ABSTRACT
The addition of the methyl-7-guanosine (m7G) “cap” on the 5' ends of coding and some non-coding RNAs is essential for their protein coding capacity and biochemical activity, respectively. It was previously considered that capping was a constitutive process that generates a complete cap on all transcripts at steady-state. However, development of new methodologies demonstrated that steady-state capping is a dynamic and regulatable feature of many coding and non-coding RNAs. Indeed, capping status of specific RNAs can flux during differentiation and development, thereby impacting on their protein-coding capacity and activity. Moreover, in some primary cancer specimens, capping can be elevated for transcripts encoding proteins involved in proliferation and survival corresponding to their increased protein levels. Overexpression of one of the capping enzymes (RNMT), the transcription factor MYC or the eukaryotic translation initiation factor eIF4E all led to increased levels of steady-state capping of selected transcripts. Additionally, transcripts can be decapped and recapped, allowing these to be sequestered until needed. This review provides a summary of the major advances in enzymatic and affinity-based approaches to quantify m7G capping. Further, we summarize the evidence for regulation of capping. Capping has emerged as a significant regulatory step in RNA metabolism which is poised to impact a myriad of biological processes.
Overview
Methyl-7 guanosine (m7G) “cap” addition to the 5' end of coding RNAs and many non-coding transcripts impacts virtually all levels of their processing, which in turn, influences the capacity for coding RNAs to be translated into protein and the biochemical activity of non-coding RNAs [Citation1–3]. For many years, capping was widely regarded as a constitutive property with the expectation that 100% of coding RNAs would be capped. Over the last 20 years, it has become evident that m7G capping is not simply a default housekeeping process, but rather is a highly regulated step in RNA processing that impacts on the biochemical functions of many transcripts [Citation2–10]. These new findings indicate that the extent of capping for a given RNA population varies and this is influenced by development, differentiation or oncogene expression [Citation7,Citation9,Citation11–13]. Moreover, once capped, RNAs can be decapped and then recapped [Citation4,Citation6,Citation14]. In this way, cap removal does not inextricably lead to RNA decay. This has led to the notion of cap homeostasis, whereby the cap status of transcripts can be regulated to impact RNA export, translation, stability etc [Citation4]. Consistent with this model, genome-wide studies suggest that uncapped RNAs are more stable than anticipated and in this way, cap status can be a means to titrate transcript activity as well as capacity to undergo other processing steps. Here, we review the consequences of dynamic capping and mechanisms involved in modulating this pathway. This review focusses on m7G capping, but there are other types of caps and these will be discussed briefly (). This review describes the RNA biochemistry related to capping, recent methodological advances to measure m7G capping and the biological impact of regulating capping.
Diversity in RNA caps
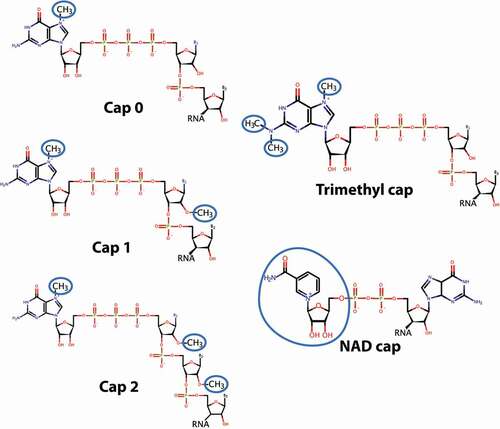
Here, we focus on the m7G cap at the 5' end of transcripts (). This is characterized by a 5'-5' pyrophosphate bond linking the m7G to the first transcribed nucleotide on the 5' end of the transcript [Citation1]. Many of the methods used to quantify capping will capture variants of this m7GpppX form because they are based on identifying 5'-5' pyrophosphate linkages. Examples of these would include unmethylated guanosine caps as well as trimethylated (TMG) caps which are methylated at positions 2, 2 and 7 on the guanosine ring (). The m7G cap, with only one methyl group on the 5' guanosine, is also referred to as Cap-0 (). RNAs can also be methylated at the 2' hydroxyl of the first transcribed nucleotide ribose referred to as Cap-1 (m7GpppXm) and on the same position in the second transcribed nucleotide ribose which is referred to as Cap-2 (m7GpppXmXm) [Citation1]. In addition, more chemically diverse caps have been identified. For instance, NAD substitutes for the m7G cap on some transcripts, particularly mitochondrial RNAs [Citation15] (). Plant viruses may have unique caps made of protein, as observed for the potyvirus protein genome linked (VPg) that is covalently attached to the 5' end of potyviral genomic RNA [Citation16]. Structural studies indicate that VPg directly binds to the cap-binding site of the eukaryotic translation initiation factor eIF4E and can recruit RNAs to the translation machinery via this interaction [Citation16]. Thus, potyvirus VPg engages eIF4E in a manner similar to the m7G cap in vitro. Whether other protein-RNA conjugates can similarly engage eIF4E for RNA export or translation remains to be examined. Also, it has been suggested that some coding RNAs could be TMG capped (), instead of m7G capped, similar to what is observed in nematode TMG mRNAs or human UsnRNAs [Citation17–19]. In this form, some TMG capped coding RNAs are able to bind eIF4E and undergo translation, as is routine in nematodes.
