling axis.
Figure 6. MiR-199a-5p promoted the progression of GC by targeting Klotho. (a) Representative images of migration and invasion assays in MGC-803 after miR-199a-5p mimics transfection. (b) The number of migrated and invaded cells in HGC-27 after miR-199a-5p inhibitor transfection. (c) The number of cell clones in MGC-803 after miR-199a-5p mimics transfection. (d) The number of cell clones in HGC-27 after miR-199a-5p inhibitor transfection. (e) The level of Klotho in 33 pairs of GC tissues and matched adjacent normal tissues. (f) In the GC tissues, the level of circ-ITCH and Klotho were positively correlated by using Pearson correlation analysis. (g) The level of circ-ITCH and Klotho were positively correlated by using Pearson correlation analysis in the GC tissues. (h) The western blot results of Klotho after mimics/inhibitor of miR-199a-5p transfection. (i) The western blot results of Klotho after circ-ITCH knockdown/overexpression. *P < 0.05, **P < 0.01, ***P < 0.001
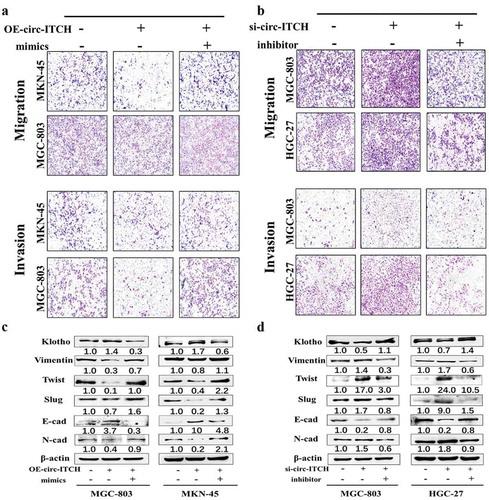
MiR-199a-5p reverses the suppressive effects of circ-ITCH on metastasis in GC cell
To further demonstrate the inhibition effect of circ-ITCH on the progression of GC through the circ-ITCH/miR-199a-5p/Klotho axis, we co-transfected circ-ITCH lentivirus vectors/siRNAs and miR-199a-5p mimics/inhibitor into GC cells. We found that the number of migrated and invaded cells decreased by circ-ITCH overexpression in MKN-45 and MGC-803 cells was reversed after co-transfected circ-ITCH lentivirus vectors and miR-199a-5p mimics ()). After co-transfecting circ-ITCH siRNAs and miR-199a-5p inhibitor in MGC-803 and HGC-27 cells, the inhibition of migration and invasion by circ-ITCH was reversed ()). Meanwhile, we also detected the level of Klotho after co-transfection. The results showed that the level change of Klotho was consistent with that of circ-ITCH, but it was reversed after miR-199a-5p mimics/inhibitor co-transfection ()). Then we detected several EMT-related proteins such as E-cad, N-cad, Slug and Twist. The results revealed that circ-ITCH overexpression inhibited the process of EMT. This effect could be reversed after miR-199a-5p overexpression. However, circ-ITCH knockdown had an opposite effect which promoted the process of EMT ()). We also detected several proliferation and apoptosis indicators after co-transfection. However, the results showed that co-translocation had no significant effect on proliferation and apoptosis of GC cells (Figure S2(E, F)). These results indicated that circ-ITCH inhibited the process of EMT and metastasis in GC, partly at least, via regulating circ-ITCH/miR-199a-5p/Klotho axis.
Figure 7. MiR-199a-5p could reverse the suppressive effects of circ-ITCH in GC cells. (a) The results of migration and invasion assays after circ-ITCH lentivirus vectors and miR-199a-5p mimics co-transfection into MKN-45 and MGC-803 cells. (b) The changes of the number of migrated and invaded cells after circ-ITCH siRNA and miR-199a-5p inhibitor transfection into MGC-803 and HGC-27 cells. (c) The western blot results of Klotho and EMT-related proteins after co-transfected circ-ITCH lentivirus vectors and miR-199a-5p mimics in MKN-45 and MGC-803 cells. (d) The western blot results of Klotho and EMT-related proteins after co-transfected circ-ITCH siRNAs and miR-199a-5p inhibitor in MGC-803 and HGC-27 cells
Discussion
CircRNAs have gained comprehensive attention due to the development of high-throughput sequencing technology and bioinformatics. CircRNAs are becoming potential biomarkers because of their abundant and stable characteristics in serum and even exosomes [Citation21]. Circ_104075 is up-regulated in hepatocellular carcinoma, acts as miR-582-3p sponge to upregulate YAP expression and potentially serves as a new diagnostic biomarker in HCC [Citation22]. In addition, the exosome-delivered ciRS-133 regulates white adipose browning by absorbing miR-133 and up-regulating PRDM16 [Citation23]. Moreover, circ-ITCH has been reported to be down-regulated in many malignant tumors, such as melanoma, osteosarcoma and prostate cancer [Citation24]. Although previous studies mention that circ-ITCH is down-regulated in GC tissues, the relation between level of exosomal circ-ITCH and diagnosis and progress of GC remain to be explored [Citation25]. In this study, we demonstrated that circ-ITCH was down-regulated in GC tissues and GC cell lines. Circ-ITCH was related to the depth of tumor invasion, suggesting that circ-ITCH can serve as a potential biomarker for GC. Moreover, we further found that circ-ITCH was enriched in serum-derived exosomes compared to serum and down-regulated in serum-derived exosomes of GC patients. Although the area under ROC curve of serum-derived exosomes was not better than that of tissues, these results suggested that the development of GC may be inhibited by the technology of engineering exosomes loaded circ-ITCH in the future. We further revealed the biological role of circ-ITCH via circ-ITCH knockdown and overexpression in GC cells. The results showed that the proliferation, migration and invasion of GC cells were significantly inhibited after circ-ITCH overexpression, whereas circ-ITCH knockdown had the opposite roles. Meanwhile, the results of qRT-PCR and western blot were consistent with the above results. Moreover, circ-ITCH has partial effect on anticancer chemotherapy. Although chemotherapy was not as effective as cisplatin, there is little damage for treatment by exosomes loaded circ-ITCH.
