ABSTRACT
The GPN proteins are a conserved family of GTP-binding proteins that are involved in the assembly and subsequent import of RNA polymerase II and III. In this study, we sought to ascertain the specificity of yeast GPN2 for RNA polymerases by screening the localization of a collection of 1350 GFP-tagged nuclear proteins in WT or GPN2 mutant cells. We found that the strongest mislocalization occurred for RNA polymerase II and III subunits and only a handful of other RNAPII associated proteins were altered in GPN2 mutant cells. Our screen identified Ess1, an Rpb1 C-terminal domain (CTD) prolyl isomerase, as mislocalized in GPN2 mutants. Building on this observation we tested for effects of mutations in other factors which regulate Rpb1-CTD phosphorylation status. This uncovered significant changes in nuclear-cytoplasmic distribution of Rpb1-GFP in strains with disrupted RNA polymerase CTD kinases or phosphatases. Overall, this screen shows the exquisite specificity of GPN2 for RNA polymerase transport, and reveals a previously unappreciated role for CTD modification in RNAPII nuclear localization.
Introduction
The assembly and nuclear transport of RNA polymerases from the cytoplasm is critical for all eukaryotes. Despite the fundamental role of RNA polymerases in all cells, the process by which they are assembled and imported into the nucleus remains poorly understood. Studies over the past several years have identified factors required for function in RNA polymerase biogenesis. In mammalian cells, the R2TP prefoldin complex [Citation1] and HSP90 have been identified as required co-factors for RNA polymerase assembly and nuclear import [Citation2], in concert with a family of three GTPases known as the GPN proteins [Citation2,Citation3]. In addition, research in the budding yeast Saccharomyces cerevisiae has also revealed a requirement for the prefoldin Bud27 [Citation4] in the assembly of all three RNA polymerases while Iwr1 and the karyopherin-like protein Rtp1 mediate nuclear import [Citation5–7].
The GPN proteins were originally identified through the physical interaction of MBDin/XAB1/GPN1 with both the DNA repair protein XPA chromatin modifier MBD2 [Citation8,Citation9]. X-ray crystallography revealed that the archaeal GPN1 ortholog PAB0955 is a GTPase that forms homodimers [Citation10], yet little was understood about the biological function of GPN1. More recently, mutation of the yeast GPN family members were shown to promote chromosomal instability [Citation11,Citation12] and defects in sister chromatid cohesion [Citation13,Citation14]. All three yeast GPN proteins, Gpn1, Gpn2 and Gpn3 have been shown to play a role in the assembly and subsequent nuclear import of RNA polymerase II (RNAPII), with Gpn2 and Gpn3 also required for correct assembly of RNA polymerase III (RNAPIII) complexes [Citation12,Citation15]. While the GTPase activity of GPN1 is required for correct RNAPII biogenesis, the precise role of each GPN protein remains unclear.
Physical interactions between GPNs and RNA polymerases are likely transient and were first observed under conditions that result in the accumulation of RNA polymerase assembly intermediates [Citation2]. Additionally, global analysis of protein complexes in yeast revealed few physical interactions for GPN proteins [Citation16]. Thus, GPNs act somewhat like molecular chaperones, aiding the biogenesis and localization of RNA polymerase complexes somehow, but not participating in the activity of the final assembled complex. Based on this idea, we sought to determine the potential substrate repertoire of the Gpn2 protein by asking whether Gpn2 regulates the assembly and subsequent nuclear import of any other protein complexes. Here we present a comprehensive screen of over 1350 GFP-tagged nuclear associated proteins for mislocalization in a gpn2-2 mutant background. These studies show strong evidence that Gpn2 function is exquisitely specific to RNAPII and RNAPIII biogenesis. While we show that not all reported players in RNA polymerase assembly have clear roles, the activity of the GPN proteins is an ancient and highly conserved factor specifically required for RNA polymerase maturation.
Methods and materials
GFP strain construction
Yeast crossing was done to generate a starter yeast strain containing the gpn2-2 mutation, a LYP1 deletion for haploid selection and a LEU2 marker under the control of a Ste2 promoter. Additionally, an HTA2::mCherry marker was introduced to provide a fluorescent nuclear marker. Three hundred and eighty-four spot arrays of GFP-tagged strains annotated as nuclear, nuclear periphery or nucleolar were prepared from the GFP tagged collection [Citation17] and crossed with the starter strain (Table S1) using a Singer ROTOR robot. Following sporulation, haploid selection was performed by plating spores onto selective media lacking leucine, histidine and uracil, supplemented with thialysine. Sporulation and subsequent tetrad dissection was used to generate all validation strains and query strains to investigate RNA polymerase subunit localization.