Enzymology of m7G Capping
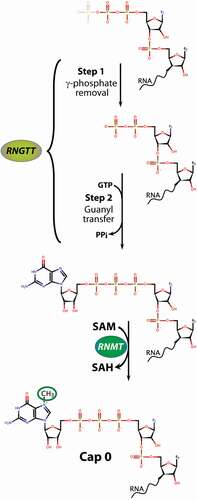
Capping is a three-step process involving RNGTT (RNA guanylyltransferase and 5' phosphatase) and RNMT (RNA guanine-7- methyltransferase) [Citation20] in mammals (). These enzymes are required for capping and cell survival [Citation2,Citation21–27]. RNGTT removes the 5' phosphate of the 5' triphosphate on the pre-mRNA or non-coding RNA using its 5' phosphatase activity [Citation28]. This produces a 5'-diphosphate-RNA which then serves as a RNGTT substrate for addition of guanosine monophosphate via a distinct 5'-5' pyrophosphate linkage. The cap guanylate is then methylated by RNMT, the only enzyme known to carry out this modification [Citation2]. To do this, it uses its methyltransferase domain and S-adenosyl methionine (SAM) as the methyl donor [Citation29,Citation30]. RNMT also binds RAM, a small protein co-factor which increases its methylation activity and can recruit RNAs to the complex [Citation29]. At steady state, localization of RNMT and RNGTT is mainly nuclear; however, these factors have also been identified in the cytoplasm suggesting that capping can occur in either compartment [Citation6,Citation23,Citation31]. In this way, RNAs that are decapped are not always fated to be degraded [Citation32]. Further, while capping is generally considered to be co-transcriptional, the cytoplasmic localization of these enzymes and the observation of re-capping activity in that compartment suggest that re-capping could also occur independent of transcription (discussed in Recapping of RNAs after Decapping section below). Thus, like splicing and polyadenylation [Citation33–35], it appears that capping can occur both co- and post-transcriptionally.
Figure 2. Biochemical pathways for m7G RNA capping. Capping is completed by two enzymes in mammals, RNGTT carries out the removal of the 5' phosphate (Step 1) and subsequent addition of the guanosine via the unique 5'-5' pyrophosphate linage (Step 2). This G-capped RNA is now a substrate for the RNMT, which uses SAM as the methyl donor to generate m7G capped RNA and S-adenosyl homocysteine (SAH). Additional methylation events depicted in are carried out by other enzymes and not described here
Methods to monitor m7G capping of specific transcripts at steady-state
The m7G cap was first identified in viral RNA. In the 1970s, caps were detected by mass spectrometry by digestion and radiolabelling of mRNA [Citation1,Citation36–40]. More recently, advanced nucleotide purification methodologies have resulted in rapid mass spectrometry cap analysis of relatively small samples of RNA, including using a method called CAP-MAP [Citation41,Citation42]. This allows quantitation of different cap structures. Cap modifications have also been detected by resolving radiolabelled nucleotides using two-dimensional thin-layer chromatography (TLC) [Citation43]. These methods of bulk RNA cap quantitation are complimentary to single gene approaches described below. These mass spectrometry approaches have proved invaluable in confirming the impact of genetic targeting of capping enzymes in cell lines and organisms [Citation44].
The development of methodologies to accurately quantify steady-state capping levels on a per transcript basis has provided major new insights into capping and evidence to support the concept of cap homeostasis and dynamic capping. Some methods can quantify fold changes in capping for a given RNA species; while others can provide the percentage of m7G- or G-capped and/or uncapped RNAs. The approaches can be roughly divided into affinity-based or enzymatic methods (). Both strategies identify the RNAs in question using RNA-Seq or RT-qPCR and for older studies, microarrays. Importantly, the approaches differ in the form of cap or uncapped RNAs detected with some approaches offering more general applicability than others. These approaches are summarized in .
Table 1. Methods to study capping on a per RNA basis
Affinity methods developed to date capture RNAs with an m7G group, regardless of methylation at other positions of the cap. These measure steady-state capping and include immunoprecipitation (IP) with anti-m7G cap antibodies in a method known as CapIP or employ pulldown approaches with purified eIF4E, an m7G cap-binding protein [Citation6,Citation9,Citation11,Citation45–47]. To ensure that RNA secondary structure or RNA binding factors do not interfere with binding to the antibody or to eIF4E, both approaches are performed using purified RNA isolated from cells rather than carrying out studies directly from cell lysates [Citation9,Citation29,Citation48]m7G cap antibodies can also recognize TMG caps. Thus, it is important to distinguish if RNAs are m7G capped rather than TMG capped. For this purpose, competition with m7G cap analogs ensure RNAs are m7G capped and in parallel competition with GpppG, ensures that RNAs are indeed methylated (GpppG will not compete for m7G capped RNAs) [Citation9]. Depletion of RNA bound to anti-m7G cap antibody after m7G cap analog treatment provides strong evidence that the RNA target is m7G capped. While CapIP readily reveals changes in capping fold between two conditions [Citation9,Citation11,Citation46], it is challenging to obtain percent occupancy of caps on a per transcript basis. Despite this limitation, many coding and non-coding RNAs were found to be regulated at the level of capping using this method [Citation11,Citation46]. Affinity approaches based on eIF4E RNA pulldowns of purified transcripts have also been employed. These utilize either purified wildtype eIF4E or the K119A eIF4E mutant protein. The K119A mutant has a higher m7G-cap affinity than wildtype eIF4E and thus is utilized to capture RNAs with increased efficiency [Citation6,Citation45]. This latter method is referred to as CapTure [Citation28]. These methods reveal differences in fold of capping between conditions.
Quantitative CapIP methods have also been developed [Citation9]. These methods rely on the determination of quantitative conditions optimizing different ratios of anti-m7G cap antibodies to purified RNA to establish conditions for maximal capture of m7G capped transcripts. These results are compared to the amount of RNA bound to IgG beads to establish the background binding. For the same samples, RNAs are monitored in the supernatant (SN) fractions of the CapIP and IgG IP. In parallel aliquots, IPs are performed in the presence of high levels of m7GpppG (relative to GpppG) to ensure specificity of the anti-m7G antibody. To obtain percentage of RNAs that are capped, levels of specific RNAs are quantified from the supernatants of the CapIP (Cap-SN) or Ctrl-IPs (Ctrl-SN) by RT-qPCR. Depletion of the RNA from the supernatant indicates RNAs are bound in the IP. Ctrl-IPs are comprised of CapIP in the presence of 100 μM m7GpppG to validate specificity and ascertain if RNAs are binding nonspecifically to the antibodies or beads. The ratio of Cap-SN to Ctrl-SN gives the percentage of uncapped RNAs. This can then be converted to a percentage of capped RNA. This method can be applied to different conditions to ascertain how percentages of capping change for candidate RNAs.
