ABSTRACT
Hepatocellular carcinoma (HCC) is generally known as one of the most common cancers in the world. Nowadays, interventional therapies such as transcatheter arterial chemoembolization (TACE) have emerged as an efficient therapy for HCC patients. Accumulating evidence has unveiled that long non-coding RNAs (lncRNAs) are crucial regulators in HCC progression. Nonetheless, the biological function of lncRNA zinc finger and SCAN domain containing 16 antisense RNA 1 (ZSCAN16-AS1) in HCC has not been systematically clarified. RT-qPCR was used to test ZSCAN16-AS1 expression in HCC cells. The biological functions of RP11-757 G1.5 on HCC cell proliferation, migration, invasion and apoptosis were investigated by colony formation, EdU, CCK-8 and transwell assays, as well as flow cytometry analysis. RNA immunoprecipitation (RIP), RNA pull-down and luciferase reporter assays were utilized to explore the specific mechanism of ZSCAN16-AS1. ZSCAN16-AS1 was significantly up-regulated in HCC cells. ZSCAN16-AS1 silence inhibited HCC cell proliferation, migration and invasion, while it accelerated HCC cell apoptosis. ZSCAN16-AS1 worked as a competing endogenous RNA (ceRNA) to regulate sperm associated antigen 9 (SPAG9) expression through sponging miR-181 c-5p. Moreover, SPAG9 could activate the c-Jun-N-terminal kinase (JNK) pathway. Taken together, our study elucidated that ZSCAN16-AS1 expedited HCC progression via modulating the miR-181 c-5p/SPAG9 axis to activate the JNK pathway, which might be a highly potential HCC therapy and treatment target.
Introduction
Hepatocellular carcinoma (HCC) is the sixth commonest cancer globally and is also the third major threat of cancer-related death [Citation1,Citation2]. Reviewing published research work, tremendous advances have been achieved in the diagnosis and treatment of HCC, including transcatheter arterial chemoembolization (TACE) therapy [Citation3]. Compared to traditional therapies, TACE is a minimally invasive approach in interventional radiology, which can limit the blood supply of tumor [Citation4]. Nevertheless, the five-year survival rate of HCC patients remains low [Citation5]. The main reasons for the poor prognosis of HCC patients are cancer recurrence and the metastasis of primary tumors in the remaining liver [Citation6]. Cancer metastasis is a complex process which involves multiple genetic changes [Citation7]. Thus, more knowledge of the molecular mechanism in the progression of HCC may be helpful to explore novel and effective diagnosis and treatment approaches for HCC.
Recently, due to the rapid development progress of microarray technology and genome sequencing technology, non-coding RNAs (ncRNAs) which are initially defined as transcriptional noise have emerged as a research hotspot [Citation8]. Among which, long non-coding RNAs (lncRNAs) are a group of ncRNAs with more than 200 transcripts [Citation9]. Although lncRNAs have limited potential to code proteins, they can regulate gene expression through various ways, such as epigenetic regulation, transcriptional regulation and post-transcriptional regulation [Citation10]. Recent studies have indicated that lncRNAs are closely correlated with many human cancers, by regulating cell proliferation, apoptosis and migration of cancers [Citation11]. It should be noted, a growing body of lncRNAs has been discovered to be aberrantly expressed in HCC and play critical roles in HCC progression [Citation12]. In addition, lncRNAs have been proved to affect gene expression through functioning as competing endogenous RNAs (ceRNAs) to sponge microRNAs (miRNAs) [Citation13]. LncRNA zinc finger and SCAN domain containing 16 antisense RNA 1 (ZSCAN16-AS1) is a brand new lncRNA, and its biological roles in cancers have never been reported. Hence, this study would focus on ZSCAN16-AS1 to dig into its regulatory mechanism in HCC.
Sperm-associated antigen 9 (SPAG9) as a new member of the cancer testis (CT) antigen family has recently been uncovered to act as an oncogenic gene in a variety of cancers, including HCC [Citation14,Citation15]. Many reports have suggested that SPAG9 is closely related to the acceleration of mitogen-activated protein kinases (MAPK) signaling pathway in cancers [Citation16]. MAPK signaling pathway mainly contains three pathways including extracellular signal-regulated kinase (ERK), c-Jun-N-terminal kinase (JNK), and p38 [Citation17]. SPAG9 has been reported to activate the JNK signaling pathway in HCC [Citation18]. However, the relation between SPAG9 and lncRNAs in HCC remains unclear.
In this research, we aimed to dig into the regulatory functions and potential mechanism of ZSCAN16-AS1 in HCC. Meanwhile, the correlation between ZSCAN16-AS1 and SPAG9 was also explored. Our findings may offer a fire-new insight into the mechanisms underlying HCC carcinogenesis.
Material and methods
Cell culture
HCC cells (Hep3B, HCCLM3 and Huh-7) were purchased from Procell (Wuhan, China). Human immortalized liver cells (THLE-3) were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Hep3B cells were grown in Eagle’s Minimum Essential Medium (ATCC; 30-2003). HCCLM3 and Huh-7 cells were grown in a DMEM medium (Hyclone, SH30022.01). THLE-3 cells were maintained in BEGM medium (Lonza/Clonetics Corporation, Walkersville). For cell culture, all mediums were added with 10% fetal bovine serum (FBS, Gibco, 10,270–106) and placed in a humid incubator with 5% CO2 at 37 °C.