Microscopy and image scoring and validation
Ninety-six well plates of query strains and controls were grown to log phase at 30°C in SC-LEU supplemented with hygromycin and thialysine. Imaging was done in 12 sample batches for paired mutant and control strains using Teflon coated multi sample slides (TekDon). Imaging was done on a Leica fluorescence microscope. Images were visually scored for obvious differences between mutant and controls using Metamorph software. Validation of hits was done for haploid strains from tetrad dissection and quantitated as described previously [Citation12]. Temperature sensitive (ts) mutant strains were obtained from the Hieter and Boone lab ts collections [Citation11,Citation18].
Cross suppression analysis
High copy plasmids representing the genes of interest were isolated from the MoBY plasmid collection [Citation19]and transformed into yeast mutants using standard lithium acetate transformation [Citation20]. Transformants were cultured overnight, serially diluted and plated on SC-LEU + G418 plates to assay complementation of the mutant growth defect.
Results and discussion
The GPN family members do not appear to exhibit functional redundancy
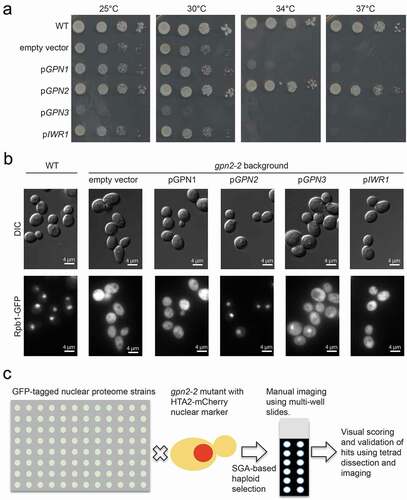
The GPN proteins sequence similarity and shared functions in RNA polymerase biogenesis raises the question of whether overexpression of any family member or related protein could complement loss of another GPN family protein. Since our focus here is GPN2, we transformed GPN2 mutant strains with 2μ plasmids from the MoBY collection [Citation19], with related genes under control of their own promoter. We chose GPN1, GPN2, GPN3, and IWR1 based on their confirmed roles in RNAPII and RNAPIII biogenesis [Citation5,Citation12]. We observed that only GPN2 could complement the temperature sensitivity of a gpn2-1 mutant and in fact, overexpression of GPN3 resulted in moderately impaired growth (). As shown in , we also observed Rpb1-GFP localization in gpn2-2 mutants containing these same plasmids and did not observe any improvement in RNAPII nuclear localization, with the exception of wild-type GPN2 expression. Thus, GPN2 must have a unique essential function not complemented by other GPN family members or regulators of RNA polymerase biogenesis like Iwr1.
Figure 1. The Gpn2 protein has a unique essential role in growth and RNAPII import in budding yeast. (a) Serial dilution plating assays of WT or gpn2-1 mutant yeast (lower five rows) carrying the indicated plasmids and grown at the indicated temperatures. Only the GPN2 insert complements the ts-allele. (b) Localization of Rpb1-GFP in gpn2-2 mutants complemented with the indicated plasmids. DIC images are shown on top, with GFP on the bottom. (c) Schematic of nuclear proteome GFP array construction. A gpn2-2 mutant with an HTA2-mCherry nuclear marker was mated to the entire array, before sporulation and haploid selection to produce an output array suitable for manual imaging on 12-well slides
In a previous study [Citation12], we observed that mutation of GPN2 did not appear to have global effects on nuclear import. However, these experiments only investigated a small sampling of nuclear proteins, leaving open the possibility that other Gpn2 substrate proteins exist. To gain a more complete understanding of the potential scope of Gpn2 functions, we surveyed the localization of over 1350 proteins with annotated nuclear, nucleolar or nuclear periphery localization (Supplementary Table S2) in a gpn2-2 mutant background (). We initially completed a visual screen for mis-localization across the entire nuclear proteome collection produced by SGA. Proteins with putative changes in localization were selected for tetrad dissection to generate strains with a clean genetic background and avoid any contamination from diploid strains that escaped the SGA selection steps. Tetrad dissected haploid strains encoding gpn2-2 and a GFP fusion of interest were then subject to manual validation by GFP fluorescence imaging to carefully assess any potential changes in localization.
