ABSTRACT
Long non-coding RNA (lncRNA) differentiation antagonizing non-protein coding RNA (DANCR) participates in the development of diverse cancers. Nevertheless, the impact of DANCR on cervical cancer (CC) remains largely unknown. This study aims to explore the effects of DANCR sponging microRNA-145-3p (miR-145-3p) on CC. Expression of KLF5, DANCR, miR-145-3p, and zinc finger E-box binding homeobox 1 (ZEB1) in CC and adjacent normal tissues was determined. Human CC cell lines were, respectively, treated with silenced DANCR or miR145-3p mimic/inhibitor. Then, the viability, migration, invasion, and apoptosis of CC cells were measured. The cell growth in vivo was observed as well. Chromatin immunoprecipitation assay was performed to analyze the binding of KLF5 and DANCR promoter. Interaction among DANCR, miR-145-3p, and ZEB1 was assessed. KLF5, DANCR, and ZEB1 were upregulated but miR-145-3p was downregulated in CC tissues. KLF5 activated DANCR expression and the high DANCR expression was related to tumor staging, infiltrating muscle depth and lymphatic metastasis of CC patients. Reduced DANCR or elevated miR-145-3p repressed malignant behaviors of CC cells. The tumor diameter and weight were also repressed by DANCR silencing or miR-145-3p elevation. The effect of DANCR knockdown on CC cells could be reversed by miR-145-3p inhibitor. MiR-145-3p was targeted by DANCR and ZEB1 was targeted by miR-145-3p. KLF5-induced overexpression of DANCR promotes CC progression via suppressing miR-145-3p to target ZEB1. This study may provide potential targets for CC treatment.
Introduction
Cervical cancer (CC) is the second most common reproductive cancer in females after breast cancer (BC), and 84% of the CC cases were reported in developing regions [Citation1]. One-third of CC patients are still diagnosed at advanced stages. The 5-year survival rate of CC patient is decreased in ~50% of advanced stage CC patients globally, and the clinical outcome is varied and difficult to predict [Citation2]. CC has high incidence and mortality rates, which result from limited screening and preventative measures, cost, late diagnosis and availability of treatments, and high human immunodeficiency virus (HIV) infection rates [Citation3].
Long non-coding RNAs (lncRNAs) are non-coding RNAs with transcripts longer than 200 nucleotides and have essential effects on the development of human diseases [Citation4]. LncRNA differentiation antagonizing non-protein coding RNA (DANCR) is a newly found oncogenic lncRNA [Citation5] that has been revealed to participate in BC [Citation6] and endometrial carcinoma (EC) [Citation7]. Interestingly, the oncogenic role of DANCR in CC has also been verified recently [Citation8]. Moreover, evidence has shown that the transcription factor is able to activate the expression of lncRNA [Citation9]. Krüppel‐like factor (KLF) is a family of transcription factors binding to the DNA domain and KLF5 is one of the family members [Citation10]. KLF5 has been reported to regulate the expression of lncRNA RP1 in BC [Citation11] and based on this phenomenon, relation between KLF5 and DANCR was studied in our research. It has been demonstrated that lncRNA can act as a competing endogenous RNA (ceRNA) to regulate mRNA by binding their common microRNAs (miRNAs) [Citation12]. The miRNAs are small non-coding RNAs that can bind to the complementary mRNA in the 3ʹ-untranslated region and negatively regulate target gene expression through degradation of the targeted RNAs or inhibition of their translation [Citation13]. MiR-145-3p is one of the miRNAs that participate in human cancer progression, such as bladder cancer [Citation14] and prostate cancer [Citation15]. In addition, it has been reported that miR-145 functions as a biomarker for the diagnosis and radiosensitivity prediction of CC [Citation16]. However, the role miR-143-3p in CC and the relation between DANCR and miR-145-3p have been rarely explored. In addition, zinc finger E-box binding homeobox 1 (ZEB1) belongs to the ZEB family and exerts critical effects on biological processes [Citation17]. The role of ZEB1 has been broadly investigated in CC [Citation18,Citation19].
We performed this study to identify the effects of KLF5/DANCR/miR-145-3p/ZEB1 axis in the development of CC, and we hypothesized that KLF5-mediated DANCR may sponge miR-145-3p to affect the progression of CC by targeting ZEB1.
Materials and methods
Ethics statement
Written informed consents were acquired from all patients before this study. The protocol of this study was confirmed by the Ethic Committee of Hainan General Hospital. Animal experiments were strictly in accordance with the Guide to the Management and Use of Laboratory Animals issued by the National Institutes of Health. The protocol of animal experiments was approved by the Institutional Animal Care and Use Committee of Hainan General Hospital.
Study subjects
A total of 112 cases of CC tissues from patients pathologically confirmed as CC in Hainan General Hospital from March 2015 to March 2018 were collected as the study group (CC), and 112 adjacent normal tissues were selected as the control group (normal). Patients in the CC group were all pathologically confirmed as primary CC. Exclusive criteria: aged >18 years; use of hormone drug in 3 months; surgical, chemoradiotherapy, or antineoplastic history; combined severe substantive organ dysfunction, including heart, kidney, and liver; combined with autoimmune disease such as systemic lupus erythematosus and rheumatoid arthritis; combined with other malignant tumor; combined with mental disease and cognitive impairment. Fresh cervical tissues harvested from all of the subjects were treated with diethyl pyrocarbonate-treated water, placed in RNase-free centrifuge tubes and stored in liquid nitrogen.
