ABSTRACT
Atrial fibrillation (AF) is the common arrhythmias. Myocardial fibrosis (MF) is closely related to atrial remodeling and leads to AF. MF is the main cause of cardiovascular diseases and a pathological basis of AF. Thus, the underlying mechanism in MF and AF development should be fully elucidated for AF therapeutic innovation. Autophagy is a highly conserved lysosomal degradation pathway, and the relationship between autophagy and MF has been previously shown. Moreover, research reported that quercetin (Que) could ameliorate MF. The current study aimed to explore the mechanism of Que in MF. The results in this study showed that in clinical AF patients and in aged rats, miR-223-3p was high-expressed, while FOXO3 and autophagy pathway related proteins, such as ATG7, p62/SQSTM1 and the ratio of LC3B-II/LC3B-I were significantly inhibited. In vivo and in vitro studies, we found that Que can effectively inhibit the expression of miR-223-3p in AF model cells and rats myocardial tissues, and meanwhile enhance the expression of FOXO3 and activate the autophagy pathway, and significantly inhibit myocardial fibrosis, and improve myocardial remodeling in atrial fibrillation. All in all, in this study, we found that Que prevents isoprenaline-induced MF by increasing autophagy via regulating miR-223-3p/FOXO3.
Introduction
Atrial fibrillation (AF) is high challenging to be treated [Citation1,Citation2]. The risk of developing AF is significantly increased with aging [Citation3,Citation4]. Epidemiological studies showed that about 5% of elderly patients aged over 65 years old will develop AF, and this number reaches 7.1% when patients aged over 85 years [Citation5]. The adverse effects of AF on cardiac functions cause the mortality of AF patients 1.5 to 2 times higher than that of non-AF patients [Citation6]. Therefore, effective prevention and treatment of AF has important clinical significance.
Cardiac structural remodeling and electrical remodeling would occur to AF patients, and are closely related to the occurrence of AF [Citation7,Citation8]. The structural remodeling and electrical remodeling can promote and influence one another, with structural remodeling serving as the basis of electrical remodeling [Citation8,Citation9]. Structural remodeling is characterized by myocardial fibrosis (MF) [Citation10,Citation11]. During the structural remodeling, cardiac fibroblasts are activated and differentiate into myofibroblasts, which will lead to a large amount of extracellular matrix deposition and further cause MF [Citation12]. Therefore, inhibition of MF is an important approach to AF prevention and treatment.
Quercetin (Que), a glycoside compound of flavonoids family, is a natural product and abundant in a variety of plants, fruits and vegetables [Citation13]. Researches proved that Que has multiple biological functions, such as anti-oxidant, anti-inflammatory, and anti-tumor effects [Citation13–15]. In an animal experiment, Que shows a significant cardioprotective effect on MF [Citation16], but the specific mechanism remains unknown. MiRNAs are involved in various biological and pathological phenomena, including in cardiovascular diseases [Citation17]. In cardiovascular biology, miR-223 has recently been studied with different cardiac pathological processes, including in cardiomyocyte hypertrophy and necrosis [Citation18,Citation19]. Liu et al. found that miR-223 could regulate MF after myocardial infarction [Citation19]. Nevertheless, the mechanism of miR-223 in CF and whether it could be regulated by Que during cardiac structural remodeling are still unclear. Increasing evidence shows that autophagy dysfunction is a leading cause of many cardiac conditions and the activation of autophagy can effectively inhibit the myocardial fibrosis caused by isoprenaline [Citation20]. The report pointed out that the forkhead box proteins O3 (FoxO3) has a potent inhibitory effect on fibroblast activation and subsequent extracellular matrix production, moreover, it can decrease the fibrosis of many organs including the heart, liver, lungs and kidneys [Citation21]. Another study found that increased activation of FoxO3 results in a significant increase in autophagy [Citation22].
Therefore, based on the previous research and findings, we hypothesized that Que may have a certain effect on MF via regulating miR-223 and FOXO3 expression, and activation of autophagy pathway. In the current study, the speculation and the underlying mechanisms were further explored by conducting a series of experiments.
Methods
Ethics Statement
The study and all animal experiments were reviewed and approved by the Ethics Committee of Dongfang Hospital, Beijing University of Chinese, and performed at Dongfang Hospital, Beijing University of Chinese. All the patients signed an informed consent, and agreed that their tissues would be used in this clinical research.
Tissue samples
Twenty-six AF cardiac tissues and twenty sinus rhythm (SR) cardiac tissues were obtained from rheumatic heart disease patients who took heart valve replacement at Dongfang Hospital, Beijing University of Chinese between March 2018 and March 2019.
Cell culture
Human embryonic kidney cell 293 T (BNCC353535) was purchased from BeNa Culture Collection (Beijing, China). The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (C11995500BT, Gbico, MA, USA) containing 10% fetal bovine serum (FBS) (10,437,010, Gbico) at 37°C with 5% CO2 in a humid atmosphere. The 293 T cells were only used for dual-luciferase reporter assays.
Luciferase reporter assays
The fragments of FOXO3-3'-UTR with wide-type (FOXO3-WT) (5'-CAAAGCAGACCCTCAAACTGACA-3') and mutant (FOXO3-Mut) (5'-CAGATGCAGACTCTCAATACAACA-3') binding sites for miR-223-3p were inserted into pmirGLO luciferase Vectors (E1330, Promega, CA, USA). MiR-223-3p mimic (M; miR10,000892-1-5) and mimic control (MC; miR1N0000002-1-5) were ordered from RIBOBIO (Guangzhou, China). The 293 T cells were seeded into 48-well plates at 3.0 × 104 cells in 300 μl medium. After let grow overnight, FOXO3-Mut or FOXO3-WT were co-transfected with miR-223-3p mimic or mimic control into 293 T cells by Lipofectamine 2000 (11,668–019, Invitrogen, MA, USA) for 48 h. Next, the cells were collected to perform Dual-Luciferase Reporter Assay (Promega). The luciferase activity of the cells was determined by GloMax fluorescence reader (Promega).
Isolation and culture of primary rat cardiac fibroblasts (RCFs)
The RCFs were extracted from one-week old Wistar rat (SLAC Laboratory Animal Technology, Shanghai, China). Before the cell extraction, the rats were anesthetized by intraperitoneal injection of 2% sodium pentobarbital (50 mg/kg) (B005, Jiancheng, Nanjing) for heart collection. Then the rats were sacrificed by cervical dislocation, at the same time, the heart blood was washed away by PBS (C0221A, Beyotime, Shanghai, China). The rat hearts were cut into 1-mm3 sections and maintained in a 10-cm culture dish. The hearts were incubated in 4 mL 0.25% EDTA- Trypsin (25,200–056, Gibco) in a humid atmosphere at 37°C with 5% CO2 for 10 min, and then added with 8 mL 10% FBS. Then the tissue sections and supernatant were collected and centrifuged for 10 min (1200 × g). After discarding the supernatant, the cells, which were RCFs, were collected and cultured in DMEM medium containing 15% FBS at 37°C with 5% CO2 in a humid atmosphere.
Establishment of AF model in RCFs
After we isolated and cultured the RCFs, the RCFs were randomly divided into five groups as follows: Control, Que-H, ISO, ISO+Que-L, and ISO+Que-H groups. Isoprenaline (ISO) (HY-B0468) was purchased from MedChemExpress (NJ, USA) and Que (Q817162) was purchased from Macklin (Shanghai, China).