Enzymatic and chemical methods have also been used to quantify the extent of capping and can be readily coupled to RNA-sequencing technologies (). One such method is referred to as Cap Quantitation (CapQ) which relies on the unique sensitivity of capped RNAs to tobacco acid pyrophosphatase (TAP) [Citation9] and their insensitivity to Alkaline Phosphatase (AP). In a mixture of capped and uncapped RNAs, AP leads to removal of 5'-phosphates on uncapped RNAs and replacement by 5' OH groups, but does not affect capped RNAs. Subsequent treatment with TAP will cleave any pyrophosphate bond and thus cleave m7G or G capped RNAs leaving a 5' monophosphate, but not alter the 5'OH RNAs originating from the uncapped population. Thus, when AP treatment is followed by TAP, the capped RNAs are converted to transcripts with a 5'-monophosphate which can be ligated to biotinylated primers, isolated and analyzed by RT-qPCR or RNA-Seq. The RNAs with 5'OH groups remain, and cannot be ligated to biotinylated primers and thus are not captured. To obtain total levels of RNAs, separate aliquots of the same samples are treated with AP and TAP followed by polynucleotide kinase (PNK) which leaves all RNAs, independently of their initial 5' end status, with a 5'-monophosphate group suitable for ligation with biotinylated oligonucleotides. In this way, the amount of capped and total RNA can be obtained from the same sample allowing a calculation of percent capping on a per transcript basis.
CapQ is derived from the Oligo-Capping and Ligation method which similarly utilize AP and TAP to identify capped RNAs [Citation49,Citation50]; but CapQ offers several advantages including calculation of percent of capped RNAs since total RNAs are also calculated from the same sample. Modification of a method referred to as SMART (Switching Mechanism at 5' end of RNA Transcript) allows selection of capped (CapSMART) or of RNAs without the m7G cap (Non-CapSMART) [Citation49]. For Non-CapSMART, RNAs are treated with AP and TAP which leaves only the capped RNAs with a 5'-monophosphate, these are then ligated to stop oligonucleotides to block template switching. For CapSMART, AP and PNK are used to obtain 5'-monophosphates on uncapped RNA which are then ligated to stop oligonucleotides to inhibit template switching, and thus only capped RNAs are observed. Another approach is RNA ligase mediated 5' rapid amplification of cDNA ends (RLM-RACE) which allows detection of uncapped RNAs by ligation to a primer to the 5'-monophosphate on uncapped RNAs [Citation7]. Chemical methods have also been used including CapTrapper whereby the diols on the 5' and 3' ends of the transcripts are oxidized and biotinylated for capture and isolation [Citation51]. CapTrapper was incorporated into the original CAGE protocols [Citation52], which have since evolved to a template switching modality, and thus more recent CAGE methods do not specifically capture capped RNAs [Citation53,Citation54]. Another strategy leverages XRN, a nuclease which degrades uncapped RNAs but not RNAs with a 5' pyrophosphate bond [Citation14]. A comparison of the XRN sensitive and insensitive populations reveals RNAs that are uncapped [Citation14]. These methods are readily amenable to RNA-Seq and indeed such studies have revealed dynamics in the population of capped RNAs [Citation7].
m7G capping as a dynamic property of both coding and non-coding RNAs
Until recently, formation of the m7G cap was considered to be a constitutive property that produced 100% correctly capped transcripts; and conversely, decapping inevitably led to RNA decay [Citation1]. However, recent studies, using the methodologies described above, indicate that capping is dynamic with different RNA populations preferentially capped and others that are incorrectly capped [Citation2,Citation5,Citation7,Citation9,Citation10]. In other words, if there are 100 transcripts of CCND1 RNAs, not the entire population will have a m7G cap [Citation11,Citation47]. About 10 years ago, studies demonstrated that impaired capping could act as an RNA surveillance mechanism [Citation5]. The most common capping error identified was non-methylation at the 7 position of the 5' guanosine [Citation5]. Stressing the relevance of this, there are cellular mechanisms in place to specifically remove RNAs without methylated caps [Citation5]. However, preferential and dynamic capping of certain populations of RNAs has been observed in plant and animal cells [Citation7,Citation9,Citation12,Citation14,Citation46,Citation48]. In this case, it appears a regulatory modality rather than a malfunction of the capping machinery.
About 15 years ago, a landmark study reported substantial uncapped RNA populations in Arabidopsis thaliana [Citation7]. To study capping, this group developed the RLM-RACE method (described above) which allowed T4 RNA ligase to ligate an oligonucleotide to the 5'-monophosphate on RNAs [Citation7]. This will capture RNAs with a 5'-monophosphate but not m7G or G-capped RNAs with a 5'-5' pyrophosphate linkage. RNAs were identified by microarray analysis. More than 90% of expressed genes had some uncapped RNAs identified; however, the relative percentage of uncapped to capped RNAs differed based on the transcript examined. Between ~500 to 1000 RNAs were enriched in their uncapped forms. Capping fluxed in a transcript-specific manner through developmental stages. Intriguingly, ~2-3% of RNAs were only identified in the uncapped form [Citation7]. These studies also discovered the first RNA elements that were linked to increased abundance of uncapped RNAs. In this case, uncapped RNAs were enriched in the presence of specific versions of AU-rich elements or with sORFs. Also, transcripts enriched in the uncapped form tended to be longer (more than 1000 nucleotides) and contain more introns [Citation7].