Quantitative real-time polymerase chain reaction (RT-qPCR)
TRIzol reagent (Invitrogen, 15596018) was used to isolate total RNAs from HCC cells. Total RNAs were reverse transcribed into cDNA with the help of a RevertAid First Strand cDNA Synthesis Kit (Sigma, 11483188001). QPCR was operated by using SYBR Green PCR Master Mix (Abcam, G013-dye). Calculation of gene expression was based on the 2−ΔΔCt method, which normalized to GAPDH or U6. Three experiments were performed.
Cell transfection
The two shRNAs against ZSCAN16-AS1 and SPAG9 were purchased from Gene Pharma (Shanghai, China), and sh-NC was used as negative controls. MiR-181 c-5p mimics, miR-181 c-5p inhibitors and pcDNA3.1-SPAG9 were acquired from Gene Pharma as well. All plasmid transfections were used Lipofectamine 3000 (Thermo Fisher, L3000015).
Colony formation
Cells (500 cells per well) were seeded in si-well plates, and then cultured for 14 days. The cells were then fixed with 4% paraformaldehyde and stained with 0.5% crystal violet. Subsequently, the number of stained cells was counted. Three experiments were performed.
5-Ethynyl-2’-deoxyuridine (EdU)
The cells were put into a 96-well plate before fixation and permeation. Then, the EdU reagent (KeyGEN BioTECH, Nanjing, China, KGA337-100) was used to stain cells based on the instructions. After fixation and staining, the images were captured using a fluorescence microscope based on the instructions. DAPI was employed to stain the cell nucleus. Three experiments were performed.
Cell counting kit-8 (CCK-8)
CCK-8 assay was operated to test the HCC cell proliferation capabilities. Prior to using CCK-8 Kit (MCE, 66,147), the transfected cells seeded onto 96-well plates which were maintained in the incubator with 5% CO2. The values of optical density were detected to reflect cell viability. Three experiments were performed.
Transwell
Invasion ability of cells was tested with the help of 24-well transwell chamber. The apical chamber was covered with Matrigel (Corning, 356,234) and added with serum-free medium. A total of 700 μL medium containing 10% FBS was put into the basolateral chamber. After 24–hour incubation, cells in the upper layer of the membrane were wiped off and those in the lower layer of the membrane were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet. The images were observed using a microscope. Cell migration assay was also measured with the same procedures, but the apical chamber was not coated with Matrigel. Three experiments were performed.
Flow cytometry analysis
The Annexin V-FITC Apoptosis Kit (Thermo Fisher, USA, V13242) was utilized in this experiment to demonstrate the apoptosis of HCCLM3 and Huh-7 cells. Supplier guidance was strictly followed. After being rinsed with PBS for two times, the transfected cells were stained by Annexin V-FITC (Beyotime Biotechnology, Shanghai, C1062L) for 10 min in a dark room. The images were observed via flow cytometry (Thermo Fisher, USA, M20036). The experiments were conducted in triplicate.
Xenograft tumor model
Four- to six-week-old BALB/c-nude mice were bought from Henan Provincial People’s Hospital. The procedures of experiments were approved by Henan Provincial People’s Hospital. The mice were stochastically divided into two groups. The HCC cells were first transfected with sh-NC and sh-ZSCAN16-AS1, respectively, and then injected subcutaneously into the right flank of nude mice. The weight and volume were monitored every 5 days. Volume = (length × width [Citation2])/2. A month later, tumors were excised from the mice sacrificed and the tumor images were shot.
Subcellular fractionation
The nuclear or cytoplasmic fractions in cells were separated with the help of a PARIS Kit (Thermo Fisher, USA, AM1921). Expression levels of ZSCAN16-AS1, GAPDH and U6 in nuclear and cytoplasm fractions were severally measured via RT-qPCR. The experiments were performed in triplicates.
Fluorescent in situ hybridization (FISH)
Cy3-labeled ZSCAN16-AS1 probe was bought from RiboBio (Guangzhou, China). The subcellular localization of ZSCAN16-AS1 was determined via the FISH Kit (Thermo Fisher, USA, F32954) conforming to manufacturer’s suggestions. Three experiments were performed.
RNA immunoprecipitation (RIP)
Transfected cells were lysed and centrifuged. Ago2 antibody containing beads was added for rotary incubation overnight at 4 °C. The mixture was washed and the purified RNA was then subjected to RT-qPCR analysis. Three experiments were repeated.
RNA pull-down assay
ZSCAN16-AS1, miR-181 c-5p wild-type (WT) and mutant (MUT) were labeled with biotin and transfected into cells, respectively. The cell lysates were then hatched with streptavidin-coupled magnetic beads at 4 °C. After being washed, the pull-down RNAs were drawn and analyzed through RT-qPCR. Three experiments were performed.
Luciferase reporter assay
Wild-type or mutant sequences toward ZSCAN16-AS1 and SPAG9 on miR-181 c-5p binding sites were cloned into pmirGLO dual luciferase vectors (Promega). The vectors were then all co-transfected with miR-181 c-5p mimics or NC mimics into cells through the Lipofectamine 3000 (Thermo Fisher, USA). The activity of the Renilla plasmid (Promega) was detected via Dual-Luciferase Reporter Assay Kit (Promega, E2920). The experiments were performed in triplicates.