Gpn2 activity is specific for RNA polymerase biogenesis and associated factors
Gpn2 seems to play a role in assembly or transport of RNAPII and RNAPIII, and its disruption leaves unincorporated subunits accumulating in the cytoplasm [Citation3,Citation12]. The gpn2-2 mutant is unique among the GPN mutants we previously screened as it has only a single-point mutation near the N-terminus (C19S) that confers hypomorphic growth and ts-phenotypes. Remarkably, among 1350 GFP fusion proteins screened manually in GPN2 and gpn2-2 cells, we observed strong mis-localization only for multiple RNAPII and RNAPIII subunits ( and Figure S1); however, the vast majority of nuclear proteins were completely unaffected by mutations in GPN2 (Supplementary Table S2). These data strongly support the tight linkage of GPN proteins to RNA polymerase biogenesis and transport as opposed to any sort of general nuclear transport or chaperone role.
Figure 2. RNAPII localization is strongly affected by gpn2-2. (a) Quantification of nuclear-to-cytoplasmic GFP signal ratios in WT or gpn2-2 mutant yeast strains bearing an RNAPII subunit GFP fusion. The GFP fusion is named on the X-axis and scores color-coded by the WT (blue) or gpn2-2 (red) genotype. Triplicates were conducted, in total n > 90, **** p < 0.0001 Student’s t-test. (b) Representative images of Rpb3-, Rpb7- and Rpb11-GFP in WT or gpn2-2 cells at the semi-permissive temperature of 30°C. For all figures HTA2-mCherry is included to mark the position of the nucleus. (c) GPN2 correlated SGA genetic interaction profiles from the Cellmap. Gene ontology terms are noted on the sides of each circle encompassing gene names that have high correlations with the genetic interaction profile of GPN2 mutants. This illustrates the strong transcriptional bias of GPN2 genetic interactions
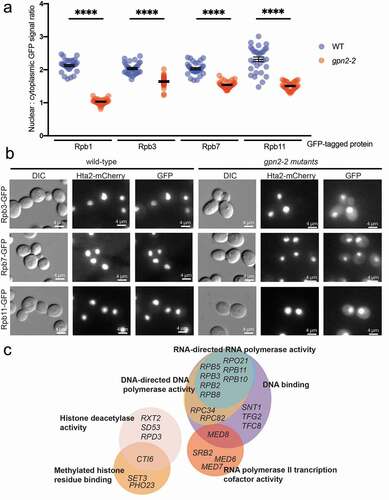
To confirm our observations by analysis of orthogonal functional data, we mined publicly available Cellmap genetic interaction data (https://thecellmap.org) to determine which yeast mutants had a synthetic genetic array (SGA) profile most similar to GPN2. We arbitrarily set a Pearson’s coefficient cutoff of 0.15, which identified 55 genes with similar SGA patterns to the GPN2 mutant included in the Cellmap study. As expected, the majority of these genes include RNA polymerase subunits and transcription associated factors, supporting a specific role of Gpn2 in driving RNA polymerase assembly and subsequent transcriptional activity ().
Gpn2 defects partially mislocalize a subset of transcription and splicing associated factors
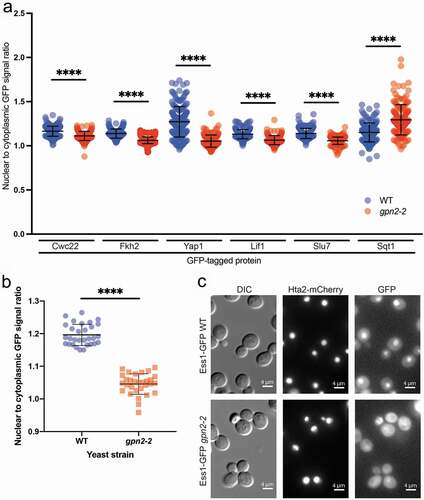
We were able to validate subtle changes in the localization of nine other proteins emerging from the primary screen, most of which have reported physical or functional connections to RNA polymerase ( and ). For the seven genes with clear changes in nuclear:cytoplasmic GFP ratios (), six had decreased nuclear intensity and one had increased nuclear intensity. One explanation for the mis-localization of these other nuclear proteins is that the cytoplasmic accumulation of RNA polymerase subunits results in the sequestration of proteins that would not normally interact with RNA polymerase until the assembled complex reaches that nucleus. In fact, the spliceosome is known to associate with the RNAPII CTD, so this may drive the mis-localization of Cwc22 and Slu7 splicing factors in the gpn2-2 strain [Citation21,Citation22], ().