Cell culture
Human CC cell lines HeLa, SiHa, and normal cervical epithelial cells H8 were both acquired from Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China). HeLa cells were incubated with Roswell Park Memorial Institute (RPMI) 1640 medium containing 10% fetal bovine serum (FBS), 100 U/mL penicillin and 100 U/mL streptomycin; SiHa cells and H8 cells were incubated in Dulbecco’s modified Eagle medium (DMEM)-F12 supplemented with 15 mmol/L hydroxyethyl piperazine ethanesulfonic acid, 0.5 mmol/L sodium pyruvate, 1.2 g/L sodium bicarbonate, 2.5 mmol/L L-glutamine, 10% FBS, 100 U/mL penicillin and 100 U/mL streptomycin (all from Gibco Company, NY, USA). After incubated for 2–3 d, cells were detached with 0.25% trypsin and cells in the logarithmic growth phase were selected for subsequent experiments.
Cell grouping
Cells were seeded at 6-well plates at 5 × 105 cells/well until reached 70% confluence. The transfection mixture was prepared based on manufactures’ information of LipofectamineTM 2000 kits (Invitrogen Inc., CA, USA), and pcDNA3.1, pcDNA3.1-KLF5, small interfering RNA (si)-negative control (NC), si-KLF5, short hairpin RNA (sh)-NC, sh-DANCR, mimic NC, miR-145-3p mimic, sh-DANCR + inhibitor NC and sh-DANCR + miR-145-3p inhibitor (all from GenePharma Co., Ltd., Shanghai, China) were transiently transfected into cells. A non-treated blank control group (the control group) was set. After 24-h culture, the original medium was replaced by medium containing 10% FBS for subsequent 48-h culture.
Colony formation assay
Cells were seeded and cultured for 18 d. Next, cells were fixed with methanol for 15 min and stained with 0.1% crystal violet dye solution for 30 min, and the colonies were counted under a microscope.
3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) assay
Cells were made into cell suspension (2–6 × 104 cells/mL) and added onto 96-well plates at 100 μL/well for 0, 24, and 48 h. Each well was appended with 20 μL MTT solution (5 mg/mL, Sigma–Aldrich Chemical Company, MO, USA) and incubated for 4 h. With the supernatant discarded, each well was supplemented with 100 μL dimethyl sulfoxide (Sigma) and placed for 10–15 min. The optical density (OD) value at 490 nm was evaluated by a microplate reader. The cell-free and medium-only well was set, and the same MTT solution and dimethyl sulfoxide were added to the well. The OD value of the well was set as the blank control to standardize the OD value of the experimental group. The experiment was repeated for three times.
Scratch test
Cells were seeded onto 6-well plates at 5 × 105 cells/well. On the second day, a 200 μL pipette tip was used to vertically scratch along the margin of a disinfected ruler. Tree parallel scratches were made on each well. The cell proliferation-induced effect was eliminated by adding serum-free medium. Cells were photographed under a microscope after 24 h and the migration rate was analyzed by the Image J software.
Transwell assay
Cells were cultured in serum-free medium for 24 h and the cell concentration was adjusted to 2.5 × 105 cells/mL. The Matrigel-coated upper chambers of Transwell (Corning Glass Works, N.Y., USA) were added with 200 μL cells and the lower chambers were appended with 750 μL DMEM complete medium for 24-h incubation at 37°C. Cells were normally fixed, stained with crystal violet dye solution and observed and photographed under an inverted microscope.
Flow cytometry
Cell apoptosis was analyzed using AnnexinV-fluoresceine isothiocyanate (FITC)/propidium iodide (PI): cells were detached using ethylene diamine tetraacetic acid-free trypsin, centrifuged at 2,000 rpm for 5 min twice, suspended with 500 μL binding buffer, mixed with 5 μL AnnexinV-FITC (BD Biosciences, NJ, USA) and incubated with 5 μL PI without light exposure for 15 min. A flow cytometer was used for detection in 1 h.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNAs in tissues and cells were extracted using Trizol method (Invitrogen) and reversely transcribed based on instructions of reverse transcription kits (TaKaRa Biotechnology Co., Ltd., Liaoning, China) to obtain complementary DNAs. Gene amplification was conducted on the Agilent MX3000P PCR amplifier (Agilent Technologies, CA, USA). Primers () were synthesized by Invitrogen and data were analyzed using 2−ΔΔCt method [Citation20]. U6 was used as the endogenous reference of miR-145-3p and glyceraldehyde phosphate dehydrogenase (GAPDH) was used as the endogenous reference of KLF5, DANCR, and ZEB1.
Table 1. Primer sequence
Western blot analysis
Total proteins in tissues and cells were extracted, and then quantified and diluted according to directions of bicinchoninic acid kits (Beyotime). The proteins were boiled with 1 × loading buffer at 100°C for 5 min and 100 μg proteins in each group were conducted with gel electrophoresis (100 g/L, Beyotime), and then transferred onto polyvinylidene fluoride membranes. The membranes were blocked with 50 g/L skim milk powder-tris buffer solution with tween blocking buffer for 1 h, incubated with primary antibodies ZEB1 (1:1,000, Proteintech Group Inc., Chicago, USA) and GAPDH (1:1,000, Santa Cruz Biotechnology, Inc, CA, USA) at 4°C overnight, and incubated with relative secondary antibody (Proteintech) for 1 h. Subsequently, the membranes were developed by enhanced chemiluminescent reagent (Pierce, Rockford, IL, USA) and scanned by the ChemiDocTM MP system (Bio-Rad Laboratories, CA, USA).