The RCFs in control group were normally cultured in complete DMEM medium at 37°C with 5% CO2 in a humid atmosphere for later use; the RCFs in Que-H group were pre-cultured in DMEM medium containing 50 μM Que for 30 min, and then cultured in complete DMEM medium for further use; the RCFs in ISO group were pre-induced by the solution containing 5 μL ISO for 72 h, and then cultured in complete DMEM medium for further use; the RCFs in ISO+Que-L or ISO+Que-H group were first cultured in DMEM medium containing 20 μM or 50 μM Que for 30 min, then induced by the solution containing 5 μL ISO for 72 h, and finally cultured in complete DMEM medium for further use.
After that, the autophagy inhibitors (3-MA) were used for reverse validation, the RCFs were randomly divided into six groups as follows: Control, 3-MA, ISO, ISO+3-MA, ISO+Que-H, ISO+Que-H + 3-MA. The RCFs in Control, ISO, ISO+Que-H group treated as above described; the RCFs in ISO+3-MA group were treated by the solution containing 5 μL ISO for 72 h with 3-MA (1 mM) treatment for 24 h; the RCFs in ISO+Que-H + 3-MA group were treated as ISO+3-MA group and follow to culture in DMEM medium containing 50 μM Que for 30 min; the RCFs in 3-MA group were treated as ISO+3-MA group without ISO.
Cell transfection
Small interfering RNA directed against FOXO3 (siFOXO3; siG151021111654-1-5) and negative control for siFOXO3 (siNC; siN0000001-1-5) were obtained from RIBOBIO. Before cell transfection, the RCFs with AF (1.0 × 106 cells) were seeded into 6-well plates containing 2 ml medium. After let grow overnight to reach a cell confluence of 20%-30%, 100 μl OPTI MEM (31,985,070, Gbico) was used to dissolve 2 μg siRNA or mimic. At the same time, 2 μl liposome 2000 (P-lipo, Promoter, Wuhan, China) was diluted by 100 μl OPTI MEM. Then the two media were mixed together and incubated for 15 min at room temperature. Finally, the previous complete media was replaced by the mixed OPTI MEM, and further added with 1.8 ml complete medium to allow cell growth for an additional 48 h.
MTT assays
MTT (B7777, APExBIO, Houston, USA) was used to detect the proliferation of AF model RCFs. After the transfection of AF model RCFs with siRNA or mimic, 1.0 × 104 cells were first maintained in 96-well plates containing 100 μl complete medium overnight and then incubated by MTT reagent (0.5 mg/ml) for 4 h. The MTT solution was replaced by 100 μl DMSO (ST038, Beyotime) in each well. Finally, the absorbance was detected at 570 nm by a microplate reader (Infinite M200 PRO, Tecan Austria GmbH, Austria).
Establishment of AF rat model
Wistar rats (age 8 weeks), and elder rats (age 22–24 months) were purchased from SLAC Laboratory Animal Technology. All the experimental animals were fed in the same animal feeding unit under 12-h dark/12-h light cycle. MiR-223-3p agomiR and agomiR-NC were obtained from Obio Technology (Shanghai, China). Comparied the autophagy levels, miR-223-3p, and FOXO3 in older and 8-week-old rats. The 8-week-old rats were randomly divided into five groups as follows: Control, ISO, ISO+Que, ISO+Que+agomiR-NC, and ISO+Que+miR-223-3p agomiR groups, with 10 rats in each group.
The rats in Control were intragastrically administrated with 200 μL normal saline once a day for 22 days; the rats in ISO group were subcutaneously injected with ISO (15 mg/kg) at multiple time points on the first day, and then given intragastric administration of 200 μL normal saline once a day for 21 days from the second day on; the rats in ISO+Que group were subcutaneously injected with ISO (15 mg/kg) at multiple points at the first day, and then given intragastric administration of 200 μL Que (25 mg/kg) once a day for 21 days from the second day on; 3 days before ISO treatment, the rats in ISO+Que+agomiR-NC or ISO+Que+miR-223-3p agomiR group were injected with agomiR-NC or miR-223-3p agomiR (200 nmol/kg) from 10 sites (1–2 mm deep) into the left ventricular free wall under anesthesia induced by 2% sodium pentobarbital (50 mg/kg), and then treated by ISO and Que in the same way as ISO+Que group. Contrast with agomiR-NC or miR-223-3p agomiR injection, the rats in ISO and ISO+Que group were injected with an equal volume of saline. 22 days later, the rats were anesthetized by intraperitoneal injection of 2% sodium pentobarbital (50 mg/kg), and then sacrificed by cervical dislocation for heart collection. Rat hearts were removed and used for subsequent experiments of RT-QPCR, HE and Masson staining, Immunohistochemical assay and Western blot assay.
RNA extraction and RT-qPCR
MiRNAs were extracted from the clinical samples, rat heart samples, and RCFs using a miRcute miRNA Isolation Kit (TianGEN, Beijing, China). In brief, the tissues were grinded in a 1.5 ml centrifugal tube with lysis buffer, and the cells were collected into 1.5 ml centrifugal tube and directly lysed by lysis buffer. Next, 200 μl chloroform (C805334, Macklin) was added into the cell solution and shaken for 1 min. After resting the cells for 5 min at room temperature, the solution was further centrifuged at 13,400 × g for 20 min. Then the precipitates were collected into a new 1.5 ml tube containing 75% ethanol (M9082, Macklin). After another centrifugation of the cells at 13,400 × g for 15 min, the miRNA sediments were diluted by RNase-free H2O (ST876, Beyotime). MRNAs were extracted from the RCFs using TRIzol reagent (15,596, Invitrogen). In brief, 3.5 × 106 RCFs were lysed by 200 µl TRIzol for 15 min at room temperature, then centrifuged for 20 min (14,000 × g). 200 µl isopropanol (H822173, Macklin, Shanghai) was mixed with the supernatant (about 200 µl) and further centrifuged for 5 min (10000 × g). Then RNase-free H2O (RF001, Real-Times, Beijing, China) was used to dilute the mRNA sediments.
The mRNAs were reverse-transcribed into cDNAs using EasyScript First-Strand cDNA Synthesis SuperMix (AE301-02, TransGen Biotech). The cDNAs with gene primers and PerfectStart Green qPCR SuperMix (AQ601-01, TransGen Biotech) were amplified in QuantStudio6 system (Applied Biosystems, CA, USA) under the following reaction conditions: at 94°C for 30 s, at 94°C for 30 s for 40 cycles, at 60°C for 30 s for 40 cycles. Finally, RNA expressions were calculated according to 2−ΔΔCT method [Citation23]. The primers sequences are shown in .
Table 1. RT-qPCR primers
HE and Masson staining
The rat heart tissues were paraffin-embedded (S25190, Yuanye, Shanghai, China), fixed on the microtome (RM2235, Leica, Solms, Germany), and sliced into 4-μm thick. Then the slices were fixed on a glass slide (80,302–3101-16-P4, ShiTai, Jiangsu, China) and deparaffinized, incubated with hematoxylin (B25380, Yuanye) for 10 min and then with eosin (G1100, Solarbio, Beijing, China) for 1 min, or masson (D026-1-2, Jiancheng) for 5 min, at room temperature. Finally, the indexes of the slices were recorded under a phase-contrast optical microscope (Axio Lab.A1 pol; Leica, Solms, Germany).