To directly monitor the m7G capped RNA population, CapIP methods were employed to monitor the enrichment of capped RNAs in mammalian cells [Citation9,Citation11,Citation46,Citation48]. In this case, the anti-m7G cap antibody will capture RNAs that have a m7G cap, and thus G-capped RNAs, despite their 5'-5' pyrophosphate linkage, would not be detected as “capped” potentially yielding a population different than the RLM 5'RACE method above. Further, these studies provide fold changes in capping on a per transcript basis using PCR strategies [Citation11,Citation46]. These studies revealed that MYC overexpression could elevate relative capping of many of its own transcriptional targets, for example, CCND1, Fbl, eIF2B1 and eIF4A1 [Citation46]. Direct elevation of the capping machinery through RNMT overexpression increased capping of several of these same targets as well increased capping of ~3-fold for MYC [Citation46]. Importantly, even though RNMT is part of the general capping machinery, the capping of all RNAs examined was not enhanced (e.g. GAPDH) characteristic of the specificity of this process. Moreover, increases in capping were normalized to total RNA levels, that is, increased capping was not simply a reflection of increased RNA levels.
The development of other methods allowed quantitative assessment of capping on a per-RNA basis as a function of eIF4E dysregulation. eIF4E is associated with many aggressive cancers [Citation55]. eIF4E acts as a m7G cap chaperone for many RNAs and is involved in translation, RNA export and 3'end processing [Citation56–59]. Both quantitative CapIP methods and in parallel, Cap quantitation (CapQ) approaches, were used to measure capping. These studies revealed that many RNA populations were much less capped at steady-state in vector control cells than anticipated. Indeed, ~30-50% of RNAs (CCND1, MYC, Mdm2, Fbl, eIF4A1, eIF2B1 and non-coding RNAs MALAT and NEAT) were capped and even RNAs that are not regulated by eIF4E, for example, ACTNB, were only 50–70% capped at steady-state [Citation9]. eIF4E overexpression, at least in part through its ability to drive expression of the capping machinery, elevated capping of subset of RNAs (Mdm2, CTNNB1, MALAT, NEAT, MYC, CCND1) to 60–100% depending on the specific RNA but did not alter other targets such as ACTNB. In support of these findings, quantitative CapIP (which depends on the presence of the m7G) and CapQ (which will identify any RNAs with a 5'-5'pyrophosphate linkage) yielded highly concordant results. Combining CapIP with RNA-Seq led to the revelation that eIF4E impacted the capping of 100s of coding and non-coding RNAs including long non-coding RNAs such as ABALON, AC006116.27 and AC006116.24. eIF4E led to increased capping of some RNAs and reduced capping of others. This suggests that there is a competition for limited resources (RNGTT, RAM, RNMT, SAM) whereby eIF4E tips the balance to favor specific transcripts. eIF4E required RNMT for its effects on capping and there was substantial overlap between eIF4E and RNMT capping targets. For example, eIF4E, like RNMT, also elevated capping of CCND1 and MYC [Citation46]. An RNA element was identified that conferred increased capping activity when fused to a LacZ reporter upon eIF4E overexpression; this element is referred to as a Cap Sensitivity Element (CapSE) [Citation9]. LacZ-CapSE transcripts were not enriched in eIF4E RIPs relative to LacZ fusion RNAs suggesting that eIF4E was not directly involved in recognition of this element. However, LacZ-CapSE was enriched in RNMT IPs relative to LacZ, supporting a role for RNMT in CapSE recognition. We note that it does not appear that eIF4E overexpression simply provides protection against decapping by directly binding to m7G caps of target RNAs [Citation9]. In support of this, eIF4E overexpression increased capping of some RNAs that were not found in eIF4E nuclear RIPs; moreover, while eIF4E interacted with LacZ-4ESE RNAs in nuclear RIPs, these did not undergo increased capping upon eIF4E overexpression [Citation9]. Thus, eIF4E elevates the capping of many RNMT capping targets and it does not need to directly interact with these RNAs in the nucleus for this to occur. This suggests that at least for some RNAs, the impact of eIF4E on capping is via its ability to increase RNMT, RAM and RNGTT protein levels.
Despite their global importance, little is known about the factors that regulate the production of the capping machinery. Overexpression of eIF4E in model systems elevates nuclear RNA export for endogenous RNGTT, RAM and RNMT (and thus increased levels of these RNAs in the cytoplasm) and additionally increases the number of ribosomes per transcripts, i.e. improves translational efficiency, for RNGTT and RNMT [Citation9]. This leads to elevated levels of these proteins upon eIF4E overexpression. By contrast, CRISPR-4E cells have reduced levels of these proteins relative to CRISPR control cells [Citation9]. eIF4E provides the first example of a factor that controls the levels of the capping machinery. As described above, this is related to the increased capping activity observed for selected RNAs by eIF4E.
Modulation of capping in cancer, differentiation and development
Regulation of capping enzymes expression has an important role in cell differentiation [Citation12]. Levels of the RNMT-RAM complex are high in embryonic stem cells, and this elevation is required for the expression of pluripotency associated genes [Citation12]. During differentiation, ERK1/2 phosphorylates RAM, targeting it for degradation. This results in repression of pluripotency-associated genes and is required for differentiation to occur. Conversely, expression of RNMT-RAM in fibroblasts enhances the efficiency of reprogramming, probably due to its impact on these pluripotency-associated genes.