Western blot
Protein was drawn from cells and transferred to PVDF membranes (Immobilon-P, IPVH00010, 0.45um) after 10% SDS-PAGE (Sangon Biotech, C671103-0125). The membranes were then blocked with 5% skimmed milk and incubated with primary antibodies against p-JNK (Abcam, 1/1000), JNK (Abcam, 1/1000), c-Jun (Abcam, 1/1000), MMP9 (Abcam, 1/1000) and GAPDH (Abcam, 1/1000) for the duration of a night at 4 °C. After being washed, membranes were incubated with horseradish peroxidase-labeled secondary antibody for 2 hours at room temperature. The protein levels were measured after adding ECL luminous solution. Three experiments were performed.
Statistical analyses
The data were shown as the mean ± standard deviation and analyzed through GraphPad Prism 6.0. Comparisons of data were analyzed by Student’s t-test, one-way analysis of variance (ANOVA) or two-way ANOVA. When the P value was less than 0.05, the differences was regarded as statistical significance. Three experiments were performed.
Results
ZSCAN16-AS1 facilitates proliferation, migration and invasion and suppresses apoptosis of HCC cells
To disclose the expression of a novel lncRNA ZSCAN16-AS1 in HCC, we analyzed its expression in liver hepatocellular carcinoma (LIHC) through GEPIA 2 website (http://gepia2.cancer-pku.cn). As presented in ), the expression of ZSCAN16-AS1 in 369 cases of LIHC tissues was dramatically higher than that in 160 cases of non-tumor tissues. Additionally, ZSCAN16-AS1 expression was also upregulated in HCC cells (Hep3B, HCCLM3 and Huh-7) compared to human immortalized liver cells (THLE-3) through RT-qPCR analysis ()). We next explored the biological role of ZSCAN16-AS1 in HCC cell growth. ZSCAN16-AS1 was stably depleted in HCCLM3 and Huh-7 cells with two specific shRNAs ()). It was observed in colony formation assays that ZSCAN16-AS1 knockdown definitely inhibited the proliferation capability of HCCLM3 and Huh-7 cells ()). Similar results were evidenced through EdU and CCK-8 assays () and S1A). Moreover, the data from transwell assays revealed that ZSCAN16-AS1 silence repressed migration and invasion capacities of HCCLM3 and Huh-7 cells ()). Subsequently, flow cytometry analysis was done to assess the apoptosis of HCCLM3 and Huh-7 cells. As presented in Figure S1B, the silence of ZSCAN16-AS1 hindered apoptosis of these HCC cells. In addition, in-vivo experiments were performed on the mice injected with transfected HCC cells. The obtained data indicated that silence of ZSCAN16-AS1 suppressed HCC tumor growth in naked mice (Figure S1C-1D). Collectively, ZSCAN16-AS1 facilitated proliferation, migration and invasion and hampered apoptosis of HCC cells.
Figure 1. ZSCAN16-AS1 facilitates proliferation, migration and invasion and suppresses apoptosis of HCC cells. A) Analysis of ZSCAN16-AS1 expression in LIHC with the help of GEPIA 2 database. B) Expression of ZSCAN16-AS1 in HCC cells (Hep3B, HCCLM3 and Huh-7) and THLE-3 cells from RT-qPCR analysis. C) Expression of ZSCAN16-AS1 in HCCLM3 and Huh-7 cells after shRNAs transfection. D-E) Effects of ZSCAN16-AS1 silence on proliferation of HCCLM3 and Huh-7 cells were measured through colony formation and EdU assays. F-G) Effects of ZSCAN16-AS1 downregulation on HCC cell migration and invasion capacities were measured through transwell assays. *P < 0.05, **P < 0.01
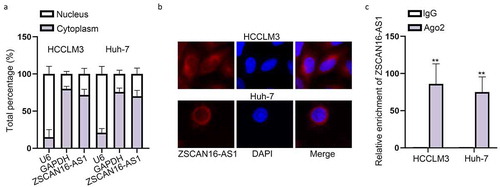
ZSCAN16-AS1 acts as a ceRNA to exert functions in HCC cells
With the aim of deciphering the potential mechanism of ZSCAN16-AS1 in HCC, we adopted subcellular fractionation and FISH assays to detect the cellular localization of ZSCAN16-AS1 in HCC cells. The results revealed that ZSCAN16-AS1 was majorly located in the cytoplasm of HCCLM3 and Huh-7 cells ()), mirroring the post-transcriptional role of ZSCAN16-AS1 in HCC progression. It has been well documented that lncRNAs can act as ceRNAs to interact with miRNAs in the development of tumors [Citation19]. Therefore, to measure the ability of ZSCAN16-AS1 to sponge miRNAs, the anti-Ago2 immunoprecipitates involving miRNAs and their interacting RNA-components were employed in HCCLM3 and Huh-7 cells for RIP assays [Citation20]. The data suggested that the amount of endogenous ZSCAN16-AS1 pulled-down via Ago2 antibody was detected by RT-qPCR and significantly higher than negative control group using IgG antibody ()), which indicated that ZSCAN16-AS1 may act as a ceRNA to sponge miRNAs. Taken together, in the next section, we tried to figure out the way ZSCAN16-AS1 as a ceRNA exerted its functions in HCC cells.
