ABSTRACT
Clear cell renal cell carcinoma (ccRCC) is a subtype of renal cell cancer with the highest mortality, infiltration, and metastasis rate, threatening human health. Despite oncogenic role of TROAP in various cancers, its function in ccRCC remains to be unraveled. The differentially expressed mRNAs (DEmRNAs) and miRNAs (DEmiRNAs) were obtained by analyzing the related data sets of ccRCC in TCGA. The expression levels of mRNAs and miRNAs in the cell were detected by qRT-PCR, while the protein levels were characterized by western blot. The viability, migratory and invasive abilities of ccRCC cells were determined by MTT, wound healing and cell invasion assays. The combination of miRNA target site prediction and dual-luciferase reporter gene assay verified the binding relationship between miR-532-3p and TROAP. Research on ccRCC displayed that TROAP expression was upregulated, while miR-532-3p was down-regulated. Besides, upregulation of TROAP could accelerate viability, migratory and invasive potentials of ccRCC cells. On the contrary, miR-532-3p could downregulate TROAP level, but TROAP upregulation reversed the viability, migration, and invasion of ccRCC cells. MiR-532-3p could attenuate the viability, migration and invasion of ccRCC cells by targeting TROAP. This may generate novel insights into molecular therapeutic targets for ccRCC.
1. Introduction
Renal cell carcinoma is originated from renal epithelial cells and is common cancer in the urinary system that accounts for 2–3% of all adult malignancies with increasing morbidity at 2–3% per decade, bringing out a serious impact on human health [Citation1–3]. Eighty percent of all renal cell carcinoma cases are clear cell renal cell carcinoma (ccRCC), which is the most common subtype of renal cell carcinoma, with the highest mortality among all renal cell carcinoma subtypes [Citation4,Citation5]. Surgical resection is currently the main treatment for ccRCC, however, 20–40% of patients still suffer from recurrence and distant metastasis after surgical resection, while the 5-year survival rate is less than 20% once metastasis occurs [Citation6,Citation7]. Therefore, it is vital to find new molecular sites for the treatment and markers for early diagnosis of ccRCC to elevate the survival rate of patients.
The earlier studies manifested that alteration of mRNA levels is essential in modulating tumor progression. For instance, FOXO3 can conspicuously stimulate cell proliferation, migration and invasion of ccRCC[Citation8]. IREB2 can be used as a tumor suppressor gene to restrain cell proliferation, migration and invasion of ccRCC[Citation9]. Trophinin-associated protein (TROAP, namely TASTIN) is a kind of soluble cytoplasmic protein, which forms a complex with trophinin and bystin, and it is involved in early embryo implantation by mediating cellular invasion and proliferation [Citation10–14]. With the deepening of research on TROAP, it is found that its expression has been up-regulated in a variety of cancers, and it can regulate the occurrence and progression of cancer. For example, Kai Li et al[Citation15]. disclosed that TROAP mRNA and protein expression levels are up-regulated in human breast cancer tissue and cell lines, and TROAP can remarkably promote cell proliferation, migration and invasion of breast cancer. Ke Jing et al[Citation10]. exhibited that TROAP is overexpressed in gastric cancer and exerts as an oncogene in gastric cancer by affecting cell proliferation and invasion. However, the regulation of TROAP in ccRCC remains to be elucidated.
Recent investigations revealed that miRNAs are aberrantly expressed in multiplex malignancies and serve as key modulators in diverse biological processes in tumors, such as cell proliferation, differentiation, invasion, and apoptosis [Citation16,Citation17]. For instance, miR-363 downregulates S1PR1 to hinder proliferation, invasion, and migration of ccRCC[Citation18–21]. MiR-1294 acts as an anti-tumor gene in ccRCC, and it restrains cell proliferation, colony formation, and invasion by partially targeting HOXA6 in ccRCC. In this study, we excavated upstream regulatory genes of TROAP by bioinformatics analysis and identified miR-532-3p as our research object. MiR-532-3p is one of miRNAs primarily found and is theorized to play a vital role in different cancers. For instance, miR-532-3p decreases in lymphoma cells and tissue and it can target β-catenin to suppress proliferation and to stimulate apoptosis of cancer cells. M. Han et al. filtered out potential biomarkers for ccRCC by microarray analysis, unearthing a significant correlation between high miR-532-3p expression and high overall survival rate of patients. Nonetheless, rarely studies were conducted on the role and potential modulatory mechanism of miR-532-3p on ccRCC.