Capping can be dysregulated in cancer in primary specimens and cell line models. One such example comes from eIF4E where capping was elevated in high-eIF4E acute myeloid leukemia (AML) patient specimens compared to blood cells from healthy volunteers [Citation60–62]. In these patient specimens, there was elevated capping of coding and non-coding transcripts (MALAT, RNMT, MYC) by ~2-3 fold relative to healthy volunteers using CapIP methods [Citation9]. Further, there were elevated protein levels for RNGTT and RNMT in these patient specimens [Citation9]. By contrast, capping of other RNAs such as ACTNB or POLR2A were the same in AML patients versus healthy volunteers, consistent with their insensitivity to eIF4E overexpression in model systems. Transcripts were normalized to corresponding total RNA levels and thus differences in RNA levels do not account for changes in levels of capping [Citation9]. These studies were the first to show that capping can be dysregulated in primary cancer specimens [Citation9]. eIF4E overexpression is known to increase transformation, this is attributed to its roles in RNA export and translation [Citation55,Citation63], but it seems likely that this could include capping as well. Consistent with the interplay between RNMT and eIF4E, genetically engineered RNMT overexpression increased capping efficiency of specific transcripts (CCND1 and MYC) increasing their protein levels with no changes in total mRNA levels [Citation46]. Through this activity, RNMT overexpression oncogenically transforms mammary epithelial cell lines in culture [Citation46].
The increased dependency of subsets of genes on high levels of capping enzymes is important when considering therapeutic targeting. The genes most and least dependent on RNMT, for example, will influence the cellular response to its inhibition. In a panel of breast cancer cell lines, RNMT inhibition was observed to selectively inhibit the proliferation of cell lines expressing oncogenic PI3KCA mutants [Citation64]. This implies that a subset of genes induced by PI3KCA are dependent on RNMT for expression, and/or that the most RNMT-dependent genes are required in PI3KCA oncogene-addicted cells [Citation64]. MYC oncogene expression has enhanced dependency on RNGTT, including dysregulated MYC [Citation65]. This dependency may be a function of the high turnover of Myc protein and RNA. MYC and N-Myc upregulate capping by promoting RNA polymerase II phosphorylation, which increased recruitment of the capping enzymes to specific genes [Citation10,Citation48,Citation65], as well as increased expression of S-adenosyl homocysteine hydrolase (SAHH), which neutralizes the inhibitory SAH bi-product of methylation reactions [Citation66].
Other oncogenic factors also regulate the capping machinery e.g. Wnt and CDK1 [Citation67]. For example, CDK1 increases cap methylation by phosphorylating the RNMT regulatory domain; this has a gene-specific impact, although what governs specificity in this case is unclear and the mechanism may involve recapping [Citation13].
Recapping of RNAs after decapping
In the early 1990s, the Maquat laboratory found that cytoplasmic β-globin RNAs were apparently re-capped after fragmentation that provided new 5' ends [Citation8]. Indeed, 5' truncated forms of β-globin bind equally well to the cap-antibody and to eIF4E as do the parent transcripts [Citation6,Citation8]. The truncated forms were restricted to the cytoplasm [Citation6]. This suggested that capping is not restricted to co-transcriptional events but also occurs post-transcriptionally in mammalian cells [Citation6,Citation8,Citation14,Citation31]. Indeed, it appears that capping can occur both in the nucleus and cytoplasm. RNMT, RAM and RNGTT are all found in the cytoplasm, albeit at lower levels at steady state than in the nucleus [Citation6,Citation14,Citation31]. The likely substrates for recapping would be RNAs that have had their caps removed by the decapping enzymes DCP1/DCP2 or DCPS or transcripts that have undergone internal endonuclease cleavage [Citation32,Citation68]. Of interest, 5' decay is slow and inefficient relative to 3' decay providing support for the ability to have a pool of RNAs ready to be recapped [Citation69]. In support of this, cap homeostasis is independent of polyA tail length [Citation70]. For many RNA substrates, 5'decapping leaves RNAs with 5'-monophosphate, and thus need a kinase in order to convert to the 5'-diphosphate which is a substrate of RNGTT. Such a kinase has been identified as a RNGTT co-factor [Citation6] and some decapping enzymes leave a 5'-diphosphate which produce suitable substrates for RNGTT (e.g. Nudt12, Nudt15, Nudt2 and Nudt3) [Citation32].
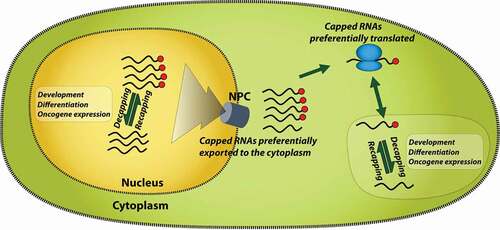
Cytoplasmic recapping on a genome-wide scale has been observed through the use of cellular fractionation and XRN sensitivity assays which identify RNAs without a 5' pyrophosphate linkage [Citation14]. In one study, XRN sensitivity was complemented with 5' RACE methods as well as exclusion of uncapped RNAs from cap affinity chromatography (comprised of eIF4E-GST bound to eIF4G to increase its affinity for the m7G cap) with all three methods yielding concordant results [Citation14]. These studies suggested that about 1000 transcripts were uncapped using microarray methods with a ~ 2–5-fold enrichment in uncapped RNAs upon overexpression of an RNGTT mutant (K294A). In these studies, which focus on cytoplasmic RNAs, it was observed that the uncapped RNAs were sequestered from polysomes suggesting that they were maintained in an inactive state as a regulatory feature [Citation14]. RNAs that were recapped were enriched in AREs in their 3' UTR [Citation14]. There are several advantages for cytoplasmic or nuclear re-capping. In the cytoplasm, uncapped RNAs could be stored in an uncapped state and then re-imported into the active translation pool [Citation14] (). Given all the necessary enzymes are present, nuclear recapping is also theoretically possible. In this way, transcripts can be stored in “stasis” waiting for further processing and export to the cytoplasm. In both cases, RNAs could be released through signaling events that would enable processes such as RNA export to the cytoplasm or reentry into the active translation pool ().