ZSCAN16-AS1 interacts with miR-181 c-5p in HCC cells
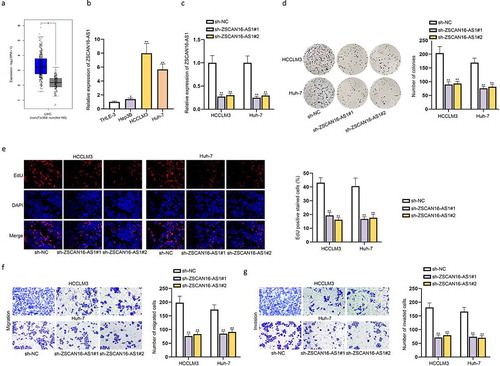
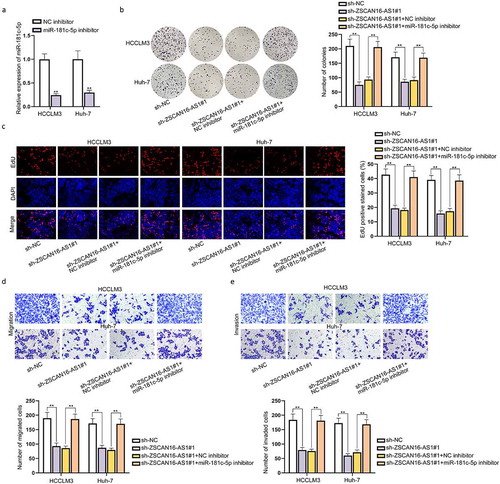
According to the starBase (http://starbase.sysu.edu.cn) website, we filtered nine potential miRNAs possibly combining with ZSCAN16-AS1 under the condition of CLIP Data ≥5 ()). We then performed RNA pull-down assays to further select the genuine miRNA interacting with ZSCAN16-AS1 in HCC cells. The data showed a large enrichment of miR-181 c-5p in the ZSCAN16-AS1 pull-down pellet compared with the control group, while no prominent variation was observed among other miRNAs in the ZSCAN16-AS1 pull-down pellet ()). Thus, we further analyzed the expression of miR-181 c-5p in HCC cells. The results displayed that miR-181 c-5p expression was down-regulated in HCC cells relative to THLE-3 cells ()). Subsequently, we overexpressed miR-181 c-5p ()) and performed gain-of-function assays to probe the role of miR-181 c-5p in HCC cell growth. Through colony formation, EdU and CCK-8 assays, we confirmed that miR-181 c-5p mimics hampered proliferation of HCCLM3 and Huh-7 cells (Figure S2A-S2C). At the same time, the cell migration and invasion were also impeded when miR-181 c-5p was overexpressed in HCCLM3 and Huh-7 cells (Figure S2D-S2E). Moreover, the outcomes of flow cytometry analysis revealed that upregulation of miR-181 c-5p promoted apoptosis of these HCC cells (Figure S2F). All these data suggested that miR-181 c-5p functioned as a tumor inhibitor in HCC progression. Furthermore, the presumptive miR-181 c-5p binding sites on ZSCAN16-AS1 were predicted by the starBase website and shown in ). In order to examine the effectiveness of these binding sites between miR-181 c-5p and ZSCAN16-AS1, we constructed luciferase reporter plasmids containing wild-type ZSCAN16-AS1 binding sites or its mutant bearing mutation in the miR-181 c-5p seed sequence, and co-transfected with miR-181 c-5p mimics into HCCLM3 and Huh-7 cells. The outcomes showed that miR-181 c-5p could bind to wild-type ZSCAN16-AS1 rather than its mutant form and the predicted binding sites of ZSCAN16-AS1 were confirmed as well ()). In conclusion, ZSCAN16-AS1 could interact with miR-181 c-5p in HCC cells.
Figure 3. ZSCAN16-AS1 interacts with miR-181 c-5p in HCC cells. A) A list of possible miRNAs combining with ZSCAN16-AS1 were predicted through starBase website. B) Enrichment of nine miRNAs in bio-ZSCAN16-AS1 groups from RNA pull down assays. C) Expression of miR-181 c-5p in HCC cells (Hep3B, HCCLM3 and Huh-7) and THLE-3 cells was quantified via RT-qPCR analysis. D) Expression of miR-181 c-5p in HCCLM3 and Huh-7 cells were detected using RT-qPCR after transfection of miR-181 c-5p mimics. E) Binding sites between ZSCAN16-AS1 and miR-181 c-5p from starBase website. F) Luciferase reporter assay results in HCCLM3 and Huh-7 cells co-transfected with miR-181 c-5p mimics and ZSCAN16-AS1-WT/MUT. **P < 0.01
ZSCAN16-AS1 regulates malignant phenotypes of HCC cells by decoying miR-181 c-5p
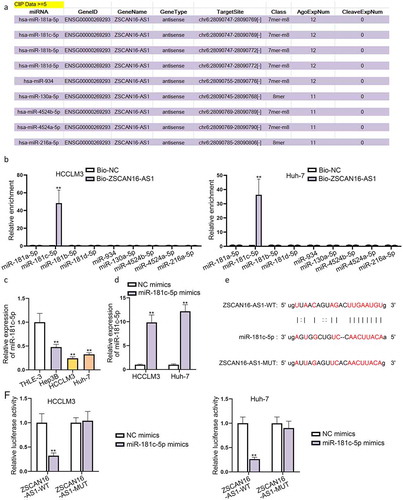
To determine the interaction between ZSCAN16-AS1 and miR-181 c-5p and their effects on HCC cell growth, we first tested the efficiency of miR-181 c-5p downregulation through RT-qPCR ()). Then, we separately transfected sh-NC, sh-ZSCAN16-AS1#1, sh-ZSCAN16-AS1#2+ NC inhibitor and sh-ZSCAN16-AS1#2+ miR-181 c-5p inhibitor into HCCLM3 and Huh-7 cells to perform a series of rescue assays. Colony formation, EdU and CCK-8 assays revelaed that downregulation of miR-181 c-5p weakened the suppressive role of ZSCAN16-AS1 knockdown on the proliferation capacity of HCCLM3 and Huh-7 cells () and S3A). Transwell assays confirmed that the lessened migration and invasion capabilities of HCCLM3 and Huh-7 cells caused by ZSCAN16-AS1 deficiency could be reversed after co-transfection of miR-181 c-5p inhibitor ()). Based on the outcomes of flow cytometry analysis, we concluded that enhanced apoptosis capabilities of these HCC cells caused by ZSCAN16-AS1 knockdown could be weakened after adding miR-181 c-5p inhibitor (Figure S3B). Taken together, ZSCAN16-AS1 regulated malignant phenotypes of HCC cells by competitively bind to miR-181 c-5p.