Cytoplasm and nuclear fractionation
The fractionation of cytoplasm and nuclear in HeLa and SiHa cells was performed based on protocols of Life Technologies’ PARIS kits (Ambion Company, Austin, TX). Cells were collected and resuspended in the cell fraction buffer, and then were incubated on ice for 10 min and centrifuged with supernatant discarded. The sediment was resuspended using Nuclei EZ storage buffer and the expression ratios of matched RNAs in cytoplasm or nuclei were analyzed by RT-qPCR with GAPDH and U6 as the controls [Citation21,Citation22].
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was conducted according to instructions of EZ-ChIPTM kits (Millipore). Cells were fixed with formalin and incubated with glycine and lysed, and the chromatin was ultrasonicated to 200–400 bp (Ouhor mechanical equipment Co., Ltd., Shanghai, China). Afterward, the fragments were precipitated by KLF5 antibody (Abcam Inc., Cambridge, MA, USA). Immunoglobulin G (Abcam) was used as the NC and RT-qPCR was used for analysis [Citation21,Citation22].
Dual luciferase reporter gene assay
Detection of luciferase reporter gene was in line with the description in a study [Citation23]. The JASPAR database (http://jaspar.genereg.net/) was used to predict the binding site of KLF5 and DANCR promoter. DANCR cyclin B1 (B1) and cyclin B2 segment sequences were synthesized by Invitrogen, and then inserted into pGL3-basic vector and co-transfected with KLF5 expression plasmid into 293T cells.
The 293T cells together with HeLa and SiHa cells were seeded onto 24-well plates and pmirGLO-DANCR-wild type (WT), pmirGLO-DANCR-mutant type (MUT), pmirGLO-ZEB1-WT and pmirGLO-ZEB1-MUT were, respectively, co-transfected with mimic-NC or miR-145-3p mimic. The relative luciferase activity was determined by a dual luciferase reporter gene detection system (Promega Corporation, Madison, WI, USA) [Citation21,Citation22].
Subcutaneous tumorigenesis in nude mice
A total of 80 BALB/C nude mice aged 4 w and obtained from Dashuo experimental animal Co., Ltd. (Sichuan, China) were fed on specific pathogen-free laminar flow racks. The nude mice were subcutaneously injected with HeLa and SiHa cell suspension (2 × 106 cells/50 µL serum) at the left armpit. The mice were divided into four groups (n = 8): the sh-NC, sh-DANCR, mimic NC, and miR-145-3p mimic groups. The non-treated control group was also set. The xenografts were harvested, measured, and weighed 5 weeks later.
Statistical analysis
All data analyses were conducted using SPSS version 21.0 software (IBM Corp. Armonk, NY, USA). The measurement data conforming to the normal distribution were expressed as mean ± standard deviation. The unpaired t-test was performed for comparisons between two groups, one-way analysis of variance (ANOVA) was used for comparisons among multiple groups and Tukey’s post hoc test was used for pairwise comparisons after one-way ANOVA. The classified variable was analyzed by Fisher’s exact probabilistic method. P value <0.05 was indicative of statistically significant difference.
Results
Relationship between DANCR expression and clinicopathological characteristics of CC patients
CC patients were separated into the high expression group (n = 67) and the low expression group (n = 45) based on the median of DANCR expression in CC tissues. According to the results of analysis, the DANCR expression was not related to age and differentiation (all P > 0.05), but associated with histological type, tumor staging, infiltrating muscle depth and lymphatic metastasis of CC patients (all P < 0.05) ().
Table 2. Relationship between DANCR expression and clinicopathological characteristics of CC patients
KLF5, DANCR, and ZEB1 are upregulated while miR-145-3p is downregulated in CC tissues and KLF5 activates DANCR in CC cells
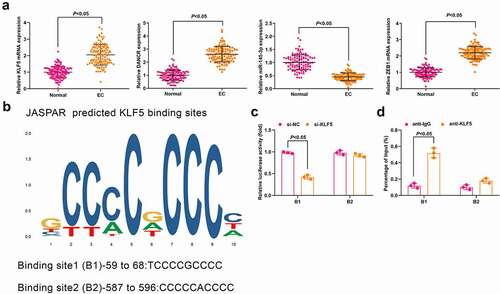
Expression of KLF5, DANCR, miR-145-3p, and ZEB1 in CC and normal tissues was determined and it came out that ()) KLF5, DANCR, and ZEB1 were upregulated while miR-145-3p was downregulated in CC tissues compared with the normal tissues (all P < 0.05). Meanwhile, the result of Pearson test implied that KLF5 expression was positively correlated with DANCR expression; DANCR expression was negatively correlated with miR-145-3p expression while positively correlated with ZEB1 expression; whereas miR-145-3p expression was negatively correlated with ZEB1 expression (Supplementary Figure 1a).