Figure 3. Model for the role of dynamic capping in RNA activity. Only nuclear RNA export and translation are shown for simplicity, but cap dynamics could impact many other facets of RNA metabolism such as splicing and polyadenylation. Dynamic capping has a major impact on transcription and has been described as a transcriptional checkpoint. NPC indicates nuclear pore complex used for RNAs to export the nucleus. Red balls indicate m7G caps
Perspectives and conclusions
Analogies to other RNA processing events furnish examples of decoupling RNA processing steps such as capping from transcription. For instance, similar to capping discussed above, splicing as well as 3' cleavage and polyadenylation (CPA) occur co-transcriptionally and/or post-transcriptionally. Indeed, RNAs can be partially processed, stored and then reenter the active transcript pool. This leaves RNAs in “stasis” until they are required for action rendering an elegant level of control that avoids the needs of further rounds of transcription and allows for fine-tuning of the composition of the final RNA product. For instance, CTN RNAs are stored in the nucleus after CPA [Citation35]. Upon certain types of stress, these RNAs undergo a second CPA step producing a transcript variant known as CAT2 mRNA which is exported to the cytoplasm and translated into the CAT2 protein [Citation35]. Similarly, RNAs with detained introns remain in the nucleus until they are required [Citation33,Citation34]. Indeed, the same RNA can undergo co-transcriptional splicing for some introns, followed later by post-transcriptional splicing for other introns [Citation34]. Studies reviewed here reveal a parallel strategy, whereby cells produce RNAs at steady state which have reduced percentage capping or even different types of caps. Dynamic m7G capping is demonstrated for specific RNAs upon overexpression of certain factors, for example, eIF4E, Myc, RNMT, or at particular developmental and differentiation stages. Capping can occur in either compartment, where in the cytoplasm it can be used to titrate the translation capacity of specific RNAs based on current cellular requirements. Alternatively, or indeed contemporaneously, uncapped RNAs could be trapped in the nucleus to await further processing and/or because uncapped RNAs are poor export substrates (). There are multiple proteins that bind the cap and initiate cap-dependent translation for selected transcripts e.g. eIF4E, eIF4E2, eIF4E3, eIF3d, CBC, and PARN [Citation71]. Thus, the capacity of eIF4E to increase capping could provide more substrates for its direct role in RNA export and/or translation, or by furnishing substrates for other cap-binding proteins. Other forms of the 5' cap can also be sensitive to cellular signals such as NAD capping can flux based on nutrient conditions [Citation32,Citation72]. Keeping RNAs in stasis is an efficient means to conserve energy in the cell, and allows for rapid responses to changes in the cellular environment. Capping could be coordinately regulated for groups of RNAs acting in the same biochemical pathway, for example, RNA regulons. Selection of groups of RNAs for regulation by capping appears to be driven by cis-acting elements in the RNA such as the CapSE, AREs, sORFs, pseudoknots, introns and likely others [Citation7,Citation9,Citation14]. In all, cap dynamics is positioned to provide sensitive control mechanisms to tune the proteome in response to biological situations.
Acknowledgments
We thank Dr Laurent Volpon for figure production.
Disclosure Statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Furuichi Y. Discovery of m(7)G-cap in eukaryotic mRNAs. Proc Jpn Acad Ser B Phys Biol Sci. 2015;91:394–409.
- Cowling VH. Regulation of mRNA cap methylation. Biochem J. 2010;425(2):295–302.
- Galloway A, Cowling VH. mRNA cap regulation in mammalian cell function and fate. Biochim Biophys Acta Gene Regul Mech. 2019;1862(3):270–279.
- Schoenberg DR, Maquat LE. Re-capping the message. Trends Biochem Sci. 2009;34(9):435–442.
- Jiao X, Xiang S, Oh C, et al. Identification of a quality-control mechanism for mRNA 5'-end capping. Nature. 2010;467(7315):608–611.
- Otsuka Y, Kedersha NL, Schoenberg DR. Identification of a Cytoplasmic Complex That Adds a Cap onto 5'-Monophosphate RNA. Mol Cell Biol. 2009;29(8):2155–2167.
- Jiao Y, Riechmann JL, Meyerowitz EM. Transcriptome-Wide Analysis of Uncapped mRNAs in Arabidopsis Reveals Regulation of mRNA Degradation. Plant Cell. 2008;20(10):2571–2585.
- Lim SK, Maquat LE. Human beta-globin mRNAs that harbor a nonsense codon are degraded in murine erythroid tissues to intermediates lacking regions of exon I or exons I and II that have a cap-like structure at the 5' termini. EMBO J. 1992;11(9):3271–3278.
- Culjkovic-Kraljacic B, Skrabanek L, Revuelta MV, et al. The eukaryotic translation initiation factor eIF4E elevates steady-state m 7 G capping of coding and noncoding transcripts. Proc Natl Acad Sci U S A. 2020;117(43):26773–26783.
- Cowling VH, Cole MD. The Myc transactivation domain promotes global phosphorylation of the RNA polymerase II carboxy-terminal domain independently of direct DNA binding. Mol Cell Biol. 2007;27:2059–2073.
- Cowling VH. Myc up-regulates formation of the mRNA methyl cap. Biochem Soc Trans. 2010;38(6):1598–1601.
- Grasso L, Suska O, Davidson L, et al. mRNA Cap Methylation in Pluripotency and Differentiation. Cell Rep. 2016;16(5):1352–1365.
- Aregger M, Kaskar A, Varshney D, et al. CDK1-Cyclin B1 Activates RNMT, Coordinating mRNA Cap Methylation with G1 Phase Transcription. Mol Cell. 2016;61(5):734–746.
- Mukherjee C, Patil D, Kennedy B, et al. Identification of cytoplasmic capping targets reveals a role for cap homeostasis in translation and mRNA stability. Cell Rep. 2012;2(3):674–684.
- Jiao X, Doamekpor SK, Bird G,et al. 5' End Nicotinamide Adenine Dinucleotide Cap in Human Cells Promotes RNA Decay through DXO-Mediated deNADding. Cell. 2017;168:1015–1027 e1010.