Table 1. Non-RNA polymerase subunits that have altered localization in a gpn2-2 mutant
Figure 3. Subtle changes in non-RNA polymerase nuclear protein localizations in gpn2-2 cells. (a) Quantification and statistical analysis of proteins with decreased or increased nuclear accumulation in gpn2-2 yeast. The GFP fusion is named on the X-axis and scores color-coded by the WT (blue) or gpn2-2 (red) genotype. (b and c) Quantification (b) and representative images (c) of Ess1-GFP fusions in WT or gpn2-2 cells. For analysis in A and B, triplicates were conducted, in total n > 90, **** p < 0.0001 Student’s t-test
YAP1, a transcription factor involved in the stress response, interacts with Bud27, a chaperone involved in RNAPII assembly, and associates with the RNAPII subunit RPB2 (Biogrid). Fkh2 is another transcription factor that we observed mislocalized and has previously reported dynamic localization [Citation23]. Thus, transcription factors could be mislocalized either due to stresses placed upon the cell in gpn2-2 mutants, or due to associations with RNA polymerase itself.
Lif1, and Ess1 were the two other proteins that had a decreased nuclear:cytoplasmic ratio confirmed following our screen. Ess1 has clear links to RNAPII that we describe below. Lif1 is a DNA repair protein that functions as part of the Ligase IV complex. Sqt1 had an increased nuclear:cytoplasmic intensity ratio, and functions as an assembly chaperone for the large ribosomal subunit Rpl10. Why Lif1 and Sqt1 were specifically affected we do not know, although we do investigate RNA polymerase I (RNAPI) localization below, which may influence ribosome biogenesis and thus Sqt1 localization. Interestingly, we also saw a notable decrease in signal intensity for the Fob1 protein (, Figure S2). Fob1 serves as a replication fork barrier between units of rDNA transcription, further supporting potential effects on ribosome biogenesis in these gpn2 mutant cells.
In addition to potential shifts from nucleus to cytoplasm, we also validated significant mis-localization of MLP2 from the nuclear envelope (Figure S2) which may have consequences for nucleocytoplasmic transport of mRNAs [Citation24]. Notably, MLP2 binds components of the spindle pole body [Citation25] and helps regulate telomere length [Citation26]. Mlp2 has also been implicated, with Mlp1, in preventing R-loop associated genome instability [Citation27]. Therefore, the defective Mlp2 localization we observe may explain part of the reported genome instability phenotype in gpn2-2 mutants [Citation11].
Overall, we validated a small set of non-RNA polymerase proteins with disrupted localization in gpn2-2 cells. We did not find a coherent pattern in these changes, except that many of the changes are in proteins linked to transcription. Therefore, we favor a model in which the nine altered proteins are being impacted indirectly either through mislocalization in complex with mislocalized RNA polymerases or through cellular stress caused by the gpn2 allele.
Phospho-regulators of the RNAPII CTD effect its nuclear localization
Ess1 is a prolyl isomerase involved in regulation of the phosphorylation of the RNAPII c-terminal domain (CTD) located on the largest subunit, Rpb1 [Citation28]. CTD phosphorylation at Tyr1, Ser2, Thr4, Ser5, or Ser7 control the transcription cycle. Our data show that there is a distinct change in the localization of Ess1-GFP in the gpn2-2 mutants ( and c). Since Ess1 binds the CTD, the retention of Ess1 in the cytoplasm may be due to binding to mis-localized Rpb1. Nevertheless, we chose to investigate whether ESS1 mutation had any effect on Rpb1 localization by testing whether an ess1-1 ts-mutant had defective Rpb1 localization. We found that Rpb1-GFP nuclear signal does exhibit a small but significant difference between wild-type and ess1 mutant backgrounds after 3 hours at the non-permissive temperature of 37°C (Figure S2C)
Since Ess1 binding with the CTD occurs most strongly when Serine-5 is phosphorylated [Citation29] and ess1-1 mutants led to changes in Rpb1-GFP, we wondered if other changes in CTD phosphorylation status might impact Rpb1 localization. In cultured mammalian cells depletion of RPAP2 results in cytoplasmic accumulation of RNAPII [Citation3]. The yeast ortholog of RPAP2, Rtr1, is a phosphatase that targets the CTD [Citation30,Citation31], so we first assessed Rpb1-GFP localization in yeast lacking RTR1 or its paralog RTR2. Both rtr1Δ and rtr2Δ resulted in increased nuclear Rbp1-GFP, suggesting that they may have a negative regulatory role in RNAPII import through phosphorylation of the CTD (Figure S3). We were unable to generate a viable double rtr1Δrtr2Δ mutant with Rpb1-GFP, possibly due to altered function with the GFP tag, so it is unclear whether the effect of deletion of both phosphatases would be additive. If CTD phosphatase loss increased nuclear Rpb1-GFP fluorescence, we reasoned that CTD kinase mutations would decrease Rpb1-GFP fluorescence. Sgv1 is an essential yeast kinase that phosphorylates the RNAPII CTD [Citation32]. Employing a series of three SGV1 ts-alleles we observed a significant decrease in Rpb1 nuclear localization in all SGV1 mutants (Figure S3).