Figure 4. ZSCAN16-AS1 regulates malignant phenotypes of HCC cells by decoying miR-181 c-5p. A) Expression of miR-181 c-5p in HCCLM3 and Huh-7 cells was detected through RT-qPCR after transfection of miR-181 c-5p inhibitor. B-C) Rescue experiments were performed in HCCLM3 and Huh-7 cells transfected with sh-NC, sh-ZSCAN16-AS1#1, sh-ZSCAN16-AS1#1+ NC inhibitor and sh-ZSCAN16-AS1#1+ miR-181 c-5p inhibitor. Colony formation and EdU assays measured the proliferation of HCCLM3 and Huh-7 cells. D-E) Transwell assays measured the migration and invasion of HCCLM3 and Huh-7 cells. **P < 0.01
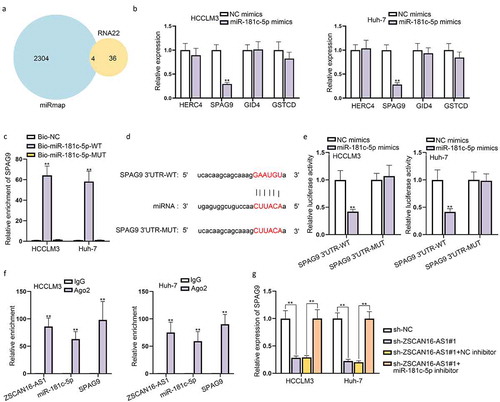
ZSCAN16-AS1 acts as a ceRNA to sponge miR-181 c-5p and further modulates SPAG9 expression
It has been reported that miRNAs function at the post-transcriptional level by usually base-pairing to the 3’-untranslated regions (UTR) of mRNAs to inhibit protein synthesis [Citation21]. Therefore, to ascertain the detailed regulatory mechanism of ZSCAN16-AS1 in HCC, we searched miRmap and RNA22 programs, and four mRNAs were predicted as downstream targets of miR-181 c-5p ()). Subsequently, we performed RT-qPCR to quantify the expression of these four mRNAs in HCCLM3 and Huh-7 cells which were transfected with miR-181 c-5p mimics, and found that only SPAG9 expression significantly declined when miR-181 c-5p increased ()). In addition, we found that SPAG9 was highly enriched in bio-miR-181 c-5p-WT groups compared to bio-NC and bio-miR-181 c-5p-MUT groups ()). Moreover, the binding sites between SPAG9 3’UTR and miR-181 c-5p were projected with the help of starBase database (), and the following luciferase reporter assays further validated that miR-181 c-5p up-regulation made a remarkable decrease of the luciferase activity in Group SPAG9 3’UTR-WT while had almost no change in GroupSPAG9 3’UTR-MUT ()). Additionally, RIP assays further validated the co-existence of ZSCAN16-AS1, miR-181 c-5p and SPAG9 in the RNA-induced silencing complex (RISC), as shown by the high combination of these three RNAs and Ago2, which verified the presence of ceRNA network among these three RNAs ()). More importantly, we found that SPAG9 expression was significantly repressed when ZSCAN16-AS1 was knocked down, and this repressive effect was offset after adding miR-181 c-5p inhibitor ()). To summarize, ZSCAN16-AS1 served as a ceRNA to competitively bind to miR-181 c-5p and further modulated SPAG9 expression.