- Coutinho De Oliveira L, Volpon L, Rahardjo AK, et al. Structural studies of the eIF4E-VPg complex reveal a direct competition for capped RNA: implications for translation. Proc Natl Acad Sci U S A. 2019;116:24056–24065.
- Yedavalli VS, Jeang KT. Trimethylguanosine capping selectively promotes expression of Rev-dependent HIV-1 RNAs. Proc Natl Acad Sci U S A. 2010;107:14787–14792.
- Liu W, Jankowska-Anyszka M, Piecyk K, et al. Structural basis for nematode eIF4E binding an m(2,2,7)G-Cap and its implications for translation initiation. Nucleic Acids Res. 2015;39:8820–8832.
- Wallace A, Filbin ME, Veo B, et al. The nematode eukaryotic translation initiation factor 4E/G complex works with a trans-spliced leader stem-loop to enable efficient translation of trimethylguanosine-capped RNAs. Mol Cell Biol. 2010;30:1958–1970.
- Mao X, Schwer B, Shuman S. Yeast mRNA cap methyltransferase is a 50-kilodalton protein encoded by an essential gene. Mol Cell Biol. 1995;15:4167–4174.
- Chu C, Shatkin AJ. Apoptosis and autophagy induction in mammalian cells by small interfering RNA knockdown of mRNA capping enzymes. Mol Cell Biol. 2008;28(19):5829–5836.
- Pillutla RC, Shimamoto A, Furuichi Y, et al. Human mRNA capping enzyme (RNGTT) and cap methyltransferase (RNMT) map to 6q16 and 18p11.22-p11.23, respectively. Genomics. 1998;54:351–353.
- Shafer B, Chu C, Shatkin AJ. Human mRNA cap methyltransferase: alternative nuclear localization signal motifs ensure nuclear localization required for viability. Mol Cell Biol. 2005;25(7):2644–2649.
- Srinivasan P, Piano F, Shatkin A. mRNA capping enzyme requirement for Caenorhabditis elegans viability. J Biol Chem. 2003;278:14168–14173.
- Tsukamoto T, Shibagaki Y, Murakoshi T, et al. Cloning and characterization of two human cDNAs encoding the mRNA capping enzyme. Biochem Biophys Res Commun. 1998;243(1):101–108.
- Yamada-Okabe T, Doi R, Shimmi O, et al. Isolation and characterization of a human cDNA for mRNA 5'-capping enzyme. Nucleic Acids Res. 1998;26:1700–1706.
- Yue Z, Maldonado E, Pillutla R, et al. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc Natl Acad Sci U S A. 1997;94(24):12898–12903.
- Chu C, Das K, Tyminski JR, et al. Structure of the guanylyltransferase domain of human mRNA capping enzyme. Proc Natl Acad Sci U S A. 2011;108:10104–10108.
- Gonatopoulos-Pournatzis T, Dunn S, Bounds R, et al. RAM/Fam103a1 is required for mRNA cap methylation. Mol Cell. 2011;44(4):585–596.
- Fabrega C, Hausmann S, Shen V, et al. Structure and mechanism of mRNA cap (guanine-N7) methyltransferase. Mol Cell. 2004;13(1):77–89.
- Trotman JB, Giltmier AJ, Mukherjee C, et al. RNA guanine-7 methyltransferase catalyzes the methylation of cytoplasmically recapped RNAs. Nucleic Acids Res. 2017;45(18):10726–10739.
- Grudzien-Nogalska E, Kiledjian M. New insights into decapping enzymes and selective mRNA decay. Wiley Interdiscip Rev RNA. 2017;8(1):e1379.
- Boutz PL, Bhutkar A, Sharp PA. Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev. 2015;29(1):63–80.
- Girard C, Will CL, Peng J, et al. Post-transcriptional spliceosomes are retained in nuclear speckles until splicing completion. Nat Commun. 2012;3(1):994.
- Prasanth KV, Prasanth SG, Xuan Z, et al. Regulating gene expression through RNA nuclear retention. Cell. 2005;123(2):249–263.
- Furuichi Y, Morgan M, Shatkin AJ, et al. Methylated, blocked 5 termini in HeLa cell mRNA. Proc Natl Acad Sci U S A. 1975;72:1904–1908.
- Furuichi Y, Shatkin AJ. A simple method for the preparation of [beta-32P]purine nucleoside triphosphase. Nucleic Acids Res. 1977;4:3341–3355.
- Salditt-Georgieff M, Jelinek W, Darnell JE, et al. Methyl labeling of HeLa cell hnRNA: a comparison with mRNA. Cell. 1976;7:227–237.
- Wei C, Gershowitz A, Moss B. N6, O2'-dimethyladenosine a novel methylated ribonucleoside next to the 5' terminal of animal cell and virus mRNAs. Nature. 1975;257:251–253.
- Wei C-M, Gershowitz A, Moss B. Methylated nucleotides block 5' terminus of HeLa cell messenger RNA. Cell. 1975;4(4):379–386.
- Wang J, Alvin Chew BL, Lai Y, et al. Quantifying the RNA cap epitranscriptome reveals novel caps in cellular and viral RNA. Nucleic Acids Res. 2019;47(20):e130.
- Galloway A, Atrih A, Grzela R, et al. CAP-MAP: cap analysis protocol with minimal analyte processing, a rapid and sensitive approach to analysing mRNA cap structures. Open Biol. 2020;10(2):190306.
- Kruse S, Zhong S, Bodi Z, et al. A novel synthesis and detection method for cap-associated adenosine modifications in mouse mRNA. Sci Rep. 2011;1(1):126.
- Pandey RR, Delfino E, Homolka D, et al. The Mammalian Cap-Specific m6Am RNA Methyltransferase PCIF1 Regulates Transcript Levels in Mouse Tissues. Cell Rep. 2020;32(7):108038.