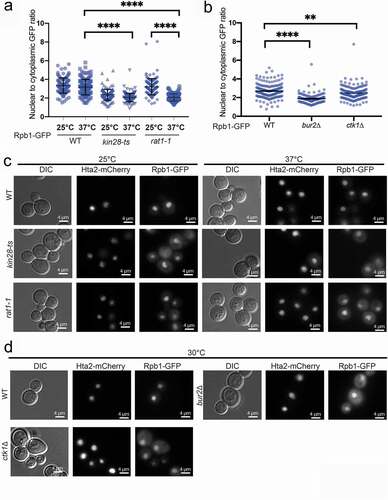
Considering the complexity and redundancy in Rpb1 CTD phosphorylation we elected to test a panel of mutants in other factors that control CTD phosphorylation to confirm our observations. Analysis of BUR2 and CTK1 deletion strains, kin28-ts and rat1-1 mutants, all of which perturb CTD phosphorylation, all showed defects in Rpb1-GFP nuclear accumulation (–d). Therefore, our data support a model in which Rpb1-CTD phosphorylation status impacts the nucleo-cytoplasmic distribution of the polymerase. Since most CTD phosphorylation presumably takes place co-transcriptionally in the nucleus this raises the possibility that RNA polymerase recycling is regulated by its CTD.
Figure 4. Significant defects in Rbp1-GFP localization result from mutation of CTD phospho-regulatory proteins. (a and b) Quantification of Rpb1-GFP nuclear-to-cytoplasmic ratios in the indicated CTD phosphoregulatory mutants. In A, results from both a permissive (25°C) and non-permissive (37°C) temperature are indicated, while in B standard growth at 30°C was used. For analysis in A and B, triplicates were conducted, in total n > 90, **** p < 0.0001, ** p < 0.01 by a Student’s t-test. (c and d) Representative images of the Rpb1-GFP in WT cells and the indicated mutant strains at the indicated temperatures
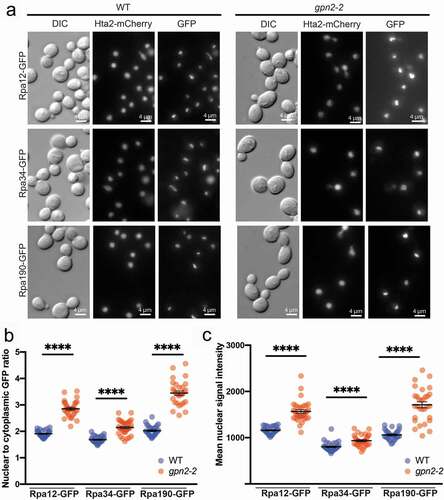
RNAPI nuclear localization increases in gpn2-2 mutants
RNAPI assembly is reported to occur in the nucleolus following nuclear import of the required subunits [Citation33]. Consistent with this we were unable to detect mislocalization of the RNAPI subunit RPA135 in GPN mutant or iwr1Δ mutant backgrounds [Citation12]. Several GFP-tagged subunits of RNAPI were included in the nuclear proteome screen and as shown in , there is no decrease in nuclear signal for Rpa12, Rpa34 or Rpa190 in a gpn2-2 mutant background. Instead, when we quantified the nuclear:cytoplasmic ratio of GFP signal, or the mean nuclear GFP intensity, gpn2-2 cells appeared to have higher levels of RNAPI subunits in the nucleus ( and c). Since gpn2-2 mutants have altered RNAPII transcription due to mis-localization, we hypothesized that defects in RNAPII activity may affect nucleolar transcription by RNAPI. We obtained a temperature sensitive RPB1 mutant called rpo21-1 and subsequently observed increased Rpa190-GFP signal at 37°C (Figure S4), similar to the gpn2-2 mutant background. These data show that defects in RNAPII function can increase RNAPI localization in the nucleolus.
Figure 5. Impacts of gpn2-2 mutation on RNAPI subunit localization. (a) Representative images of the indicated RNAPI subunit GFP fusion in WT or gpn2-2 cells. (b) Quantification of the nuclear:cytoplasmic ratio of the indicated GFP fusion in WT or gpn2-2 cells. (c) Quantification of the total nuclear signal intensity for the indicated GFP fusion in WT or gpn2-2 cells. For B and C, triplicates were conducted, in total n > 90, **** p < 0.0001 by a Student’s t-test
Perspective
Molecular chaperone proteins can be generalists or specialists; interacting with a wide range of non-native substrate proteins or being dedicated to a single protein substrate. The assembly and transport roles of GPN-family proteins seem to fall into the highly-specialized category. Our data confirm that the GPN-family members cannot complement the loss of GPN2 function, supporting a unique essential function for each family member. In addition, direct tests of >1300 nuclear protein localizations in gpn2-2 mutants found strong effects only on RNA polymerase subunits, with a handful of weaker effects on a small number of proteins. Thus, Gpn2 protein has a unique essential function that is almost completely restricted to its effects on
RNAPII and RNAPIII assembly and nuclear import. Gpn2 does not have a general nuclear transport function. Our data are consistent with previous reports that Gpn2 has an essential function in the assembly of an Rpb3 sub-complex of RNAPII [Citation34]. Since Gpn2 mutants also have RNAPIII defects it remains to be determined whether an analogous Rpc40 sub-complex exists during assembly of polymerase III.