Figure 1. KLF5, DANCR and ZEB1 are upregulated while miR-145-3p is downregulated in CC tissues and KLF5 activates DANCR in CC cells. (a) Expression of KLF5, DANCR, miR-145-3p and ZEB1 in CC and normal tissues was determined using RT-qPCR; (b) binding site of KLF5 of DANCR promoter was predicted using JASPAR; (c) KLF5 binding DANCR promoter region was assessed using luciferase report gene assay; (d) Relative enrichment of KLF5 in DANCR promoter was detected by ChIP assay; n = 112 in Fig. A, N = 3 in Fig. C, D; the measurement data conforming to the normal distribution were expressed as mean ± standard deviation and unpaired t-test was performed for comparisons between two groups. B1, cyclin B1; B2, cyclin B2
The expressions of KLF5, DANCR, miR-145-3p, and ZEB1 in CC cells Hela, Siha and normal cervical epithelial cells H8 were detected by RT-qPCR (Supplementary Figure 1B). The results showed that KLF5, DANCR, and ZEB1 were significantly enriched in CC cells, whereas miR-145-3p was markedly reduced (all P < 0.05).
In consideration of the high expression level of DANCR in CC tissues, we next aimed to reveal the potential mechanisms of DANCR in CC. Evidence has shown that the transcription factor is able to activate the expression of lncRNA [Citation22]. Thus, we searched the JASPAR algorithm and two high-score binding sites of KLF5 regulating diverse cellular processes were found to distribute in DANCR promoter ()). For further confirmation, we synthesized pcDNA3.1, pcDNA3.1-KLF5, si-NC, and si-KLF5. It was found that pcDNA3.1-KLF5 transfection upregulated KLF5 and si-KLF5 transfection downregulated KLF5 (both P < 0.05; Supplementary Figure 1C). Moreover, it was observed in RT-qPCR that siRNA-mediated KLF5 silencing significantly inhibited endogenous DANCR expression, while pcDNA3.1-KLF5-mediated KLF5 enrichment elevated DANCR expression (both P < 0.05; Supplementary Figure 1D).
Dual-luciferase reporter gene assay was performed to further confirm the specific-binding sites, and we found that si-KLF5 reduced the luciferase activity in DANCR cyclin B1 area of CC cells (P < 0.05; )). Results of ChIP assay indicated that KLF5 was able to bind to the B1 binding sites in DANCR ()). These findings suggested that KLF5 activated DANCR expression in CC cells.
Inhibited DANCR restricts CC cell malignant behaviors
It was detected using RT-qPCR that in HeLa and SiHa cells that the expression of DANCR was decreased in the sh-DANCR group compared with the sh-NC group (P < 0.05) ()).
Figure 2. Inhibited DANCR restricts CC cell malignant behaviors. (a) DANCR expression in HeLa and SiHa cells was determined using RT-qPCR; (b) Colony formation ability of HeLa and SiHa cells was determined using colony formation assay; (c) Viability of HeLa and SiHa cells was assessed using MTT assay; (d) Migration ability of HeLa and SiHa cells was assessed using scratch test; (e) Invasion ability of HeLa and SiHa cells was assessed using Transwell assay; (f) Apoptosis rate of HeLa and SiHa cells was assessed using flow cytometry; repetitions = 3; * P < 0.05 vs the sh-NC group; the measurement data conforming to the normal distribution were expressed as mean ± standard deviation, one-way ANOVA was used for comparisons among multiple groups and Tukey’s post hoc test was used for pairwise comparisons after one-way ANOVA
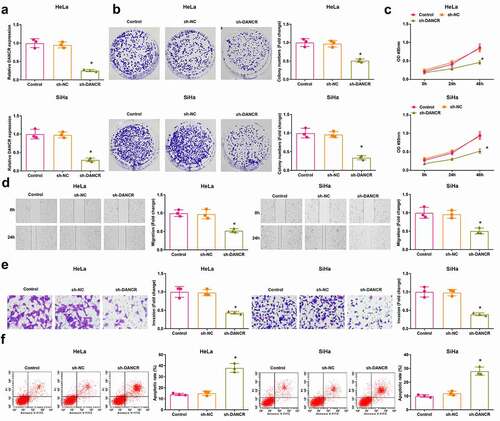
The viability of cells was assessed using colony formation assay and MTT assay, the scratch test and Transwell assay were used to determine the cell migration and invasion, and the apoptosis was detected using flow cytometry. The results (–f)) reflected that cell viability, migration, and invasion of HeLa and SiHa cells were suppressed, while the cell apoptosis was promoted in the sh-DANCR group versus the sh-NC group (all P < 0.05). These data revealed a suppressive role of DANCR inhibition in CC cell growth.
Elevated miR-145-3p inhibits CC cell malignant behaviors
Expression of miR-145-3p in HeLa and SiHa cells was detected by RT-qPCR and we found that versus the mimic NC group, the miR-145-3p mimic group had higher miR-145-3p expression (P < 0.05) ()).