In this study, we unveiled the expression of TROAP on ccRCC cells and investigated the role and potential mechanism of TROAP on ccRCC. The results manifested that miR-532-3p repressed cell viability, migration, and invasion of cancer cells via targeting TROAP, which may generate novel insights into ccRCC therapy.
2. Materials and methods
2.1. Bioinformatics analysis
The expression profiles of mature miRNAs (Normal: 71, Tumor: 545), mRNAs (Normal: 72, Tumor: 539) were downloaded from The Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/) along with clinical data. Differential analysis was performed on expression of miRNAs and mRNAs in the normal group and the tumor group using the “edgeR” package with |logFC|>1.5 and padj<0.05 as thresholds. The survival curves of differentially expressed mRNAs (DEmRNAs) were obtained based on the clinical information of samples, and the relationship between target gene expression and clinical staging was analyzed. The starBase (http://starbase.sysu.edu.cn/) and miRWalk (http://mirwalk.umm.uni-heidelberg.de/) databases were employed to predict the upstream target genes of the target mRNA, and predicted target genes were combined with differentially expressed miRNAs (DEmiRNAs) to obtain a set of miRNAs that bound to the target mRNA. Correlation analysis was performed for final screening.
2.2. Cell culture
Human renal tubular epithelial cell line HKC (No. BNCC338628), ccRCC cell lines 786-O (No. BNCC338472), A498 (No. BNCC338630), Caki-2 (No. BNCC340136) were all accessed from BeNa Culture Collection (BNCC). CM9-1 medium containing 90% Dulbecco’s Modified Eagle Medium (DMEM)-H/F12 and 10% fetal bovine serum (FBS) was used to cultivate HKC cell line. CM1-1 medium consisting of 90% DMEM-H and 10% FBS was used to cultivate 786-O cell line. A498 cell line was cultured with CM2-1 medium, which was composed of 90% Roswell Park Memorial Institute-1640 (RPMI-1640) and 10% FBS. Caki-2 cell line was cultured in the medium with 90% McCoy’s 5a and 10% FBS. All cell lines were cultured in a humidified incubator at 37°C with 5% CO2. After 48 h of transfection, the cells could be harvested for subsequent experiments.
2.3. Cell transfection
ccRCC cells were seeded into a 6-well plate. After cells were cultured overnight, the miR-532-3p-mimic ((miR-mimic)), NC-mimic, oe-TROAP, and oe-NC were transfected into the cells using the Lipofectamine 2000 kit (Invitrogen, Carlsbad, CA, USA). The above plasmids used for transfection were all purchased from GeneChem Company (Shanghai, China). Subsequent experiments could be conducted 48 h after transfection.
2.4. Real-time quantitative polymerase chain reaction (qRT-PCR)
Total RNA was extracted using TRIzol reagent (ComWin Biotech, Beijing, China) according to the protocols, and then it was reverse-transcribed with TransScript First-Strand cDNA Synthesis SuperMix Kit (TransGen Biotech, Beijing, China) to obtain complementary DNA (cDNA). qRT-PCR was conducted using TransStart Green qPCR SuperMix (TransGen Biotech, Beijing, China). The relative expression of miRNA and mRNA was calculated and expressed by 2−ΔΔCT method. U6 and GAPDH were used as the internal references for miR-532-3p and TROAP, respectively. The qRT-PCR program was run by Applied Biosystems 7500 detection system. The primer sequences used in qRT-PCR were presented in .