One of the clearest non-RNAPII/III related hits from our screen was Ess1-GFP, which was displaced to the cytoplasm in our gpn2-2 mutant. Since Ess1 binds the CTD of Rpb1 we tested a range of CTD modifiers and found that many either increased or decreased nuclear:cytoplasmic ratios of Rpb1-GFP. This suggests that the CTD phosphorylation status is an important contributor to the distribution of RNAPII in cells. Whether the CTD and associated modifications play a role in assembly and import, or turnover and recycling of RNA polymerase subunits is not clear. Human RPAP2 regulates the nuclear import of RNAPII in human cells [Citation3]; however, loss of the yeast orthologs Rtr1 and Rtr2 actually increased nuclear intensity of Rpb1-GFP. Rtr1/2 are phosphatases for the CTD which should increase CTD phosphorylation. Similarly, rat1-1 alleles have been reported to lead to hyper-phosphorylated CTD, but this allele had the opposite effects in our assay, decreasing nuclear Rpb1-GFP, along with mutations in any of the CTD kinases tested. Thus, we speculate that timing and target residues of these kinases and phosphatases on the CTD is likely important to determine any effects of the CTD on RNAPII import or export.
In our previous study, we demonstrated that gpn2-2 mutants have an increase in the nuclear to cytoplasmic ratio for Rpa135-GFP [Citation12]. In extending this work to other RNAPI subunits we show that defects in gpn2-2 activity result in accumulation of RNAPI at the nucleolus, although we cannot state whether they are part of an assembled complex. This phenotype may be due to altered transcription by RNAPII as an rpo21-1 mutant allele of RPB1 exhibited the same phenotype. Previous work has shown that RNAPI accumulates in soybean nuclei upon inhibition of RNAPII activity [Citation35] and a recent paper revealed a role for RNAPII in transcription of rRNA genes [Citation36]. Thus there may be some interplay between RNAPI and RNAPII where increased RNAPI in the nucleus is a compensatory mechanism for reduced RNAPII activity. Notably, stress conditions have been shown to cause increased homodimerization of inactive RNAPI subunits, which could possibly affect GFP signal intensity in gpn2-2 mutants [Citation37]. Finally, another contributing factor could be altered stoichiometry of RNAPI subunits since four subunits, Rpb5, Rpb8, Rpb10 and Rpb12 are shared by all three RNA polymerases. The sequestration of RNAPII subunits in the cytoplasm could alter the availability of these proteins for integration into RNAPI, leading to accumulation of unassembled RNAPI in the nucleolus. Additional experiments will be required to dissect the indirect effects of disrupting one polymerase on the others.
Several recent publications have investigated the role of GPN family proteins in cancers, including breast cancer [Citation38,Citation39] and small cell lung carcinoma [Citation40]. However, data from the CBio Cancer Genomics Portal [Citation41] indicates that mutations and copy number changes in the GPN proteins are rare in cancers. The essential nature of the GPN family members may limit their potential as a therapeutic target in human cancers; however, we cannot rule out that they may be identified as contributing factors in rare disease. Mutations associated with the genetic disorder Treacher-Collins syndrome have been shown to affect the stability of RNA polymerases I and III [Citation42]. Perhaps alterations of GPN proteins could also contribute to rare disease phenotypes through their key role in RNA polymerase assembly and transcription. Regardless of their involvement in human disease, their ubiquitous presence in eukaryotes and essential function makes the GPN proteins a fascinating and still poorly understood protein family. Based on the data presented here, we suggest that the future of GPN research should focus on the biochemical mechanisms behind GPN activity in promoting RNA polymerase assembly.
Supplemental Material
Download Zip (18.7 MB)Acknowledgments
This work was supported by operating grants to P.C.S. from the Natural Sciences and Engineering Research Council of Canada (NSERC), and the Canadian Institutes of Health Research (CIHR). P.C.S. is a Michael Smith Foundation for Health Research Scholar and a CIHR New Investigator.