Figure 3. Elevated miR-145-3p restricts CC cell malignant behaviors. (a) miR-145-3p expression in HeLa and SiHa cells was determined using RT-qPCR; (b) Colony formation ability of HeLa and SiHa cells was determined using colony formation assay; (c) Viability of HeLa and SiHa cells was assessed using MTT assay; (d) Migration ability of HeLa and SiHa cells was assessed using scratch test; (e) Invasion ability of HeLa and SiHa cells was assessed using Transwell assay; (f) Apoptosis rate of HeLa and SiHa cells was assessed using flow cytometry; repetitions = 3; * P < 0.05 vs the mimic NC group; the measurement data conforming to the normal distribution were expressed as mean ± standard deviation and unpaired t-test was performed for comparisons between two groups

The results of colony formation assay, MTT assay, scratch test, Transwell assay and flow cytometry suggested that (–f)) reflected that cell viability, migration, and invasion of HeLa and SiHa cells were suppressed, and the cell apoptosis was facilitated in the miR-145-3p mimic group compared with the mimic NC group (all P < 0.05). The above findings indicated that miR-145-3p served as a tumor suppressor during the development of CC.
MiR-145-3p is targeted by DANCR and ZEB1 is targeted by miR-145-3p
Accumulating evidence has indicated that lncRNAs serve as ceRNAs to interact with miRNAs, thus affecting miRNA expression in cytoplasm [Citation24,Citation25]. We extracted RNA in cytoplasm and nuclei to assess the expression of DANCR and it was discovered that DANCR mainly expressed in cytoplasm ()). The complementary-binding sites of DANCR and miR-145-3p, and miR-145-3p and ZEB1 were predicted by a bioinformatic tool (https://cm.jefferson.edu/rna22/Precomputed/) ()). It was confirmed by dual-luciferase reporter gene assay that miR-145-3p was a target gene of DANCR and ZEB1 was a target gene of miR-145-3p ().
Figure 4. MiR-145-3p is targeted by DANCR and ZEB1 is targeted by miR-145-3p. (a) Subcellular localization analysis of DANCR; (b) Binding sites of DANCR and miR-145-3p, and miR-145-3p and ZEB1 were predicted; (c) Target relationships between DANCR and miR-145-3p, and miR-145-3p and ZEB1 in HeLa cells were confirmed by dual luciferase reporter gene assay; (d) Target relationships between DANCR and miR-145-3p, and miR-145-3p and ZEB1 in SiHa cells were confirmed by dual luciferase reporter gene assay; (e) Expression of miR-145-3p in HeLa and SiHa cells was assessed by RT-qPCR; (f,g) ZEB1 expression in HeLa and SiHa cells was assessed by RT-qPCR and Western blot analysis; * P < 0.05 vs the sh-NC group, + P < 0.05 vs the mimic NC group; repetitions = 3; the measurement data conforming to the normal distribution were expressed as mean ± standard deviation, unpaired t-test was performed for comparisons between two groups, one-way ANOVA was used for comparisons among multiple groups and Tukey’s post hoc test was used for pairwise comparisons after one-way ANOVA
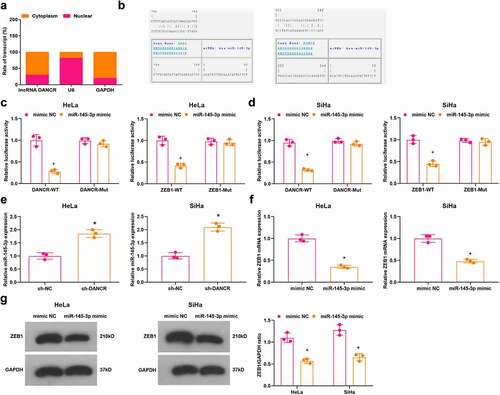
Outcomes of RT-qPCR and Western blot analysis implied that (–g)) in HeLa and SiHa cells, the sh-DANCR group had elevated expression of miR-145-3p compared with the sh-NC group (all P < 0.05); miR-145-3p mimic group had down-regulated ZEB1 expression versus the mimic NC group (both P < 0.05).
These results implied that DANCR targets miR-145-3p, which interacts with ZEB1. DANCR inhibition promotes miR-145-3p expression to inhibit ZEB1 expression.
DANCR/miR-145-3p/ZEB1 axis affects CC progression
Results of RT-qPCR and Western blot analysis indicated that in HeLa and SiHa cells, ZEB1 was upregulated in the sh-DANCR + miR-145-3p inhibitor group contrasted to the sh-DANCR + inhibitor NC group (P < 0.05) ()).
Figure 5. MiR-145-3p inhibition reverses the role of DANCR silencing in CC cells. (a) ZEB1 expression in HeLa and SiHa cells was determined using RT-qPCR and Western blot analysis; (b) Colony formation ability of HeLa and SiHa cells was determined using colony formation assay; (c) Viability of HeLa and SiHa cells was assessed using MTT assay; (d) Migration ability of HeLa and SiHa cells was assessed using scratch test; (e) Invasion ability of HeLa and SiHa cells was assessed using Transwell assay; (f) Apoptosis rate of HeLa and SiHa cells was assessed using flow cytometry; repetitions = 3; # P < 0.05 vs the sh-DANCR + inhibitor NC group; the measurement data conforming to the normal distribution were expressed as mean ± standard deviation and unpaired t-test was performed for comparisons between two groups
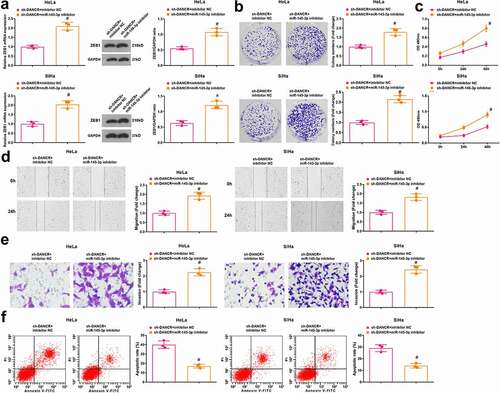
The viability, colony formation ability, migration, invasion, and apoptosis of HeLa and SiHa cells were determined and we found that (–f)) compared with the sh-DANCR + inhibitor NC group, the viability, colony formation ability, migration, and invasion were enhanced while apoptosis was inhibited in cells of the sh-DANCR + miR-145-3p inhibitor group (all P < 0.05).