Immunohistochemical assay
After HE and Masson staining, the tissue slices were incubated with antigen repair solution (p0081, Beyotime) for 10 min at room temperature, and then with endogenous peroxidase blocker (BF06060, Biodragon, Beijing, China) for another 10 min at room temperature. The tissues were blocked by 5% FBS for 1 h at room temperature, and incubated with antibody of Col1A1 (67,288-1-Ig, 1:2500, Proteintech, Wuhan, China) or Fibronectin (66,042-1-Ig, 1:500, Proteintech) overnight at 4°C. Then, corresponding secondary antibody (G-21,234, 1:500, Thermo Scientific) was further incubated with all the slices for 30 min and treated by DBA reagent (SFQ004, 4A Biotech, Beijing, China) for 30 min. Next, the slices were processed by hematoxylin (B25380, Yuanye) for 10 min. Finally, the indexes of the slices were recorded under a phase-contrast optical microscope (Axio Lab.A1 pol; Leica, Solms, Germany). The ratio of positive cell number to the total number was counted from 5 randomly selected fields.
Western blot
Total proteins were isolated from tissues and cells using NP-40 (P0013F, Beyotime). In brief, 250 µl NP-40 was incubated with 3.5 × 106 cells and 20 mg tissues for 15 min at 4°C. After centrifugation for 30 min (14,000 × g), the total protein (supernatant) was collected, and the concentration was detected by BCA kit (P0009, Beyotime). Next, for protein separation, 40 µg total protein and 2 µl marker (PR1910, Solarbio, Beijing, China) were added into the ach lane on SDS-PAGE gels (P0052A, Beyotime) at 35 volts for 10 min, at 100 volts for 90 min, and then the proteins were transferred to PVDF membranes (ISEQ00010, Millipore, MA, USA), which were then incubated with methanol (M813895, Macklin). Subsequently, the membranes were first incubated with 5% nonfat milk for 1 h at room temperature, and then with relative first antibodies as follows: ATG7 (1:2000, ab133528, 77 kD, Abcam, CA, USA), LC3B I/II (1:2000, ab48394, 17/19kD, Abcam), p62/SQSTM1 (1:3000, ab56416, 61kD, Abcam), Cyclin D1 (1:2000, ab16663, 36kD, Abcam), Fibronectin (1:1000, ab268020, 285kD, Abcam), Col1A1 (1:1000, ab6308, 130kD, Abcam), FOXO3 (1:2500, ab23683, 90kD, Abcam), and β-actin (1:5000, 42kD, ab8226, Abcam). The next day, HRP-conjugated secondary antibodies (goat anti-mouse IgG secondary antibody (1:5000, ab205719, Abcam) and goat anti-rabbit IgG secondary antibody (1:5000, ab205718, Abcam)) were further incubated with the membranes for 1 h at room temperature. SuperSignal West Pico Chemiluminescent Substrate (34,078, Thermo Scientific) was reacted with membranes for signal development. Image Lab™ Software (version 3.0) was used for densitometric analysis and data quantification in Western blot (Bio-Rad Laboratories Inc., Hercules, CA, USA).
Statistical analysis
Student’s t-test and one-way ANOVA were conducted to analyze the data using SPSS software (version 18.0). LSD and Dunnet’s were used as post-hoc tests. Pearson correlation analysis was used to analyze the correlation between the expressions of miR-223-3p and FOXO3 in the cardiac tissues of AF patients. The data were indicated as Mean ± standard deviation. All the experiments were conducted three times. A statistically significant result was defined when P < 0.05.
Results
Up-regulated miR-223-3p affected autophagy-related gene expressions by targeting FOXO3 that was low-expressed in AF myocardial tissues
The expressions of miR-223-3p and FOXO3 between AF and SR groups were compared (, b), and the data revealed that miR-223-3p expression was obviously higher and FOXO3 expression was lower in AF group than those in SR group (P < 0.001). As shown in , we also found that the expression of miR-223-3p was negatively correlated with that of FOXO3 in AF cardiac tissues (r = –0.535, P < 0.001). The expressions of autophagy-related genes in AF and SR groups were further determined, and we observed that the expressions of ATG7 and LC3B-II were obviously higher but the expression of p62/SQSTM1 was lower in SR group than those in AF group (P < 0.001) (), with a higher ratio of LC3B-II/LC3B-I in SR group than in AF group (P < 0.001) (). These results demonstrated that the progression of autophagy was inhibited in AF tissues. FOXO3 was predicted to be a target of miR-223-3p because FOXO3-3'-UTR contained a target sequence base pairing with miR-223-3p (). Dual-luciferase reporter was conducted to verify the prediction. As shown in , luciferase activity of 293 T cells co-transfected with FOXO3-WT and miR-223-3p mimic was reduced, as compared with the MC group (P < 0.001). However, after the co-transfection the 293 T cells with FOXO3-MUT and mimic, no difference in the luciferase was detected, as compared with the MC group. These findings verified that FOXO3 could be targeted by miR-223-3p.