Table 1. Information of primer sequences
2.5. Western blot
The cultured ccRCC cells were lysed with radioimmunoprecipitation assay (RIPA) lysis buffer (containing protease inhibitors) and centrifuged to extract total proteins, and the protein concentration was quantified with the bicinchoninic acid (BCA) protein assay kit (Thermo Fisher, Waltham, USA). Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was conducted on a total of 30 μg of proteins for 1–2 h, the proteins were then transferred to a polyvinylidene fluoride (PVDF) membrane (EMD Millipore, Bedford, Massachusetts, USA). After blocking the membrane, the primary antibody TROAP (Proteintech, Wuhan, China) or GAPDH (Proteintech) was added according to the experimental requirements and the membrane was incubated at 4°C for 12 h. Afterward, the membrane was incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG H&L (HRP) (abcam, Cambridge, UK) for 1 h at room temperature. Protein levels were displayed by an electrochemiluminescence (ECL) kit (Solarbio, Beijing, China).
2.6. MTT assay
ccRCC cells (2 × 103 cells/well) were seeded into a 96-well plate. Then, after 0, 24, 48, 72 h, of inoculation, 20 μL of MTT (5 mg/ml) solution (Shanghai Zeye Biotechnology Co., Ltd., China) was added to each well, and cells were incubated at 37°C with 5% CO2 for 4 h. Next, the supernatant was aspirated, and 150 μL dimethyl sulfoxide (DMSO) was added to dissolve the precipitate. The optical density (OD) value of each well was measured by a microplate reader (Molecular Devices, Sunnyvale, CA, USA) at 490 nm. The experiment was repeated three times.
2.7. Wound healing assay
First, 5 × 105 ccRCC cells were inoculated into a 6-well plate. When cell density reached about 90%, a 200 mL sterile pipette tip was used to draw a straight line in each well. Afterward, cells were cultured in 2 mL of fresh medium without FBS in a cell incubator at 37°C with 5% CO2. Then, an inverted microscope was employed to photograph the scratched area at the time of scratching and after 48 h of incubation. The experiment was repeated three times. The healing status was expressed by wound healing rate. Wound healing rate = (initial scratch width – 48 h scratch width)/initial scratch width.
2.8. Cell invasion assay
First, 2 × 104 ccRCC cells were inoculated in the upper chamber of Transwell chamber (sigma; China) coated with matrigel (BD, USA). Then, 650 μL of DMEM medium containing 20% FBS was added to the lower chamber. Cells were incubated for 24 h in an incubator. Then the culture medium was discarded and uninvaded cells on the filter membrane were removed with a cotton swab. Next, invaded cells were fixed with 4% paraformaldehyde for 15 min, and stained with 0.1% crystal violet for 10 min. Five fields were randomly selected with an inverted microscope to photograph cells and the cells passing through the membrane were counted. The experiment was repeated in triplicate.
2.9. Dual-luciferase reporter gene assay
The wild-type (Wt) or mutant-type (Mut) sequence of the 3ʹuntranslated region (3ʹ-UTR) of TROAP was cloned into the psiCHECK-2 dual-luciferase vector (Promega, Madison, WI). Subsequently, the luciferase plasmid containing Wt or Mut sequence and miR-mimic was co-transfected into ccRCC cells Caki-2. Simultaneously, the plasmid and NC-mimic were also co-transfected into Caki-2 cells. After 48 h of transfection, the firefly and renilla luciferase activities were measured. The luciferase activity was detected by the luciferase reporter analysis system (Promega, USA). The experiment was repeated in three sets to obtain stable results.
2.10. Statistical analysis
The experimental data obtained in this study were processed by GraphPad Prism 6 (GraphPad Software Inc., San Diego, CA, USA). The measurement data were presented as mean ± standard deviation. The comparison between the two groups was carried out using t-test,, and the comparison among multiple groups was by one way analysis of variance (ANOVA). P < 0.05 indicated a significant difference.