The above data indicated that the tumor suppressor role of silenced DANCR in CC is reversed by miR-145-3p inhibition, and the DANCR/miR-145-3p/ZEB1 axis affects CC progression.
Silenced DANCR or elevated miR-145-3p decelerates CC cell growth in vivo
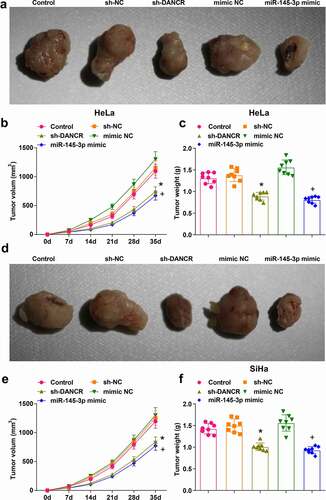
The tumor growth was observed through nude-mouse transplanted tumor models and the effects of DANCR and miR-145-3p on HeLa and SiHa cells in nude mice were evaluated. Five weeks after the subcutaneous tumorigenesis, the diameter and weight of xenografts were measured. The outcomes indicated that (–f)) relative to the sh-NC or mimic NC group, the diameter, and weight of xenografts were both reduced in the sh-DANCR or miR-145-3p mimic (all P < 0.05). Thus, we concluded that silenced DANCR or elevated miR-145-3p decelerated CC cell growth in vivo.
Figure 6. Silenced DANCR or elevated miR-145-3p decelerates CC cell growth in vivo. (a) Subcutaneous xenografts harvested from nude mice injected with HeLa cells; (b) Volume of HeLa xenografts 5 weeks after tumorigenesis; (c) Weight of HeLa xenografts 5 weeks after tumorigenesis; (d) Subcutaneous xenografts harvested from nude mice injected with SiHa cells; (e) Volume of SiHa xenografts 5 weeks after tumorigenesis; (f) Weight of SiHa xenografts 5 weeks after tumorigenesis; *, P < 0.05 vs the sh-NC group, +, P < 0.05 vs the mimic NC group; n = 8; the measurement data conforming to the normal distribution were expressed as mean ± standard deviation, one-way ANOVA was used for comparisons among multiple groups and Tukey’s post hoc test was used for pairwise comparisons after one-way ANOVA
Discussion
CC is the fourth commonly diagnosed cancer and the fourth leading cause of cancer-related deaths in female [Citation26]. The present research focused on the role of lncRNA DANCR sponging miR-145-3p in the progression of CC by regulating ZEB1. Results in our study indicated that KLF5-activated DANCR was capable of downregulating miR-145-3p to promote progression of CC by elevating ZEB1.
DANCR expression in CC tissues and adjacent normal tissues was assessed and we have found that DANCR expression was higher in CC tissues. As recently reported, DANCR is broadly enriched in CC tissues and cells [Citation27], and it has also been identified that DANCR is highly expressed in endometrial carcinoma tissues in relation to the normal control tissues [Citation7]. Based on the dysregulation of DANCR in CC, we investigated the relationship between DANCR expression and clinicopathological characteristics of CC patients. The data indicated that DANCR expression was associated with tumor staging, infiltrating muscle depth and lymphatic metastasis of CC patients. Similarly, Chen et al. have found that high DANCR expression is associated with lymph node metastasis and TNM stage of patients with pancreatic ductal adenocarcinoma [Citation28]. Furthermore, in gastric cancer, DANCR expression is related to TNM stage, lymphatic metastasis, and invasion depth [Citation29]. The CC cells were treated with sh-DANCR to observe the role of DANCR knockdown in biological processes of CC cells. It was found that inhibition of DANCR suppressed viability, migration and invasion, and promoted apoptosis of CC cells. In accordance with this finding, a literature has revealed that DANCR silencing inhibits proliferation, migration, and invasion of glioma cells [Citation5], and it has also been verified that downregulation of DANCR restrains proliferation, migration and invasion, and facilitates apoptosis of osteosarcoma cells [Citation30]. Moreover, the inhibitive role of silenced DANCR in CC cell growth in vivo has been uncovered in our study as well. Consistently, Wang et al. have found that DANCR silencing decelerates cholangiocarcinoma cell growth in vivo [Citation31]. In addition, lncRNAs are known to act as ceRNAs to sponge miRNAs, thus influencing the expression of target mRNAs [Citation24]. DANCR has been demonstrated to sponge miR-214-5p in pancreatic cancer [Citation32] and miR-216-5p in hepatocellular carcinoma [Citation33]. However, the role of DANCR sponging miR-145-3p in CC has not been studied yet.