Figure 1. Up-regulated miR-223-3p regulated expressions of autophagy-related genes by targeting FOXO3 that was low-expressed in AF myocardial tissues. (a). MiR-223-3p expression in myocardial tissues was determined by RT-qPCR, U6 was used as an internal control. (b). FOXO3 expression in myocardial tissues was determined by RT-qPCR, β-actin was used as an internal control. (c) The correlation of miR-223-3p with FOXO3 was analyzed by Pearson correlation analysis. (d-f) The expressions of ATG7, LC3B-I, LC3B-II, and p62/SQSTM1 in the myocardial tissues were determined by Western blot, β-actin was used as an internal control. (^^^P < 0.001, vs. AF). (g). FOXO3-3'-UTR containing binding sites of miR-223-3p was predicted by TargetScan. (h). Luciferase assay validated that miR-223-3p targeted FOXO3 in 293 T cells, (^^^P < 0.001, vs. MC). All the experiments were conducted three times. (AF: atrial fibrillation, SR: sinus rhythm, M: miR-223-3p mimic, MC: mimic control)
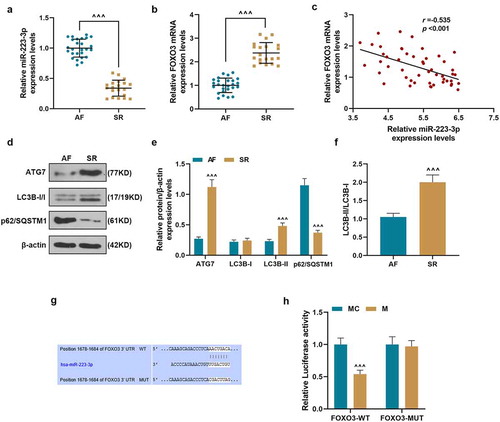
Que inhibited the cell proliferation, reduced miR-223-3p expression, and regulated the expressions of proliferation-related and autophagy-related genes in ISO-induced RCFs
For detecting the effect of Que on MF, RCFs was treated by Que and then treated with ISO or not. As shown in , after culture for 48 and 72 h, the proliferation of RCFs was significantly decreased by 50 μM Que (Que-H) but increased by ISO, as compared with control group (P < 0.001). After treatment with Que-H and Que-L (20 μM), the cell proliferation of RCFs induced by ISO was significantly decreased after cell culture for 48 and 72 h, as compared with ISO group (P < 0.001, P < 0.01, respectively). These results indicated that Que could reverse the significantly decreased proliferation of RCFs induced by ISO. Next, we detected the effect of Que on the expressions of proliferation-related genes in ISO-induced RCFs at transcriptional and translational levels. As shown in , the expressions of Cyclin D1, Fibronectin, and Col1A1 in RCFs were reduced by Que-H but increased by ISO, as compared with control group (P < 0.001), while in ISO+Que-H and ISO+Que-L groups, the above expressions increased by ISO were reversed by Que, as compared with ISO group (P < 0.001). The expression of miR-223-3p () was determined, and the result was similar with the changes of the expression of proliferation-related genes, indicating that Que could inhibit the expression of miR-223-3p in ISO-reduced RCFs. Furthermore, as shown in , the expressions of FOXO3, ATG7, and LC3B-II in RCFs were increased by Que-H but reduced by ISO, as compared with control group (P < 0.001), while in ISO+Que-H and ISO+Que-L groups, these expressions decreased by ISO were all further reversed by Que, as compared with ISO group (P < 0.001). Among all groups, the expressions of LC3B-I and p62/SQSTM1 showed opposite change to LC3B-II, but the ratio of LC3B-II/LC3B-I was consistent with LC3B-II among all groups (). In order to verify the effect of Que on autophagy, the autophagy inhibitor 3-MA was used for reverse verification. The results as shown in , that 3-MA can promote the effect of ISO on cyclin D1, Fibronectin, ColA1, LC3B-II and P62/SQSTM1 expression (P < 0.05), and 3-MA can significantly reverse the inhibitory effect of Que on cyclin D1, Fibronectin, ColA1, and P62/SQSTM1 expression, and reverse the effect of Que on LC3B-II expression (P < 0.001). All these results revealed that Que inhibited the proliferation of RCFs, reduced miR-223-3p expression and regulated the expressions of proliferation-related and autophagy in ISO-reduced RCFs.
Figure 2. Que inhibited the cell proliferation and miR-223-3p expression and regulated the expressions of proliferation-related and autophagy-related genes in ISO-reduced RCFs. (a). The cell proliferation of RCFs after induction by ISO or treatment with Que was detected by MTT assays. (b). The mRNA expressions of Cyclin D1, Fibronectin, and Col1A1 of RCFs after the induction with ISO or treatment with Que was detected by RT-qPCR, β-actin was used as an internal control. (c-d) The protein expressions of Cyclin D1, Fibronectin, and Col1A1 of RCFs after the induction with ISO or treatment with Que were detected by Western blot, β-actin was used as an internal control. (e) The expressions of miR-223-3p of RCFs after induction with ISO or treatment with Que were detected by RT-qPCR, U6 was used as an internal control. (f-i) The protein expressions of FOXO3, ATG7, LC3B-I, LC3B-II, and p62/SQSTM1 of RCFs after the induction with ISO or treatment with Que were detected by Western blot, β-actin was used as an internal control. All the experiments were conducted three times. (**P < 0.01, ***P < 0.001, vs. Control, ^^P < 0.01, ^^^P < 0.001, vs. ISO). (Que: quercetin, ISO: isoprenaline)
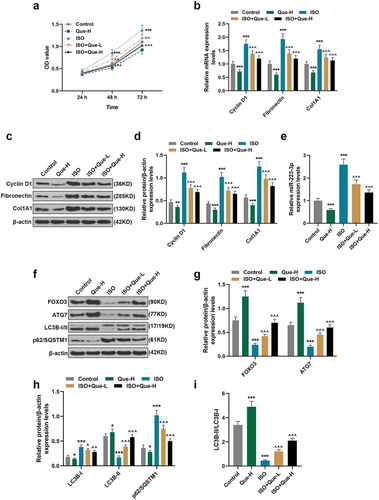
Figure 3. Que promoted autophagy in ISO-reduced RCFs. (a-b). The protein expressions of Cyclin D1, Fibronectin, and Col1A1of RCFs after the induction with ISO or 3-MA only, treatment with ISO and 3-MA, treatment with ISO and Que and treatment with ISO, Que and 3-MA were detected by Western blot, β-actin was used as an internal control. (c-e) The protein expressions of LC3B-I, LC3B-II, and p62/SQSTM1 of RCFs after the induction with ISO or 3-MA only, treatment with ISO and 3-MA, treatment with ISO and Que and treatment with ISO, Que and 3-MA were detected by Western blot, β-actin was used as an internal control. All the experiments were conducted three times. (***P < 0.001, vs. Control, ^P < 0.05, ^^P < 0.01, ^^^P < 0.001, vs. ISO, ##P < 0.01, ###P < 0.001, vs. ISO+Que-H). (Que: quercetin, ISO: isoprenaline, 3-MA: 3-Methyladenine)
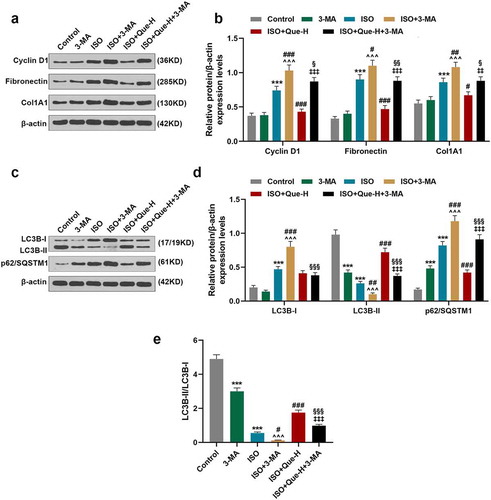
MiR-223-3p overexpression and FOXO3 knockdown reversed the inhibitory effects of Que on the significantly decreasing the proliferation of RCFs and the expressions of proliferation-related and autophagy-related genes in ISO-reduced RCFs
MiR-223-3p mimic and siFOXO3 were transfected into RCFs and the ISO-reduced RCFs. As shown in , MiR-223-3p mimic significantly up-regulated miR-223-3p, and siFOXO3 greatly down-regulated FOXO3 (P < 0.001). As shown in , miR-223-3p expression was reduced by Que-H compared with ISO group RCFs, and greatly increased miR-223-3p mimic in ISO+Que-H + M group as compared with ISO+Que-H group (P < 0.001). siFOXO3 showed no effect on the expression of miR-223-3p in ISO+Que-H+ siFOXO3 group. As for the cell proliferation of RCFs (), the cell proliferation of ISO-reduced RCFs was significantly reduced by Que-H in comparison with ISO group (P < 0.001), but miR-223-3p mimic and siFOXO3 significantly reversed the inhibitory effect of Que on the cell proliferation in ISO+Que-H + M and ISO+Que-H+ siFOXO3 groups in comparison with ISO+Que-H group (P < 0.01). In addition, the expressions of proliferation-related genes at transcriptional and translational levels were detected, from , it can be observed that Que significantly inhibited the expressions of Cyclin D1, Fibronectin, and Col1A1 in ISO-reduced RCFs compared with ISO group (P < 0.001), but miR-223-3p mimic and siFOXO3 greatly reversed the inhibitory effect of Que on the above expressions in ISO+Que-H + M and ISO+Que-H+ siFOXO3 groups compared with ISO+Que-H group (P < 0.001). Furthermore, the expressions of FOXO3 and autophagy-related proteins were determined, as shown in , the expressions of FOXO3, ATG7, and LC3B-II were increased and the expressions of LC3B-I and p62/SQSTM1 were reduced by Que in comparison with ISO group (P < 0.001), but in ISO+Que-H + M and ISO+Que-H+ siFOXO3 groups, miR-223-3p mimic and siFOXO3 greatly reversed the regulatory effects of Que on the above gene expressions as compared with ISO+Que-H group (P < 0.001). Moreover, the ratio of LC3B-II/LC3B-I was consistent with LC3B-II among all groups (). All these results revealed that miR-223-3p overexpression and FOXO3 knockdown reversed the inhibitory effects of Que on cell proliferation and the expressions of proliferation-related and autophagy-related genes in ISO-reduced RCFs.