The function of circRNAs is largely determined by their location. CircRNAs located in the cytoplasm usually act as miRNA or protein sponge. For example, ciRS-7 contains more than 70 conserved miR-7 target sites, which can regulate the expression of many downstream target genes [Citation26]. CircMBL can be strongly and specifically bound by MBL, and the binding of circMBL and MBL proteins affects each other depending on the binding sites [Citation27]. CircRNAs located in the nucleus, most of them are ciRNAs or EIciRNAs, regulate the parental gene transcription through interaction with U1 snRNA or Pol II [Citation13]. In present study, we identified the location of circ-ITCH using nucleo-cytoplasmic separation assay. The results showed that circ-ITCH was mainly located in the cytoplasm, suggesting that circ-ITCH may act as a miRNA sponge. Next, we found that circ-ITCH had the potential binding site with miR-199a-5p through bioinformatics prediction, which was verified by dual-luciferase reporter assay. Overexpression of miR-199a-5p promoted migration and invasion of GC cells, whereas knockdown of miR-199a-5p had opposite results. Meanwhile, we also verified the expression level of miR199a-5p in GC tissues and cell lines. The results demonstrated that miR-199a-5p highly expressed both in GC tissues and cell lines and it was negatively correlated with the expression of circ-ITCH. Thus, we hypothesized that circ-ITCH acted as miR-199a-5p sponge and inhibited the progression of gastric cancer by reducing its expression. Then, we performed co-transfection in GC cell lines. The results showed that overexpression of miR-199a-5p could reverse the inhibitory effect of circ-ITCH on GC cells, confirming our hypothesis. Whereas, the function of miR-199a-5p in GC was mainly reflected in metastasis
, but had no significant effect on proliferation and apoptosis, indicating that circ-ITCH may act partly through miR-199a-5p in GC.
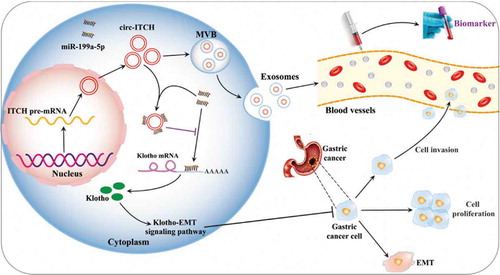
Klotho, an anti-aging factor [Citation28], has been found to function as a tumor suppressor in recent years. Many pathways such as the FGF, IGF-1 R, and Wnt signaling pathways are reported to be involved in the function of Klotho [Citation29]. Klotho can inhibit IGF-1/insulin pathways and regulate the expression of Bax/Bcl-2 to suppress growth and promote apoptosis in A549 cells [Citation30]. Klotho is the downstream target gene of miR-199a-5p [Citation20,Citation31]. To further explore how miR-199a-5p regulates in GC, we verified whether circ-ITCH/miR-199a-5p/Klotho pathway regulated in the progression of GC cells. Our results showed that the expression of Klotho was decreased after circ-ITCH knockdown, but increased after circ-ITCH overexpression. However, the level of Klotho was reversed after miR-199a-5p co-transfection. In short, the increase and decrease of Klotho expression were consistent with circ-ITCH. It is well known that Klotho-mediated regulation of cell EMT is a way to regulate tumor progression [Citation32]. In our study, the expression of EMT-related proteins was also changed. The level of E-cad was increased while N-cad, Twist and Slug were decreased when circ-ITCH was overexpressed and these results could be reversed after miR-199a-5p mimics transfection. However, circ-ITCH knockdown had opposite effects. Taken together, circ-ITCH increases the expression of Klotho by acting as mir-199a-5p sponge, and eventually affects the progression of GC through the EMT pathways ().