3. Results
3.1. TROAP is highly expressed in ccRCC
The edgeR package was used to perform differential analysis on mRNA between the normal group and the tumor group in TCGA database to obtain 3633 DEmRNAs (). Related studies have denoted that TROAP is up-regulated in a variety of tumor tissue, and is related to dismal prognosis of cancer, which can promote proliferation and migration of cancer cells [Citation15,Citation22], but its expression and mechanism in ccRCC remain to be explored. Accordingly, TROAP was selected as the subject of this study from several mRNAs with significant differences. TROAP was prominently highly expressed in ccRCC tissue ()). At the same time, the survival analysis of patients with ccRCC demonstrated that patients with high expression of TROAP in tumor tissue had a worse prognosis ()). the expression level of TROAP was significantly different in different G, stage, T, M, and N stages of tumor ()) with a gradually increasing trend. These results indicated that TROAP was up-regulated in ccRCC, which was likely to be a factor leading to an unfavorable prognosis in patients with ccRCC.
Figure 1. The expression level of TROAP is up-regulated in ccRCC (a): Volcano map of DEmRNAs in ccRCC, the red dots represent up-regulated mRNAs, and the green dots represent down-regulated mRNAs; (b): The expression of TROAP in ccRCC; (c): Survival curves of TROAP expression on the prognosis of patients; (d-h): Box plots of TROAP expression in different G, stage, T, M, and N stages of ccRCC, respectively; (i): The results of qRT-PCR detection of mRNA expression of TROAP in human renal tubular epithelial cell line HKC and three ccRCC cell lines including A498; (j): The results of western blot of protein expression of TROAP in human renal tubular epithelial cell line HKC and three ccRCC cell lines including A498; * p<0.05
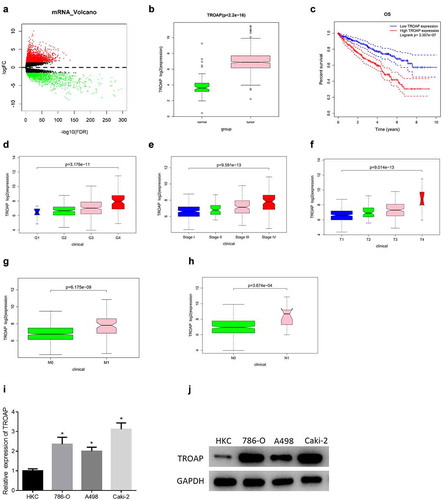
Therefore, in order to verify that TROAP expression was differentially up-regulated in ccRCC, the mRNA and protein expression levels of TROAP in human renal tubular epithelial cell line HKC and ccRCC cell lines such as A498 were measured. The results illustrated that the mRNA and protein expression levels of TROAP in ccRCC cell lines ((786-O, A498, Caki-2)) were remarkably higher than those in human renal tubular epithelial cell line HKC (, )). Caki-2 cell line with the highest expression was selected for subsequent experiments. By combining bioinformatics analysis of TROAP expression in ccRCC and experimental detection of TROAP expression in cell lines, it was confirmed that TROAP was highly expressed in ccRCC.
3.2. TROAP promotes cell viability, migration and invasion of ccRCC
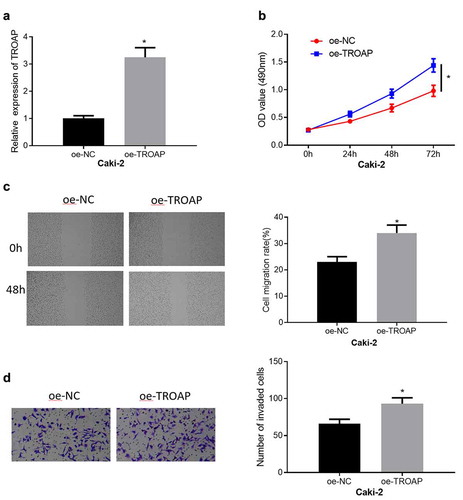
To further confirm that TROAP was an unfavorable factor for the malignant progression of ccRCC, the effect of TROAP on the migration, invasion and viability of Caki-2 cells was observed. First, oe-TROAP was transfected into Caki-2 cells, and then qRT-PCR was used to prove that transfection of oe-TROAP could over-express TROAP in Caki-2 cells ()). Subsequent functional experiments reflected the effect of overexpression of TROAP on the malignant phenotype of Caki-2 cells. Overexpression of TROAP played a significant role in promoting cell viability ()). In addition, the results of cell migration and invasion assays manifested that overexpression of TROAP could also markedly stimulate the migration and invasion of Caki-2 cells (, )). Through these experiments, it was fully confirmed that TROAP was an oncogene in ccRCC, which promoted the viability, migration and invasion of cancer cells.