Figure 4. MiR-223-3p overexpression and FOXO3 knockdown reversed the inhibitory effect of Que on cell proliferation and the expressions of proliferation-related and autophagy-related genes in ISO-reduced RCFs. (a). The expression of miR-223-3p after transfection with miR-223-3p mimic was detected by RT-qPCR, U6 was used as an internal control. (b).The expression of FOXO3 protein in RCFs after transfection with siFOXO3 were detected by Western blot, β-actin was used as internal control. (c).The expression of FOXO3 mRNA in RCFs after transfection with siFOXO3 were detected by RT-qPCR, β-actin was used as internal control. (d). The expression of miR-223-3p in ISO-induced RCFs after treatment with Que and transfection with miR-223-3p mimic or siFOXO3 was detected by RT-qPCR, U6 was used as an internal control. (e). The cell proliferation of ISO-induced RCFs after treatment with Que and transfection with miR-223-3p mimic or siFOXO3 was detected by MTT assay. (f). The mRNA expressions of Cyclin D1, Fibronectin, and Col1A1 of ISO-induced RCFs after treatment with Que and transfection with miR-223-3p mimic or siFOXO3 was detected by RT-qPCR, β-actin was used as an internal control. (g-h) The protein expressions of Cyclin D1, Fibronectin, and Col1A1 of ISO-induced RCFs after treatment with Que and transfection with miR-223-3p mimic or siFOXO3 was detected by Western blot, β-actin was used as an internal control. (i-l) The protein expressions of FOXO3, ATG7, LC3B-I, LC3B-II, and p62/SQSTM1 of ISO-induced RCFs after treatment with Que and transfection with miR-223-3p mimic or siFOXO3 was detected by Western blot, β-actin was used as an internal control. All experiments were conducted three times. (***P < 0.001, vs. ISO, ^^P < 0.01, ^^^P < 0.001, vs. ISO+Que-H). (Que: quercetin, ISO: isoprenaline, M: miR-223-3p mimic, MC: mimic control, siFOXO3: small interfering RNA directed against FOXO3, siNC: siRNA negative control)
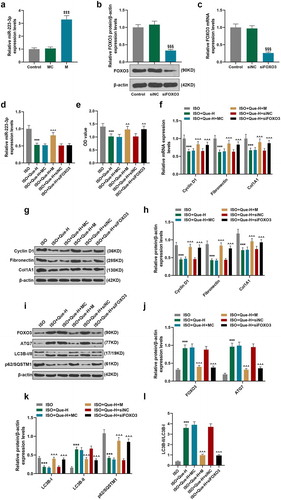
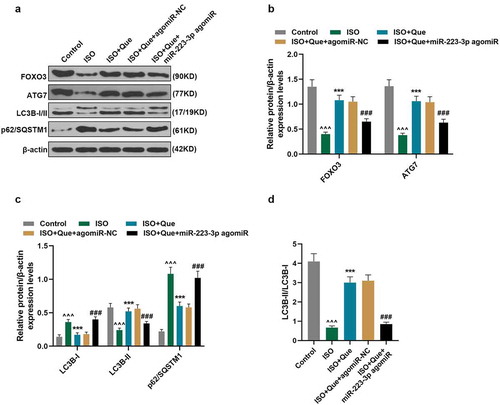
MiR-223-3p overexpression reversed the inhibitory effects of Que on MF and the expressions of Fibronectin and Col1A1 in ISO-reduced AF model rats
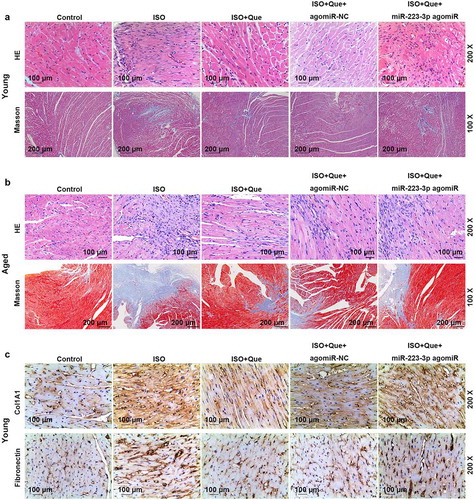
To compared the autophagy-related genes, miR-223-3p and FOXO3 between young and aged rats, we measured the miR-223-3p level and autophagy-related genes and FOXO3 proteins, and we found that the expression of miR-223-3p and p62/SQSTM1 were increased and FOXO3, ATG 7 and LC3B-II/LC3B-I were decreased in aged rats, as compared with control group (P < 0.001) (). To verify the current findings on cell model in vivo, ISO-reduced AF model was established in rats, which were further processed by Que and miR-223-3p agomiR. As shown in , the expression of miR-223-3p was increased in ISO-reduced AF model rats as compared with control group (P < 0.001), but decreased by Que decreased compared the ISO group (P < 0.001). In ISO+Que+miR-223-3p agomiR and miR-223-3p agomiR were observed to be able to greatly reverse the inhibitory effect of Que on miR-223-3p expression in comparison with ISO+Que group (P < 0.001). Furthermore, HE and Masson staining was performed to detect the changes in MF, as shown in , myocardial fibers showed an orderly arrangement, the morphology of myocardial cells was normal, and there were few corresponding collagen fibers in young control group. Slight myocardial fibrosis was observed in aged rats compared with younger rats in control group. But, the same tendency was observed in younger and aged rats: in ISO group, myocytes arranged disorderly, myocardial fibrosis and collagen fibers significantly increased. In ISO+Que, myocardial fibers regularly arranged, the proliferation of collagen fibers and myocardial fibrosis was greatly reduced, indicating that Que prevented ISO-induced cardiomyocyte hypertrophy and fibrous matrix deposition. In ISO+Que+miR-223-3p agomiR group, miR-223-3p agomiR noticeably reversed the inhibitory effect of Que on the MF in ISO-reduced AF model rats. The expressions of related proteins were determined in young rats, as shown in , the expressions of Col1A1 and Fibronectin were obviously increased in ISO group but decreased in ISO+Que group, and miR-223-3p agomiR greatly reversed the effect of Que on the expressions of Co1A 1 and Fibronectin in ISO+Que+miR-223-3p agomiR group.