In conclusion, we have demonstrated that circ-ITCH is significantly down-regulated in GC tissues, serum exosomes of GC patients, and low circ-ITCH expression is related to invasion depth of GC. Circ-ITCH overexpression inhibits the proliferation, migration and invasion of GC cells. Mechanically, circ-ITCH acts as the sponge of miR-199a-5p and regulates the miR-199a-5p/Klotho axis, which can affect the EMT process of GC. Our findings suggest that circ-ITCH plays a critical role in the progression of gastric cancer and serves as a potential biomarker.
Ethics declarations
All patients gave their written informed consent to participate in this study. And the study was approved by the Medical Ethics Committee of Jiangsu University ((IRB protocol number: 2,020,161).
Supplemental Material
Download PDF (8.6 MB)Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
- Nie Y, Wu K, Yu J, et al. A global burden of gastric cancer: the major impact of China. Expert Rev Gastroenterol Hepatol. 2017;11:651–661.
- Shah MA. Gastrointestinal cancer: targeted therapies in gastric cancer-the dawn of a new era. Nat Rev Clin Oncol. 2014;11:10–11.
- Song Z, Wu Y, Yang J, et al. Progress in the treatment of advanced gastric cancer. Tumour Biol. 2017;39:1010428317714626.
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157.
- Dong W, Dai ZH, Liu FC, et al. The RNA-binding protein RBM3 promotes cell proliferation in hepatocellular carcinoma by regulating circular RNA SCD-circRNA 2 production. EBioMedicine. 2019;45:155–167.
- Zhang X, Wang S, Wang H, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18:20.
- Xie M, Yu T, Jing X, et al. Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR-582-3p/HUR/VEGF axis and suppressing HSP90 degradation. Mol Cancer. 2020;19:112.
- Chen J, Li Y, Zheng Q, et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer Lett. 2017;388:208–219.
- Yang C, Yuan W, Yang X, et al. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol Cancer. 2018;17:19.
- Wang X, Wang R, Wu Z, et al. Circular RNA ITCH suppressed prostate cancer progression by increasing HOXB13 expression via spongy miR-17-5p. Cancer Cell Int. 2019;19:328.
- Lin C, Xu X, Yang Q, et al. Circular RNA ITCH suppresses proliferation, invasion, and glycolysis of ovarian cancer cells by up-regulating CDH1 via sponging miR-106a. Cancer Cell Int. 2020;20:336.
- Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–691.
- Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. 2016;32:309–316.
- Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227.
- Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314.
- Chen J, Shin VY, Siu MT, et al. miR-199a-5p confers tumor-suppressive role in triple-negative breast cancer. BMC Cancer. 2016;16:887.
- Li Y, Wang D, Li X, et al. MiR-199a-5p suppresses non-small cell lung cancer via targeting MAP3K11. J Cancer. 2019;10:2472–2479.
- Wang C, Ba X, Guo Y, et al. MicroRNA-199a-5p promotes tumour growth by dual-targeting PIAS3 and p27 in human osteosarcoma. Sci Rep. 2017;7:41456.
- He XJ, Ma YY, Yu S, et al. Up-regulated miR-199a-5p in gastric cancer functions as an oncogene and targets klotho. BMC Cancer. 2014;14:218.
- Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984.
- Zhang X, Xu Y, Qian Z, et al. circRNA_104075 stimulates YAP-dependent tumorigenesis through the regulation of HNF4a and may serve as a diagnostic marker in hepatocellular carcinoma. Cell Death Dis. 2018;9:1091.
- Zhang H, Zhu L, Bai M, et al. Exosomal circRNA derived from gastric tumor promotes white adipose browning by targeting the miR-133/PRDM16 pathway. Int J Cancer. 2019;144:2501–2515.
- Li Y, Ge YZ, Xu L, et al. Circular RNA ITCH: A novel tumor suppressor in multiple cancers. Life Sci. 2020;254:117176.
- Ghasemi S, Emadi-Baygi M and Nikpour P. Down-regulation of circular RNA ITCH and circHIPK3 in gastric cancer tissues. Turk J Med Sci. 2019;49:687–695.
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388.
- Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66.
- Kuro-o M, Matsumura Y, Aizawa H, et al. Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature. 1997;390:45–51.
- Zhou X, Wang X. Klotho: a novel biomarker for cancer. J Cancer Res Clin Oncol. 2015;141:961–969.
- Chen B, Wang X, Zhao W, et al. Klotho inhibits growth and promotes apoptosis in human lung cancer cell line A549. J Exp Clin Cancer Res. 2010;29:99.
- Wu C, Lv C, Chen F, et al. The function of miR-199a-5p/Klotho regulating TLR4/NF-kappaB p65/NGAL pathways in rat mesangial cells cultured with high glucose and the mechanism. Mol Cell Endocrinol. 2015;417:84–93.
- Chen B, Huang S, Pisanic Ii TR, et al. Rab8 GTPase regulates Klotho-mediated inhibition of cell growth and progression by directly modulating its surface expression in human non-small cell lung cancer. EBioMedicine. 2019;49:118–132.