Disclosure Statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Zhao R, Kakihara Y, Gribun A, et al. Molecular chaperone Hsp90 stabilizes Pih1/Nop17 to maintain R2TP complex activity that regulates snoRNA accumulation. J Cell Biol. 2008 February 11;180(3):563–578.
- Boulon S, Pradet-Balade B, Verheggen C, et al. HSP90 and its R2TP/Prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II. Mol Cell. 2010 September 24;39(6):912–924.
- Forget D, Lacombe AA, Cloutier P, et al. The protein interaction network of the human transcription machinery reveals a role for the conserved GTPase RPAP4/GPN1 and microtubule assembly in nuclear import and biogenesis of RNA polymerase II. Mol Cell Proteomics. 2010 December 01;9(12):2827–2839.
- Miron-Garcia MC, Garrido-Godino AI, Garcia-Molinero V, et al. The prefoldin bud27 mediates the assembly of the eukaryotic RNA polymerases in an rpb5-dependent manner. PLoS Genet. 2013;9(2):e1003297.
- Czeko E, Seizl M, Augsberger C, et al. Iwr1 directs RNA polymerase II nuclear import. Mol Cell. 2011 April 22;42(2):261–266.
- Esberg A, Moqtaderi Z, Fan X, et al. Iwr1 protein is important for preinitiation complex formation by all three nuclear RNA polymerases in Saccharomyces cerevisiae. PLoS One. 2011;6(6):e20829.
- Gomez-Navarro N, Peiro-Chova L, Iwr EF. facilitates RNA polymerase II dynamics during transcription elongation. Biochim Biophys Acta Gene Regul Mech. 2017 July 01;1860(7):803–811.
- Nitta M, Saijo M, Kodo N, et al. A novel cytoplasmic GTPase XAB1 interacts with DNA repair protein XPA. Nucleic Acids Res. 2000 November 01;28(21):4212–4218.
- Lembo F, Pero R, Angrisano T, et al. MBDin, a novel MBD2-interacting protein, relieves MBD2 repression potential and reactivates transcription from methylated promoters. Mol Cell Biol. 2003 March 01;23(5):1656–1665.
- Gras S, Chaumont V, Fernandez B, et al. Structural insights into a new homodimeric self-activated GTPase family. EMBO Rep. 2007 June 01;8(6):569–575.
- Ben-Aroya S, Coombes C, Kwok T, et al. Toward a comprehensive temperature-sensitive mutant repository of the essential genes of Saccharomyces cerevisiae. Mol Cell. 2008 April 25;30(2):248–258.
- Minaker SW, Filiatrault MC, Ben-Aroya S, et al. Biogenesis of RNA polymerases II and III requires the conserved GPN small GTPases in Saccharomyces cerevisiae. Genetics. 2013 March 01;193(3):853–864.
- Alonso B, Chaussinand G, Armengaud J, et al. A role for GPN-loop GTPase yGPN1 in sister chromatid cohesion. Cell Cycle. 2011 June 01;10(11):1828–1837.
- Alonso B, Beraud C, Meguellati S, et al. Eukaryotic GPN-loop GTPases paralogs use a dimeric assembly reminiscent of archeal GPN. Cell Cycle. 2013 February 01;12(3):463–472.
- Staresincic L, Walker J, Dirac-Svejstrup AB, et al. GTP-dependent binding and nuclear transport of RNA polymerase II by Npa3 protein. J Biol Chem. 2011 October 14;286(41):35553–35561.
- Krogan NJ, Cagney G, Yu H, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006 March 30;440(7084):637–643.
- Huh WK, Falvo JV, Gerke LC, et al. Global analysis of protein localization in budding yeast. Nature. 2003 October 16;425(6959):686–691.
- Li Z, Vizeacoumar FJ, Bahr S, et al. Systematic exploration of essential yeast gene function with temperature-sensitive mutants. Nat Biotechnol. 2011 April 01;29(4):361–367.
- Ho CH, Magtanong L, Barker SL, et al. A molecular barcoded yeast ORF library enables mode-of-action analysis of bioactive compounds. Nat Biotechnol. 2009 April 01;27(4):369–377.
- Gietz RD, Woods RA. Yeast transformation by the LiAc/SS Carrier DNA/PEG method. Methods Mol Biol. 2006;313:107–120.
- David CJ, Boyne AR, Millhouse SR, et al. The RNA polymerase II C-terminal domain promotes splicing activation through recruitment of a U2AF65-Prp19 complex. Genes Dev. 2011 May 01;25(9):972–983.
- David CJ, Manley JL. The RNA polymerase C-terminal domain: a new role in spliceosome assembly. Transcription. 2011 October 01;2(5):221–225.