Figure 5. ZSCAN16-AS1 acts as a ceRNA to sponge miR-181 c-5p and further regulates SPAG9 expression. A) Possible mRNAs combined with miR-181 c-5p from RNA22 and miRmap programs. B) Expression of SPAG9 in HCCLM3 and Huh-7 cells after miR-181 c-5p increase from RT-qPCR analysis. C) Combination between SPAG9 and miR-181 c-5p from RNA pull down assays. D) Binding sites between SPAG9 3’UTR and miR-181 c-5p were obtained from starBase website. E) Luciferase activities of SPAG9 3’UTR-WT/MUT were detected in HCCLM3 and Huh-7 cells after miR-181 c-5p overexpression. F) RIP assays detected the enrichment of ZSCAN16-AS1, miR-181 c-5p and SPAG9 in Ago2. G) Expression of SPAG9 in HCCLM3 and Huh-7 cells transfected with sh-NC, sh-ZSCAN16-AS1#1, sh-ZSCAN16-AS1#1+ NC inhibitor and sh-ZSCAN16-AS1#1+ miR-181 c-5p inhibitor from RT-qPCR analysis. **P < 0.01
SPAG9 activates JNK pathway which may affect malignant behaviors of HCC cells
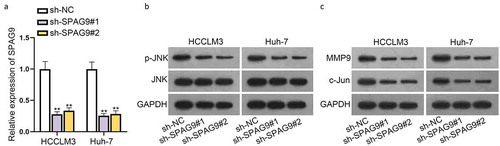
It has been reported that SPAG9 can regulate the JNK activity [Citation18]. Herein, we also silenced the expression of SPAG9 ()) and used western blot to detect the impacts of SPAG9 on JNK activation. We discovered that the level of p-JNK protein dropped in HCCLM3 and Huh-7 cells after SPAG9 deletion ()). We also measured the JNK downstream target gene levels after SPAG9 deletion, such as MMP9 and c-Jun. We confirmed that SPAG9 repression caused a considerable decrease of c-Jun and MMP9 expression ()). According to previous research work, JNK has been reported to correlate with the multiple malignant behaviors of cancer cells [Citation22]. In this study, to figure out the effect of JNK on HCC cells, we first tested the efficiency of JNK depletion (Figure S4A). Then, CCK-8 and transwell assays were implemented to test the proliferation, migration and invasion capacities of HCCLM3 and Huh-7 cells. As presented in Figure S4B-4D, we concluded that JNK depletion could hamper malignant behaviors of these HCC cells. Aside from that, flow cytometry analysis was done and the collected data indicated that HCC cell apoptosis was promoted due to JNK depletion (Figure S4E). Overall, these data suggested that SPAG9 could activate the JNK pathway in HCC cells and JNK could further exert influences on malignant behaviors of HCC cells.
Figure 6. SPAG9 activates JNK pathway which may affect malignant behaviors of HCC cells. A) Expression of SPAG9 in HCCLM3 and Huh-7 cells after shRNAs transfection. B-C) Protein levels of p-JNK, JNK, MMP9 and c-Jun were measured in HCCLM3 and Huh-7 cells after SPAG9 deletion using western blot. **P < 0.01
ZSCAN16-AS1 regulates SPAG9 to activate the JNK pathway and facilitate HCC cell growth
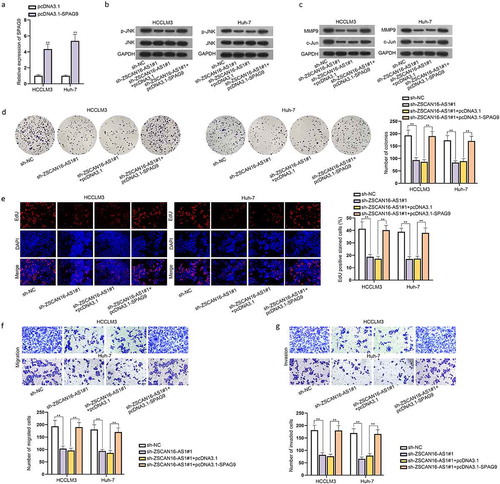
Before exploring the regulatory relationship between ZSCAN16-AS1 and SPAG9, we searched on GEPIA 2 database to find out the correlation of these two genes (Figure S5A). To test whether ZSCAN16-AS1 modulated SPAG9 to activate the JNK pathway and affect HCC cell growth, we first examined the efficiency of SPAG9 overexpression ()). Then, we transfected sh-NC, sh-ZSCAN16-AS1#1, sh-ZSCAN16-AS1#1+ pcDNA3.1 and sh-ZSCAN16-AS1#1+ pcDNA3.1-SPAG9 into HCCLM3 and Huh-7 cells to perform rescue experiments, respectively. Based on western blot data, we found that ZSCAN16-AS1 silence led to an apparent decline of the protein levels of p-JNK, MMP9 and c-Jun, while this suppressive effect was reversed after co-transfection of pcDNA3.1-SPAG9 ()). Subsequently, we performed colony formation, EdU and CCK-8 assays, finding that SPAG9 upregulation restored the reduced proliferation ability of HCCLM3 and Huh-7 cells caused by ZSCAN16-AS1 silence () and S5B). Meanwhile, the outcomes indicated that migration and invasion capacities of HCCLM3 and Huh-7 cells were suppressed after ZSCAN16-AS1 depletion and were further neutralized by overexpressing SPAG9 ()). The promoted apoptosis of these HCC cells led by ZSCAN16-AS1 silence was recovered by overexpressing SPAG9 (Figure S5C). In summary, ZSCAN16-AS1 regulated SPAG9 to activate the JNK pathway and facilitate HCC cell growth.