MiR-145-3p expression in CC tissues and adjacent normal tissues was assessed as well, and it was found that the CC tissues had lower miR-145-3p expression than normal tissues. In line with our finding, Matsushita et al. have elucidated that miR-145-3p is lowly expressed in bladder cancer tissues [Citation14], and this downregulation has been identified in prostate cancer tissues as well [Citation15]. As an anti-tumor miRNA in CC, miR-145-3p was found to constrain viability, migration and invasion, and facilitate apoptosis of CC cells in our study. Similarly, it has been validated that miR-145-3p suppresses proliferation and promotes apoptosis of osteosarcoma cells [Citation34]. The tumor repressor role of miR-145-3p has also been clarified in a research that miR-145-3p is able to restrict malignant behaviors of tumor cells [Citation35]. In addition, we have found that the transfection of miR-145-3p inhibitor reversed the effect of DANCR silencing on CC cells. A literature has revealed that reduction of miR-145-5p abolished lncRNA SNHG1 inhibition-induced suppression of viability, proliferation, migration, and invasion in non-small cell lung cancer cells [Citation36]. Besides, miRNAs could negatively regulate target gene expression [Citation13], and we discovered that ZEB1 was a target gene of miR-145-3p and was elevated in CC tissues. Although miR-145 has been reported to target ZEB1 in pancreatic cancer [Citation37], the target relationship between miR-145-3p and ZEB1 in CC remains unknown. Moreover, the frequent overexpression of ZEB1 in CC tissues has been studied [Citation18,Citation19].
Aim of this study is to investigate the role of KLF5/DANCR/miR-145-3p/ZEB1 axis in the development of CC, and we found that the inhibition of DANCR elevated miR-145-3p to suppress malignant behaviors of CC cells by reducing ZEB1, thus decelerating CC development. Findings in our research may be help for exploring molecular mechanisms in CC.
Conflicts of interest
No potential conflict of interest was reported by the author(s).
Supplemental Material
Download Zip (794.6 KB)Acknowledgments
We acknowledge and appreciate our colleagues for their valuable suggestions and technical assistance for this study.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Ngune I, Kalembo F, Loessl B, et al. Biopsychosocial risk factors and knowledge of cervical cancer among young women: a case study from Kenya to inform HPV prevention in Sub-Saharan Africa. PLoS One. 2020;15(8):e0237745.
- Park S, Kim J, Eom K, et al. microRNA-944 overexpression is a biomarker for poor prognosis of advanced cervical cancer. BMC Cancer. 2019;19(1):419.
- Ifediora CO, Azuike EC. Knowledge and attitudes about cervical cancer and its prevention among female secondary school students in Nigeria. Trop Med Int Health. 2018;23(7):714–723.
- Lin PC, Huang HD, Chang CC, et al. Long noncoding RNA TUG1 is downregulated in non-small cell lung cancer and can regulate CELF1 on binding to PRC2. BMC Cancer. 2016;16:583.
- Feng L, Lin T, Che H, et al. Long noncoding RNA DANCR knockdown inhibits proliferation, migration and invasion of glioma by regulating miR-135a-5p/BMI1. Cancer Cell Int. 2020;20(1):53.
- Zhang KJ, Tan XL, Guo L. The long non-coding RNA DANCR regulates the inflammatory phenotype of breast cancer cells and promotes breast cancer progression via EZH2-dependent suppression of SOCS3 transcription. Mol Oncol. 2020;14(2):309–328.
- Sun J, Gao S, Lu C. Knockdown of differentiation antagonizing non-protein coding RNA exerts anti-tumor effect by up-regulating miR-214 in endometrial carcinoma. Mol Cell Biochem. 2019;460(1–2):9–15.
- Tian W, Lei N, Guo R, et al. Long non-coding RNA DANCR promotes cervical cancer growth via activation of the Wnt/beta-catenin signaling pathway. Cancer Cell Int. 2020;20:61.
- Dong H, Wang W, Mo S, et al. SP1-induced lncRNA AGAP2-AS1 expression promotes chemoresistance of breast cancer by epigenetic regulation of MyD88. J Exp Clin Cancer Res. 2018;37(1):202.
- Wang Z, Qiu X, Zhang H, et al. KLF5 influences cell biological function and chemotherapy sensitivity through the JNK signaling pathway in anaplastic thyroid carcinoma. J Biochem Mol Toxicol. 2020;34(5):e22469.
- Jia X, Shi L, Wang X, et al. KLF5 regulated lncRNA RP1 promotes the growth and metastasis of breast cancer via repressing p27kip1 translation. Cell Death Dis. 2019;10(5):373.
- Zhang Z, Li M, Zhang Z. lncRNA MALAT1 modulates oxaliplatin resistance of gastric cancer via sponging miR-22-3p. Onco Targets Ther. 2020;13:1343–1354.
- Yan Y, Dang HW, Zhang X, et al. The protective role of MiR-206 in regulating cardiomyocytes apoptosis induced by ischemic injury by targeting PTP1B. Biosci Rep. 2020;40(1).
- Matsushita R, Yoshino H, Enokida H, et al. Regulation of UHRF1 by dual-strand tumor-suppressor microRNA-145 (miR-145-5p and miR-145-3p): inhibition of bladder cancer cell aggressiveness. Oncotarget. 2016;7(19):28460–28487.
- Zheng Y, Chen C-J, Lin Z-Y, et al. Circ_KATNAL1 regulates prostate cancer cell growth and invasiveness through the miR-145-3p/WISP1 pathway. Biochem Cell Biol. 2020;98(3):396–404.