Figure 5. Expression of miR-223-3p and autophagy-related proteins in young and aged rats. (a). The expression of miR-223-3p in young and aged rats was detected by RT-qPCR, U6 was used as an internal control. (b-d). The protein expressions of FOXO3, ATG7, LC3B-I, LC3B-II, and p62/SQSTM1 of young and aged rats were detected by Western blot, β-actin was used as an internal control. (e-f) The expression of miR-223-3p in ISO-reduced young and aged AF model rats was detected by RT-qPCR, U6 was used as an internal control. All experiments were conducted three times. (‡‡‡P < 0.001, ‡‡P < 0.01, ‡P < 0.05, vs. Young, ^^^P < 0.001, vs. Control, ***P < 0.001, vs. ISO, ###P < 0.001, vs. ISO+Que). (Que: quercetin, ISO: isoprenaline, AF: atrial fibrillation)
Figure 6. MiR-223-3p overexpression reversed the inhibitory effects of Que on the MF and the expressions of Fibronectin and Col1A1 in ISO-reduced AF model rats. (a-b). HE and Masson staining were conducted to observe the changes in cardiac hypertrophy and fibrosis of ISO-reduced AF model rat. (c). The expressions of Col1A1 and Fibronectin were detected by immunohistochemical. All experiments were conducted three times. (Que: quercetin, ISO: isoprenaline, AF: atrial fibrillation)
MiR-223-3p overexpression reversed the effect of Que on inhibiting the expressions of autophagy-related genes in ISO-reduced AF model rats
We then tested the underlying mechanism in young rats. As for autophagy, as shown in , the expressions of FOXO3, ATG7, and LC3B-II were reduced and the expressions of LC3B-I and p62/SQSTM1 were increased in ISO-reduced AF model rats in comparison with control rats (P < 0.001). In ISO+Que group, Que obviously increased the expressions of FOXO3, ATG7, and LC3B-II and decreased the expressions of LC3B-I and p62/SQSTM1 in comparison with ISO group (P < 0.001). Moreover, in ISO+Que+miR-223-3p agomiR group, miR-223-3p agomiR significantly reversed the effects of Que on the expressions of these autophagy-related genes. The results also showed that the ratio of LC3B-II/LC3B-I was consistent with that of LC3B-II among all the groups (), indicating that Que promoted autophagy in ISO-reduced AF model rats, while miR-223-3p agomiR reversed such an effect of Que.
Figure 7. MiR-223-3p overexpression reversed the inhibitory effects of Que on autophagy-related genes expressions in ISO-reduced AF model rats. (a-d). The protein expressions of FOXO3, ATG7, LC3B-I, LC3B-II, and p62/SQSTM1 of ISO-reduced AF model rats were detected by Western blot, β-actin was used as an internal control. All experiments were conducted three times. (^^^P < 0.001, vs. Control, ***P < 0.001, vs. ISO, ###P < 0.001, vs. ISO+Que). (Que: quercetin, ISO: isoprenaline, AF: atrial fibrillation)
Discussion
MF is an important pathological change of cardiac dysfunction caused by the continuous progression of various cardiovascular diseases (such as AF, myocardial infarction, hypertension, etc.), and it is also a major cause of myocardial remodeling [Citation12,Citation24]. Therefore, prevention or improvement of MF is crucial for the prevention of ventricular remodeling and restoration of cardiac function.
In recent years, Que has been increasingly studied for its strong biological activities and extensive pharmacological properties [Citation14]. In addition to the effects of anti-oxidant, anti-inflammation, and anti-tumor of Que, research also reported that it also has a strong cardioprotective effect on ISO-induced MF [Citation16]. ISO, which could mimic the activity of the β-adrenergic receptor, will cause myocardial necrosis. Iso-induced cardiac hypertrophy is a reliable and well-defined prototype associated with arrhythmia, myocardial cell loss, and fibrosis during the development of heart failure [Citation25]. In order to verify the finding, MF model induced by ISO was established in cells and rats. Our data revealed that Que inhibited the cell proliferation of ISO-induced RCFs and ameliorated the MF in ISO-induced rats, which confirmed the inhibitory of Que on MF. Nevertheless, the specific mechanisms should be further investigated.
MiRNAs are involved in various biological and pathological processes, including in cardiovascular diseases [Citation17,Citation26]. MiR-223 has been widely studied in cardiovascular disease [Citation18,Citation19]. Liu et al. proved that miR-223 regulates MF after myocardial infarction [Citation19]. In this study, in order to investigate whether miR-223 regulated MF in AF, AF and SR cardiac tissues were clinically collected and detected for the expression of miR-223. The results showed that the expression of miR-223-3p, a subtype of miR-223, was high-expressed in AF cardiac tissues, suggesting that miR-223-3p may have crucial effect on AF progression. MiR-223-3p also has crucial regulatory effects on various cells in many diseases, such as in type 2 diabetes, Kawasaki disease and colon cancer [Citation27–29]. However, the effect of miR-223-3p on AF has not been reported. MiR-223 could regulate the proliferation, invasion, inflammation, autophagy of various cells [Citation30–34], and has all the functions of miR-223 except for affecting autophagy [Citation35,Citation36]. Therefore, the effect of miR-223-3p on autophagy of cardiac cells was examined in the present study, and we speculated that the effects of miR-223-3p on cardiac cells were regulated by Que.
Studies increasingly found that promoting autophagy can achieve the purpose of inhibiting MF and improving heart function. In a rat model of acute myocardial infarction, atorvastatin could improve the survival of cardiomyocytes and reduce the fibrosis and infarct area, which may be related to autophagy induced by AMPK/mTOR pathway [Citation37]. Another study found that supplementing the diet with the natural polyamine spermidine can extend the life of mice by inducing autophagy and reverse aging-related cardiac dysfunctions [Citation38]. In addition, the autophagy mechanism includes autophagy-related (ATG) protein components, ATG7, known as E1-like activating enzyme, which could facilitate the generation of LC3-phosphatidylethanolamine (PE) [Citation39,Citation40]. Therefore, it is necessary to study the potential role of autophagy in the treatment of AF and MF. As one of the proteins that interact with LC3B, protein p62/SQSTM1 plays an important role in the process of autophagy like LC3B [Citation41,Citation42]. Increased expression of LC3B-II and decreased expression of p62/SQSTM1 are indicative of an enhanced autophagy [Citation43]. LC3B is believed to be the first mammalian protein to be located in the autophagosome membrane [Citation44]. Our subsequent experimental results revealed that Que inhibited the expression of miR-223-3p, which then increased the cell proliferation of ISO-induced RCFs, and reversed the effect of Que on the ISO-induced RCFs. The effect of miR-223-3p on ISO-induced RCFs was related to the inhibition of the cells autophagy by decreasing ATG7 and LC3B-I/II expression and increasing that of p62/SQSTM1. In addition, the autophagy inhibitor 3-MA was used in this study, and the results verified our hypothesis that the use of 3-MA partially reversed Que’s effect on cells. Furthermore, these results in vitro were further confirmed by our in vivo experiments, in which miR-223-3p was found to promote MF and cardiac hypertrophy of ISO-induced rats. These results confirmed our speculation that Que could inhibit the MF by increasing autophagy via regulating miR-223-3p. It should be noted that the down-stream mechanism is required to be further investigated.