Figure 7. ZSCAN16-AS1 regulates SPAG9 to activate the JNK pathway and facilitate HCC cell growth. A) Expression of SPAG9 in HCCLM3 and Huh-7 cells was detected using RT-qPCR after transfection of pcDNA3.1-SPAG9. B-C) Rescue experiments were performed in HCCLM3 and Huh-7 cells transfected with sh-NC, sh-ZSCAN16-AS1#1, sh-ZSCAN16-AS1#1+ pcDNA3.1 and sh-ZSCAN16-AS1#1+ pcDNA3.1-SPAG9. Western blot was performed to detect the protein levels of p-JNK, MMP9, JNK and c-Jun in HCCLM3 and Huh-7 cells under different conditions. D-E) Colony formation and EdU assays measured the proliferation of HCCLM3 and Huh-7 cells. F-G) Transwell assays measured the migration and invasion of HCCLM3 and Huh-7 cells. **P < 0.01
Discussion
Accumulating evidences have demonstrated that lncRNAs are tumor-associated biological molecules which can function as oncogenes or tumor inhibitors to take part in the progression of HCC. LncRNA CDKN2BAS has been identified to correlate with HCC metastasis and poor prognosis in HCC patients and has been proven to engage in miR-153-5p/ARHGAP18 axis [Citation23]. LncRNA SNHG8 accelerates the tumorigenesis and indicates tumor recurrence in HCC via targeting miR-149-5p [Citation24]. Moreover, lncRNA RUNX1-IT1 suppresses HCC cell proliferation via modulating the MAPK pathway [Citation25]. In this study, a novel lncRNA ZSCAN16-AS1 was discovered, which was seen to be highly expressed in HCC cells. The relevant loss-of-function assays unveiled that subdual of ZSCAN16-AS1 obviously restricted proliferation, migration and invasion of HCC cells in vitro and facilitated HCC cell apoptosis. These results suggested the essential roles of ZSCAN16-AS1 in HCC.
LncRNAs have been well documented to exert their biological functions through ceRNAs mode [Citation26]. The lncRNA-miRNA-mRNA axis has been widely reported in the HCC progression [Citation27]. This research attempted to verify the possible molecular mechanism of ZSCAN16-AS1 in HCC cells. Based on previous studies, miR-181 c-5p has been identified to play crucial roles in several types of cancers, such as pancreatic cancer [Citation28] and cervical squamous cell carcinoma [Citation29]. In this study, bioinformatics analysis and mechanism dissection revealed thatmiR-181 c-5p could combine with ZSCAN16-AS1. In our research, we further validated that overexpressing miR-181 c-5p greatly restrained cell proliferation, migration and invasion and promoted cell apoptosis in HCC. More importantly, miR-181 c-5p downregulation could counteract the repressive effects of ZSCAN16-AS1 silence on HCC cell proliferation, migration, invasion and apoptosis. All these data suggested the miR-181 c-5p worked as a tumor in HCC progression.
Based on the ceRNA hypothesis, we further investigated the downstream mRNA targeted by miR-181 c-5p. SPAG9 exerts oncogenic influences on many cellular processes and cancers, which has been reported to be a promising therapeutic target for several cancers [Citation30,Citation31]. SPAG9 is a positive regulator in osteosarcoma progression and is involved in metastasis [Citation32]. SPAG9 can function as a diagnostic biomarker for colorectal cancer [Citation33]. Notably, the overexpression of SPAG9 is correlated with poor prognosis and tumor progression in HCC [Citation34]. In this study, we found that SPAG9 was the target gene of miR-181 c-5p. Moreover, we confirmed that ZSCAN16-AS1 could elevate SPAG9 expression via sponging miR-181 c-5p in HCC cells, which supported the ceRNA hypothesis.
It has been reported that SPAG9 can regulate the JNK signaling pathways to get involved in cancer progression, including HCC [Citation18,Citation35]. The JNK signaling pathway is a part of the MAPK pathway, which has been demonstrated to promote cell proliferation, differentiation and migration in cancers [Citation36]. Moreover, c-Jun and MMP9, two major downstream molecules of the JNK pathway, are important participators in JNK-mediated tumor growth [Citation37]. In our study, we further confirmed that SPAG9 could activate the JNK signaling pathway and SPAG9 silence significantly lessened the protein levels of p-JNK, c-Jun and MMP9. Moreover, our study proved that ZSCAN16-AS1 could regulate SPAG9 to activate the JNK pathway and impact on HCC cell growth.
In conclusion, the regulatory mechanism of ZSCAN16-AS1/miR-181 c-5p/SPAG9/JNK in HCC might be a promising target for HCC treatment. Aside from that, it has been reported that SPAG9 can also activate the p38 MAPK pathway [Citation38]. Thus, the interaction between SPAG9 and the p38 MAPK pathway in HCC remains to be dug into in the future.
Supplemental Material
Download Zip (15.1 MB)Acknowledgement
All supports are sincerely acknowledged.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
Not applicable.
Supplementary material
Supplemental data for this article can be accessed here.
References
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90.
- Bruix J, Sherman M. Management of hepatocellular carcinoma. Hepatology. 2005;42:1208–1236. (Baltimore, Md)
- Chen J, Wu G. Retrospective analysis of Chinese patients with hepatocellular carcinoma (HCC) undergoing transcatheter arterial chemoembolization (TACE) with or without prophylactic antibiotic therapy. J Coll Physicians Surg Pak. 2018;28:914–918.
- Lu W, Jin XL, Yang C, et al. Comparison of efficacy between TACE combined with apatinib and TACE alone in the treatment of intermediate and advanced hepatocellular carcinoma: a single-center randomized controlled trial. Cancer Biol Ther. 2017;18(6):433–438.
- Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362(9399):1907–1917. . (London, England)
- Chen J, Cao SW, Cai Z, et al. Epithelial-mesenchymal transition phenotypes of circulating tumor cells correlate with the clinical stages and cancer metastasis in hepatocellular carcinoma patients. Cancer Biomarkers. 2017;20(4):487–498.
- Wan L, Pantel K, Kang Y. Tumor metastasis: moving new biological insights into the clinic. Nat Med. 2013;19(11):1450–1464.
- Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18(1):5–18.
- Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981.
- Gil N, Ulitsky I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat Rev Genet. 2020;21(2):102–117.