- Wei H, Wen-Ming C, Jun-Bo J. Plasma miR-145 as a novel biomarker for the diagnosis and radiosensitivity prediction of human cervical cancer. J Int Med Res. 2017;45(3):1054–1060.
- Viswanathan K, Siddiqui MT, Borczuk AC. The diagnostic utility of zinc E-box 1 (ZEB1) transcription factor for identification of pulmonary sarcomatoid carcinoma in cytologic and surgical specimens. J Am Soc Cytopathol. 2020;9(1):55–61.
- Xu J, Wang H, Wang H, et al. The inhibition of miR-126 in cell migration and invasion of cervical cancer through regulating ZEB1. Hereditas. 2019;156(1):11.
- Chen G, Huang P, Xie J, et al. microRNA211 suppresses the growth and metastasis of cervical cancer by directly targeting ZEB1. Mol Med Rep. 2018;17(1):1275–1282.
- Ayuk SM, Abrahamse H, Houreld NN. The role of photobiomodulation on gene expression of cell adhesion molecules in diabetic wounded fibroblasts in vitro. J Photochem Photobiol B. 2016;161:368–374.
- Wang ZY, Duan Y, Wang P. SP1-mediated upregulation of lncRNA SNHG4 functions as a ceRNA for miR-377 to facilitate prostate cancer progression through regulation of ZIC5. J Cell Physiol. 2020;235(4):3916–3927.
- Quan X, Zhao M, Yang X, et al. AP2gamma mediated downregulation of lncRNA LINC00511 as a ceRNA suppresses trophoblast invasion by regulating miR-29b-3p/Cyr61 axis. Biomed Pharmacother. 2019;120:109269.
- Wu J, Zhao W, Wang Z, et al. Long non-coding RNA SNHG20 promotes the tumorigenesis of oral squamous cell carcinoma via targeting miR-197/LIN28 axis. J Cell Mol Med. 2019;23(1):680–688.
- Xian D, Zhao Y. LncRNA KCNQ1OT1 enhanced the methotrexate resistance of colorectal cancer cells by regulating miR-760/PPP1R1B via the cAMP signalling pathway. J Cell Mol Med. 2019;23(6):3808–3823.
- Li Y, Wang H, Huang H. Long non-coding RNA MIR205HG function as a ceRNA to accelerate tumor growth and progression via sponging miR-122-5p in cervical cancer. Biochem Biophys Res Commun. 2019;514(1):78–85.
- Su HC, Wu S-C, Yen L-C, et al. Gene expression profiling identifies the role of Zac1 in cervical cancer metastasis. Sci Rep. 2020;10(1):11837.
- Liang H, Zhang C, Guan H, et al. LncRNA DANCR promotes cervical cancer progression by upregulating ROCK1 via sponging miR-335-5p. J Cell Physiol. 2019;234(5):7266–7278.
- Chen L, Liu J, Tang T, et al. lncRNA differentiation antagonizing nonprotein coding RNA overexpression accelerates progression and indicates poor prognosis in pancreatic ductal adenocarcinoma. Onco Targets Ther. 2018;11:7955–7965.
- Pan L, Liang W, Gu J, et al. Long noncoding RNA DANCR is activated by SALL4 and promotes the proliferation and invasion of gastric cancer cells. Oncotarget. 2018;9(2):1915–1930.
- Pan Z, Wu C, Li Y, et al. LncRNA DANCR silence inhibits SOX5-medicated progression and autophagy in osteosarcoma via regulating miR-216a-5p. Biomed Pharmacother. 2020;122:109707.
- Wang N, Zhang C, Wang W, et al. Long noncoding RNA DANCR regulates proliferation and migration by epigenetically silencing FBP1 in tumorigenesis of cholangiocarcinoma. Cell Death Dis. 2019;10(8):585.
- Yao Z, Chen Q, Ni Z, et al. Long non-coding RNA differentiation antagonizing nonprotein coding RNA (DANCR) promotes proliferation and invasion of pancreatic cancer by sponging miR-214-5p to regulate E2F2 expression. Med Sci Monit. 2019;25:4544–4552.
- Wang J, Pu J, Zhang Y, et al. DANCR contributed to hepatocellular carcinoma malignancy via sponging miR-216a-5p and modulating KLF12. J Cell Physiol. 2019;234(6):9408–9416.
- Wu G, Yu W, Zhang M, et al. MicroRNA-145-3p suppresses proliferation and promotes apotosis and autophagy of osteosarcoma cell by targeting HDAC4. Artif Cells Nanomed Biotechnol. 2018;46(sup2):579–586.
- Yamada Y, Koshizuka K, Hanazawa T, et al. Passenger strand of miR-145-3p acts as a tumor-suppressor by targeting MYO1B in head and neck squamous cell carcinoma. Int J Oncol. 2018;52(1):166–178.
- Lu Q, Shan S, Li Y, et al. Long noncoding RNA SNHG1 promotes non-small cell lung cancer progression by up-regulating MTDH via sponging miR-145–5p. FASEB J. 2018;32(7):3957–3967.
- Gao Y, Zhang Z, Li K, et al. Linc-DYNC2H1-4 promotes EMT and CSC phenotypes by acting as a sponge of miR-145 in pancreatic cancer cells. Cell Death Dis. 2017;8(7):e2924.