- Choi YH, Hagedorn CH. Purifying mRNAs with a high-affinity eIF4E mutant identifies the short 3’ poly(A) end phenotype. Proc Natl Acad Sci U S A. 2003;100(12):7033–7038.
- Cowling VH. Enhanced mRNA cap methylation increases cyclin D1 expression and promotes cell transformation. Oncogene. 2010;29(6):930–936.
- Cowling VH, Cole MD. Myc Regulation of mRNA Cap Methylation. Genes Cancer. 2010;1(6):576–579.
- Cole MD, Cowling VH. Specific regulation of mRNA cap methylation by the c-Myc and E2F1 transcription factors. Oncogene. 2009;28(9):1169–1175.
- Machida RJ, Lin -Y-Y, Oudejans C. Four Methods of Preparing mRNA 5' End Libraries Using the Illumina Sequencing Platform. PLoS One. 2014;9(7):e101812.
- Kazuo M, Sumio S. Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene. 1994;138(1–2):171–174.
- Carninci P, Kvam C, Kitamura A, et al. High-efficiency full-length cDNA cloning by biotinylated CAP trapper. Genomics. 1996;37(3):327–336.
- Shiraki T, Kondo S, Katayama S, et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc Natl Acad Sci U S A. 2003;100(26):15776–15781.
- Salimullah M, Mizuho S, Plessy C, et al. NanoCAGE: a high-resolution technique to discover and interrogate cell transcriptomes. Cold Spring Harb Protoc. 2011;2011(1):5559.
- Kodzius R, Kojima M, Nishiyori H, et al. CAGE: cap analysis of gene expression. Nat Methods. 2006;3(3):211–222.
- Carroll M, Borden KLB. The oncogene eIF4E: using biochemical insights to target cancer. J Interferon Cytokine Res. 2013;33(5):227–238.
- Davis MR, Delaleau M, Borden KLB. Nuclear eIF4E Stimulates 3'-End Cleavage of Target RNAs. Cell Rep. 2019;27(5):1397–1408.
- Volpon L, Culjkovic-Kraljacic B, Sohn HS, et al. A biochemical framework for eIF4E-dependent mRNA export and nuclear recycling of the export machinery. RNA. 2017;23(6):927–937.
- Culjkovic-Kraljacic B, Borden KLB. Aiding and abetting cancer: mRNA export and the nuclear pore. Trends Cell Biol. 2013;23(7):328–335.
- Borden KL. The eukaryotic translation initiation factor eIF4E wears a “cap” for many occasions. Translation (Austin). 2016;4:e1220899.
- Topisirovic I, Guzman ML, McConnell MJ, et al. Aberrant eukaryotic translation initiation factor 4E-dependent mRNA transport impedes hematopoietic differentiation and contributes to leukemogenesis. Mol Cell Biol. 2003;23(24):8992–9002. .
- Assouline S, Culjkovic B, Cocolakis E, et al. Molecular targeting of the oncogene eIF4E in acute myeloid leukemia (AML): a proof-of-principle clinical trial with ribavirin. Blood. 2009;114(2):257–260. .
- Volpon L, Culjkovic-Kraljacic B, Osborne MJ, et al. Importin 8 mediates m 7 G cap-sensitive nuclear import of the eukaryotic translation initiation factor eIF4E. Proc Natl Acad Sci U S A. 2016;113(19):5263–5268.
- Volpon L, Osborne MJ, Borden KLB. Biochemical and Structural Insights into the Eukaryotic Translation Initiation Factor eIF4E. Current Protein & Peptide Science. 2019;20(6):525–535.
- Dunn S, Lombardi O, Lukoszek R, et al. Oncogenic PIK3CA mutations increase dependency on the mRNA cap methyltransferase, RNMT, in breast cancer cells. Open Biol. 2019;9(4):190052.
- Lombardi O, Varshney D, Phillips NM, et al. c-Myc deregulation induces mRNA capping enzyme dependency. Oncotarget. 2016;7(50):82273–82288.
- Fernandez-Sanchez ME, Gonatopoulos-Pournatzis T, Preston G, et al. S-adenosyl homocysteine hydrolase is required for Myc-induced mRNA cap methylation, protein synthesis, and cell proliferation. Mol Cell Biol. 2009;29(23):6182–6191.
- Posternak V, Ung MH, Cheng C, et al. MYC Mediates mRNA Cap Methylation of Canonical Wnt/β-Catenin Signaling Transcripts By Recruiting CDK7 and RNA Methyltransferase. Mol Cancer Res. 2017;15(2):213–224.
- Mercer TR, Dinger ME, Bracken CP, et al. Regulated post-transcriptional RNA cleavage diversifies the eukaryotic transcriptome. Genome Res. 2010;20(12):1639–1650.
- Song M-G, Li Y, Kiledjian M. Multiple mRNA decapping enzymes in mammalian cells. Mol Cell. 2010;40(3):423–432.
- Kiss DL, Oman KM, Dougherty JA, et al. Cap homeostasis is independent of poly(A) tail length. Nucleic Acids Res. 2016;44(1):304–314.
- Borden KLB, Volpon L. The diversity, plasticity, and adaptability of cap-dependent translation initiation and the associated machinery. RNA Biol. 2020;17(9):1239–1251.
- Walters RW, Matheny T, Mizoue LS, et al. Identification of NAD + capped mRNAs in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2017;114(3):480–485.
- Bochnig P, Reuter R, Bringmann P, et al. A monoclonal antibody against 2,2,7-trimethylguanosine that reacts with intact, class U, small nuclear ribonucleoproteins as well as with 7-methylguanosine-capped RNAs. Eur J Biochem. 1987;168(2):461–467.
- Moteki S, Price D. Functional coupling of capping and transcription of mRNA. Mol Cell. 2002;10(3):599–609.