- Dastidar RG, Hooda J, Shah A, et al. The nuclear localization of SWI/SNF proteins is subjected to oxygen regulation. Cell Biosci. 2012 August 29;2(1):30.
- Strambio-de-castillia C, Blobel G, Rout MP. Proteins connecting the nuclear pore complex with the nuclear interior. J Cell Biol. 1999 March 08;144(5):839–855.
- Niepel M, Strambio-de-castillia C, Fasolo J, et al. The nuclear pore complex-associated protein, Mlp2p, binds to the yeast spindle pole body and promotes its efficient assembly. J Cell Biol. 2005 July 18;170(2):225–235.
- Hediger F, Dubrana K, Gasser SM. Myosin-like proteins 1 and 2 are not required for silencing or telomere anchoring, but act in the Tel1 pathway of telomere length control. J Struct Biol. 2002 December 01;140(1–3):79–91.
- Garcia-Benitez F, Gaillard H, Aguilera A. Physical proximity of chromatin to nuclear pores prevents harmful R loop accumulation contributing to maintain genome stability. Proc Natl Acad Sci U S A. 2017 October 10;114(41):10942–10947.
- Hani J, Schelbert B, Bernhardt A, et al. Mutations in a peptidylprolyl-cis/trans-isomerase gene lead to a defect in 3ʹ-end formation of a pre-mRNA in Saccharomyces cerevisiae. J Biol Chem. 1999 January 01;274(1):108–116.
- Namitz KEW, Zheng T, Canning AJ, et al. Structure analysis suggests Ess1 isomerizes the carboxy-terminal domain of RNA polymerase II via a bivalent anchoring mechanism. Commun Biol. 2021 March 25;4(1):398.
- Gibney PA, Fries T, Bailer SM, et al. Rtr1 is the Saccharomyces cerevisiae homolog of a novel family of RNA polymerase II-binding proteins. Eukaryot Cell. 2008 June 01;7(6):938–948.
- Mosley AL, Pattenden SG, Carey M, et al. Rtr1 is a CTD phosphatase that regulates RNA polymerase II during the transition from serine 5 to serine 2 phosphorylation. Mol Cell. 2009 April 24;34(2):168–178.
- Irie K, Nomoto S, Miyajima I, et al. SGV1 encodes a CDC28/cdc2-related kinase required for a G alpha subunit-mediated adaptive response to pheromone in S. Cerevisiae Cell. 1991 May 31;65(5):785–795.
- Dundr M, McNally JG, Cohen J, et al. Quantitation of GFP-fusion proteins in single living cells. J Struct Biol. 2002 December 01;140(1–3):92–99.
- Zeng F, Hua Y, Liu X, et al. Gpn2 and Rba50 Directly Participate in the Assembly of the Rpb3 Subcomplex in the Biogenesis of RNA Polymerase II. 10.1128/MCB.00091,18.Print 2018 Jul 1.. Mol Cell Biol. 2018 June 14;38(13).
- Guilfoyle TJ, Lin CY, Chen YM, et al. Enhancement of soybean RNA polymerase I by auxin. Proc Natl Acad Sci U S A. 1975 January 01;72(1):69–72.
- Abraham KJ, Khosraviani N, Chan JNY, et al. Nucleolar RNA polymerase II drives ribosome biogenesis. Nature. 2020 September 01;585(7824):298–302.
- Torreira E, Louro JA, Pazos I, et al. The dynamic assembly of distinct RNA polymerase I complexes modulates rDNA transcription. Elife. 2017 March 06;6. doi:10.7554/eLife.20832
- Barbosa-Camacho AA, Mendez-Hernandez LE, Lara-Chacon B, et al. The Gpn3 Q279* cancer-associated mutant inhibits Gpn1 nuclear export and is deficient in RNA polymerase II nuclear targeting. FEBS Lett. 2017 November 01;591(21):3555–3566.
- Lara-Chacon B, Guerrero-Rodriguez SL, Ramirez-Hernandez KJ, et al. Gpn3 is essential for cell proliferation of breast cancer cells independent of their malignancy degree. Technol Cancer Res Treat. 2019 January 01;18:1533033819870823. .
- Huang F, Huffman KE, Wang Z, et al. Guanosine triphosphate links MYC-dependent metabolic and ribosome programs in small-cell lung cancer. J Clin Invest. 2021 January 04;131(1). DOI:10.1172/JCI139929
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012 May 01;2(5):401–404.
- Walker-Kopp N, Jackobel AJ, Pannafino GN, et al. Treacher Collins syndrome mutations in Saccharomyces cerevisiae destabilize RNA polymerase I and III complex integrity. Hum Mol Genet. 2017 November 01;26(21):4290–4300.