- Chi Y, Wang D, Wang J, et al. Long non-coding RNA in the pathogenesis of cancers. Cells. 2019;8(9):8.
- Zhang H, Liao Z, Liu F, et al. Long noncoding RNA HULC promotes hepatocellular carcinoma progression. Aging (Albany NY). 2019;11(20):9111–9127.
- Bai Y, Long J, Liu Z, et al. Comprehensive analysis of a ceRNA network reveals potential prognostic cytoplasmic lncRNAs involved in HCC progression. J Cell Physiol. 2019;234(10):18837–18848. .
- Yan Q, Lou G, Qian Y, et al. SPAG9 is involved in hepatocarcinoma cell migration and invasion via modulation of ELK1 expression. Onco Targets Ther. 2016;9:1067–1075.
- Xiao C, Fu L, Yan C, et al. SPAG9 is overexpressed in osteosarcoma, and regulates cell proliferation and invasion through regulation of JunD. Oncol Lett. 2016;12(4):2674–2679. .
- Li D, Yang M, Liao A, et al. Linc00483 as ceRNA regulates proliferation and apoptosis through activating MAPKs in gastric cancer. J Cell Mol Med. 2018;22(8):3875–3886. .
- Duan Y, Li J, Jing X, et al. Fucoidan induces apoptosis and inhibits proliferation of hepatocellular carcinoma via the p38 MAPK/ERK and PI3K/Akt signal pathways. Cancer Manag Res. 2020;12:1713–1723.
- Lou G, Dong X, Xia C, et al. Direct targeting sperm-associated antigen 9 by miR-141 influences hepatocellular carcinoma cell growth and metastasis via JNK pathway. J Exp Clin Cancer Res. 2016;35(1):14. .
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352.
- Vm K, Gm B. MicroRNA expression: protein participants in microRNA regulation. Methods Mol Biol. 2017;1617:27–37. (Clifton, NJ)
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79(1):351–379.
- Chen F. JNK-induced apoptosis, compensatory growth, and cancer stem cells. Cancer Res. 2012;72(2):379–386.
- Chen J, Huang X, Wang W, et al. LncRNA CDKN2BAS predicts poor prognosis in patients with hepatocellular carcinoma and promotes metastasis via the miR-153-5p/ARHGAP18 signaling axis. Aging (Albany NY). 2018;10(11):3371–3381.
- Dong J, Teng F, Guo W, et al. lncRNA SNHG8 promotes the tumorigenesis and metastasis by sponging miR-149-5p and predicts tumor recurrence in hepatocellular carcinoma. Cell Physiol Biochem. 2018;51(5):2262–2274.
- Yan PH, Wang L, Chen H, et al. LncRNA RUNX1-IT1 inhibits proliferation and promotes apoptosis of hepatocellular carcinoma by regulating MAPK pathways. Eur Rev Med Pharmacol Sci. 2019;23(19):8287–8294.
- Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272–283.
- Li B, Mao R, Liu C, et al. FAL1 promotes cell proliferation and migration by acting as a CeRNA of miR-1236 in hepatocellular carcinoma cells. Life Sci. 2018;197:122–129.
- Gao ZQ, Wang JF, Chen DH, et al. Long non-coding RNA GAS5 antagonizes the chemoresistance of pancreatic cancer cells through down-regulation of miR-181c-5p. Biomed Pharmacothe. 2018;97:809–817.
- Li N, Cheng C, Wang T. MiR-181c-5p mitigates tumorigenesis in cervical squamous cell carcinoma via targeting glycogen synthase kinase 3β interaction protein (GSKIP). Onco Targets Ther. 2020;13:4495–4505.
- Sun HF, Wang WD, Feng L. Effect of SPAG9 on migration, invasion and prognosis of prostate cancer. Int J Clin Exp Pathol. 2017;10(9):9468–9474.
- Luo S, Ren B, Zou G, et al. SPAG9/MKK3/p38 axis is a novel therapeutic target for liver cancer. Oncol Rep. 2019;41(4):2329–2336.
- Yang X, Zhou W, Liu S. SPAG9 controls the cell motility, invasion and angiogenesis of human osteosarcoma cells. Exp Ther Med. 2016;11(2):637–644.
- Tavakoli Koudehi A, Mahjoubi B, Mirzaei R, et al. AKAP4, SPAG9 and NY-ESO-1 in Iranian colorectal cancer patients as probable diagnostic and prognostic biomarkers. Asian Pac J Cancer Prev. 2018;19(2):463–469.
- Xie C, Fu L, Liu N, et al. Overexpression of SPAG9 correlates with poor prognosis and tumor progression in hepatocellular carcinoma. Tumour Biol. 2014;35(8):7685–7691.
- Li R, Gunarta IK, Suzuki R, et al. JLP-JNK signaling protects cancer cells from reactive oxygen species-induced cell death. Biochem Biophys Res Commun. 2018;501(3):724–730. .
- Xu R, Hu J. The role of JNK in prostate cancer progression and therapeutic strategies. Biomed Pharmacothe. 2020;121:109679.
- Cai J, Du S, Wang H, et al. Tenascin-C induces migration and invasion through JNK/c-Jun signalling in pancreatic cancer. Oncotarget. 2017;8(43):74406–74422.
- Miao ZF, Wang ZN, Zhao TT, et al. Overexpression of SPAG9 in human gastric cancer is correlated with poor prognosis. Virchows Arch. 2015;467(5):525–533.