As an autophagy regulator, FOXO3 is widely reported to regulate autophagy in many diseases. Zhou et al. demonstrated that FOXO3 promotes autophagy after cerebral ischemia/reperfusion [Citation22]; Gso et al. found that FOXO3 inhibits gastric adenocarcinoma cell growth via promoting autophagy [Citation45]; FOXO3 also regulates kidney tubular autophagy after urinary tract obstruction [Citation46].Study has shown that FOXO3 may regulate ATG7 at the transcriptional level and further to regulate autophagy [Citation39]. Our study proves that FOXO3 can also regulate ATG7 of RCFs cells and regulate autophagy. In addition, FOXO3 targeted by miR-223-3p enhances the functions of immune cells [Citation47]. The latest report showed that miR-223-3p expression can enhance the autophagy of mesenchymal stem cells by targeting FOXO3 [Citation48]. Consistent with previous research, we also verified that miR-223-3p targeted FOXO3 could inhibit the expression of FOXO3, thus suppressing autophagy and further promoting MF in ISO-induced MF rats. Moreover, these effects of miR-223-3p on ISO-induced MF rats could be further regulated by Que. However, there are still some limitations in our study, such as the study of elderly rats needs to be explored in future studies.
Based on the current findings, we concluded that Que could prevent ISO-induced MF by increasing autophagy via regulating miR-223-3p/FOXO3.
Availability of Data and Materials
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Acknowledgments
Not applicable
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Witassek F, Springer A, Adam L, et al. Health-related quality of life in patients with atrial fibrillation: the role of symptoms, comorbidities, and the type of atrial fibrillation. PLoS One. 2019;14(12):e0226730. PubMed PMID: 31869399.
- Moazzami K, Shao IY, Chen LY, et al. Atrial Fibrillation, Brain Volumes, and Subclinical Cerebrovascular Disease (from the Atherosclerosis Risk in Communities Neurocognitive Study [ARIC-NCS]). Am J Cardiol. 2020 Jan 15;125(2):222–228. PubMed PMID: 31771759.
- Loring Z, Shrader P, Allen LA, et al. Guideline-directed therapies for comorbidities and clinical outcomes among individuals with atrial fibrillation. Am Heart J. 2020 Jan;219:21–30. PubMed PMID: 31710841; eng.
- Wingerter R, Steiger N, Burrows A, et al. Impact of Lifestyle Modification on Atrial Fibrillation. Am J Cardiol. 2020 Jan 15;125(2):289–297. PubMed PMID: 31761147.
- Yamac AH, Kucukbuzcu S, Ozansoy M, et al. Altered expression of micro-RNA 199a and increased levels of cardiac SIRT1 protein are associated with the occurrence of atrial fibrillation after coronary artery bypass graft surgery. Cardiovasc Pathol. 2016 May-Jun;25(3):232–236. PubMed PMID: 26952538.
- Reddy V, Taha W, Kundumadam S, et al. Atrial fibrillation and hyperthyroidism: a literature review. Indian Heart J. 2017 Jul - Aug;69(4):545–550. PubMed PMID: 28822529; PubMed Central PMCID: PMCPMC5560908.
- Liu Z, Finet JE, Wolfram JA, et al. Calcium/calmodulin-dependent protein kinase II causes atrial structural remodeling associated with atrial fibrillation and heart failure. Heart Rhythm. 2019 Jul;16(7):1080–1088. PubMed PMID: 30654134; PubMed Central PMCID: PMCPMC6800146.
- Molina CE, Abu-Taha IH, Wang Q, et al. Profibrotic, Electrical, and Calcium-Handling Remodeling of the Atria in Heart Failure Patients With and Without Atrial Fibrillation. Front Physiol. 2018;9(1383). PubMed PMID: 30356673; PubMed Central PMCID: PMCPMC6189336. eng. 10.3389/fphys.2018.01383
- Liaquat MT, Makaryus AN. Cardiac Electrical and Structural Remodeling. StatPearls. Treasure Island (FL): StatPearls Publishing StatPearls Publishing LLC; 2019.
- Molkentin JD, Bugg D, Ghearing N, et al. Fibroblast-Specific Genetic Manipulation of p38 Mitogen-Activated Protein Kinase In Vivo Reveals Its Central Regulatory Role in Fibrosis. Circulation. 2017 Aug 8;136(6):549–561. PubMed PMID: 28356446; PubMed Central PMCID: PMCPMC5548661.
- Christensen G, Herum KM, Lunde IG. Sweet, yet underappreciated: proteoglycans and extracellular matrix remodeling in heart disease. Matrix Biol. 2019 Jan;75-76:286–299. PubMed PMID: 29337052.
- Jiang L, Li L, Ruan Y, et al. Ibrutinib promotes atrial fibrillation by inducing structural remodeling and calcium dysregulation in the atrium. Heart Rhythm. 2019 Sep;16(9):1374–1382. PubMed PMID: 30959203.
- Tang SM, Deng XT, Zhou J, et al. Pharmacological basis and new insights of quercetin action in respect to its anti-cancer effects. Biomed Pharmacother. 2020 Jan;121:109604. PubMed PMID: 31733570.
- Giordani B, Basnet P, Mishchenko E, et al. Utilizing Liposomal Quercetin and Gallic Acid in Localized Treatment of Vaginal Candida Infections. Pharmaceutics. 2019 Dec 20;12(1):9. PubMed PMID: 31861805.
- Ince E. The protective effect of quercetin in the alcohol-induced liver and lymphoid tissue injuries in newborns. Mol Biol Rep. 2020 Jan;47(1):451–459. PubMed PMID: 31673888.
- Li M, Jiang Y, Jing W, et al. Quercetin provides greater cardioprotective effect than its glycoside derivative rutin on isoproterenol-induced cardiac fibrosis in the rat. Can J Physiol Pharmacol. 2013 Nov;91(11):951–959. PubMed PMID: 24117263; eng.
- Su Q, Lv X. Revealing new landscape of cardiovascular disease through circular RNA-miRNA-mRNA axis. Genomics. 2019 Oct 15;112(2):1680–1685. PubMed PMID: 31626900; eng.
- Taibi F, Metzinger-Le Meuth V, Massy ZA, et al. miR-223: an inflammatory oncomiR enters the cardiovascular field. Biochim Biophys Acta. 2014 Jul;1842(7):1001–1009. PubMed PMID: 24657505.
- Liu X, Xu Y, Deng Y, et al. MicroRNA-223 Regulates Cardiac Fibrosis After Myocardial Infarction by Targeting RASA1. Cell Physiol Biochem. 2018;46(4):1439–1454. PubMed PMID: 29689569.
- Wang L, Yuan D, Zheng J, et al. Chikusetsu saponin IVa attenuates isoprenaline-induced myocardial fibrosis in mice through activation autophagy mediated by AMPK/mTOR/ULK1 signaling. Phytomedicine. 2019 May;58:152764. PubMed PMID: 31005723; eng.
- Xin Z, Ma Z, Hu W, et al. FOXO1/3: potential suppressors of fibrosis. Ageing Res Rev. 2018 Jan;41:42–52. PubMed PMID: 29138094; eng.
- Zhou H, Wang X, Ma L, et al. FoxO3 transcription factor promotes autophagy after transient cerebral ischemia/reperfusion. Int J Neurosci. 2019 Aug;129(8):738–745. PubMed PMID: 30595062; eng.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001 Dec;25(4):402–408. PubMed PMID: 11846609; eng.
- Saturska H, Shulhai Capital AC, Levchuk R, et al. Medical and social issues of cardiovascular diseases and their solution based on the experimental study of myocardial fibrosis. Wiadomosci lekarskie (Warsaw, Poland: 1960). 2019;72(1):35–39. PubMed PMID: 30796859.
- Ulla A, Mohamed MK, Sikder B, et al. Coenzyme Q10 prevents oxidative stress and fibrosis in isoprenaline induced cardiac remodeling in aged rats. BMC Pharmacol Toxicol. 2017 Apr 20;18(1):29. PubMed PMID: 28427467; PubMed Central PMCID: PMCPMC5399319. eng.
- Kabekkodu SP, Shukla V, Varghese VK, et al. Clustered miRNAs and their role in biological functions and diseases. Biol Rev Camb Philos Soc. 2018 Nov;93(4):1955–1986. PubMed PMID: 29797774; eng.
- Parrizas M, Mundet X, Castano C, et al. miR-10b and miR-223-3p in serum microvesicles signal progression from prediabetes to type 2 diabetes. J Endocrinol Invest. 2019 Nov 12;43(4):451–459. PubMed PMID: 31721085.
- Wang X, Ding YY, Chen Y, et al. Corrigendum: miR-223-3p Alleviates Vascular Endothelial Injury by Targeting IL6ST in Kawasaki Disease. Front Pediatr. 2019;7:449. PubMed PMID: 31763338; PubMed Central PMCID: PMCPMC6848949.
- Chai B, Guo Y, Cui X, et al. MiR-223-3p promotes the proliferation, invasion and migration of colon cancer cells by negative regulating PRDM1. Am J Transl Res. 2019;11(7):4516–4523. PubMed PMID: 31396355; PubMed Central PMCID: PMCPMC6684915.
- Zhou Y, Chen E, Tang Y, et al. miR-223 overexpression inhibits doxorubicin-induced autophagy by targeting FOXO3a and reverses chemoresistance in hepatocellular carcinoma cells. Cell Death Dis. 2019 Nov 6;10(11):843. PubMed PMID: 31695022; PubMed Central PMCID: PMCPMC6834650.
- Wu W, Shan Z, Wang R, et al. Overexpression of miR-223 inhibits foam cell formation by inducing autophagy in vascular smooth muscle cells. Am J Transl Res. 2019;11(7):4326–4336. PubMed PMID: 31396338; PubMed Central PMCID: PMCPMC6684933.
- Shu Y, Wang Y, Lv WQ, et al. ARRB1-promoted NOTCH1 degradation is suppressed by oncomiR miR-223 in T cell acute lymphoblastic leukemia. Cancer Res. 2019 Dec 10 PubMed PMID: 31822496; eng. DOI:10.1158/0008-5472.can-19-1471.
- Fang C, Xu L, He W, et al. Long noncoding RNA DLX6-AS1 promotes cell growth and invasiveness in bladder cancer via modulating the miR-223-HSP90B1 axis. Cell Cycle. 2019 Dec;18(23):3288–3299. PubMed PMID: 31615303.
- Guan YZ, Sun C, Wang HL, et al. MiR-223-5p inhibitor suppresses microglia inflammation and promotes Nrg-1 in rats of spinal cord injury. Eur Rev Med Pharmacol Sci. 2019 Nov;23(22):9746–9753. PubMed PMID: 31799641; eng.
- Xiao W, Wang X, Wang T, et al. MiR-223-3p promotes cell proliferation and metastasis by downregulating SLC4A4 in clear cell renal cell carcinoma. Aging (Albany NY). 2019 Jan 22;11(2):615–633. PubMed PMID: 30668544; PubMed Central PMCID: PMCPMC6366987. eng.
- Dong HC, Li PN, Chen CJ, et al. Sinomenine Attenuates Cartilage Degeneration by Regulating miR-223-3p/NLRP3 Inflammasome Signaling. Inflammation. 2019 Aug;42(4):1265–1275. PubMed PMID: 30847744; eng.
- Wang W, Wang H, Geng QX, et al. Augmentation of autophagy by atorvastatin via Akt/mTOR pathway in spontaneously hypertensive rats. Hypertens Res. 2015 Dec;38(12):813–820. PubMed PMID: 26224487; eng.
- Eisenberg T, Abdellatif M, Schroeder S, et al. Cardioprotection and lifespan extension by the natural polyamine spermidine. Nat Med. 2016 Dec;22(12):1428–1438. PubMed PMID: 27841876; PubMed Central PMCID: PMCPMC5806691. eng.
- Yu S, Yu M, He X, et al. KCNQ1OT1 promotes autophagy by regulating miR-200a/FOXO3/ATG7 pathway in cerebral ischemic stroke. Aging Cell. 2019 Jun;18(3):e12940. PubMed PMID: 30945454; PubMed Central PMCID: PMCPMC6516167. eng.
- Yuan Y, Zhao J, Gong Y, et al. Autophagy exacerbates electrical remodeling in atrial fibrillation by ubiquitin-dependent degradation of L-type calcium channel. Cell Death Dis. 2018 Aug 29;9(9):873. PubMed PMID: 30158642; PubMed Central PMCID: PMCPMC6115437. eng.
- Pankiv S, Clausen TH, Lamark T, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007 Aug 17;282(33):24131–24145. PubMed PMID: 17580304; eng.
- Komatsu M, Waguri S, Koike M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007 Dec 14;131(6):1149–1163. PubMed PMID: 18083104; eng.
- Gyongyosi A, Zilinyi R, Czegledi A, et al. The Role of Autophagy and Death Pathways in Dose-dependent Isoproterenolinduced Cardiotoxicity. Curr Pharm Des. 2019;25(19):2192–2198. PubMed PMID: 31258063; PubMed Central PMCID: PMCPMC6806536. eng.
- Kabeya Y, Mizushima N, Ueno T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000 Nov 1;19(21):5720–5728. PubMed PMID: 11060023; PubMed Central PMCID: PMCPMC305793. eng.
- Gao Y, Qi W, Sun L, et al. FOXO3 Inhibits Human Gastric Adenocarcinoma (AGS) Cell Growth by Promoting Autophagy in an Acidic Microenvironment. Cell Physiol Biochem. 2018;49(1):335–348. PubMed PMID: 30138933.
- Li L, Zviti R, Ha C, et al. Forkhead box O3 (FoxO3) regulates kidney tubular autophagy following urinary tract obstruction. J Biol Chem. 2017 Aug 18;292(33):13774–13783. PubMed PMID: 28705935; PubMed Central PMCID: PMCPMC5566530.
- Wei Y, Chen S, Sun D, et al. miR-223-3p promotes autoreactive Th17 cell responses in experimental autoimmune uveitis (EAU) by inhibiting transcription factor FOXO3 expression. FASEB J. 2019 Dec;33(12):13951–13965. PubMed PMID: 31645142.
- Long C, Cen S, Zhong Z, et al. FOXO3 is targeted by miR-223-3p and promotes osteogenic differentiation of bone marrow mesenchymal stem cells by enhancing autophagy. Hum Cell. 2020 Sep 13 PubMed PMID: 32920731; eng. DOI:10.1007/s13577-020-00421-y.