Figure 2. TROAP promotes cell viability, migration and invasion of ccRCC (a): The results of qRT-PCR detection of TROAP mRNA expression in cells after transfected with oe-TROAP; (b): The effect of overexpressed TROAP on cell viability was detected by MTT assay; (c): The effect of overexpression of TROAP on cell migration was assessed through wound healing assay (40×); (d): The effect of overexpressed TROAP on cell invasion was measured by cell invasion assay (100×); * p<0.05
3.3. MiR-532-3p can target and suppress the expression of TROAP
In order to further study the mechanism of TROAP to regulate cell viability of ccRCC, the miRNAs that may bind and regulate TROAP were predicted. Differential analysis of the miRNA expression data of ccRCC in TCGA database was conducted by edgeR to obtain 82 DEmiRNAs, among which 51 were differentially up-regulated, and the other 31 were down-regulated ()). Then the upstream regulatory genes of TROAP were predicted via miRWalk and starBase databases. The prediction results were compared with 31 down-regulated miRNAs to screen out miRNAs in both groups, that was, two DEmiRNAs that might target TROAP, namely miR-532-3p and miR-138-5p ()). Next, Pearson correlation analysis was performed on TROAP with miR-532-3p and miR-138-5p, and it was found that miR-532-3p was negatively correlated with TROAP with a higher correlation coefficient than miR-138-5p and TROAP ()). The down-regulation trend of miR-532-3p in TCGA dataset of ccRCC was presented in ). Therefore, it was believed that miR-532-3p was likely to be able to bind and regulate TROAP, and subsequent verification had been carried out.
Figure 3. MiR-532-3p can target and suppress the expression of TROAP (a): Volcano map of DEmiRNAs in ccRCC dataset, the red dots represent up-regulated miRNAs, and the green dots represent down-regulated miRNAs; (b): Venn diagram of the predicted upstream target miRNAs of TROAP and down-regulated DEmiRNAs in TCGA database; (c): Pearson correlation analysis of miRNAs (miR-532-3p; miR-138-5p) and TROAP expression in ccRCC; (d): Expression of miR-532-3p in ccRCC; (e): The results of qRT-PCR of miR-532-3p expression in normal tissue cell line and cancer cell lines such as A498; (f): The efficiency of overexpression of miR-532-3p in cancer cells was measured through qRT-PCR; (g): The results of qRT-PCR of TROAP mRNA expression level with overexpressed miR-532-3p in cancer cells; (h): The results of western blot of TROAP protein level with overexpressed miR-532-3p in cancer cells; (i): The binding sites of miR-532-3p and TROAP, and verification results of dual-luciferase reporter gene assay of targeted binding relationship; * p < 0.05
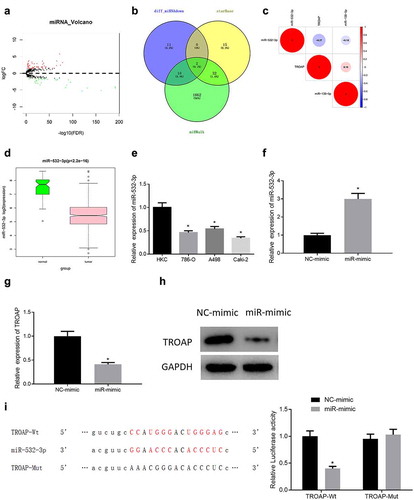
To verify that miR-532-3p could regulate TROAP, the expression of miR-532-3p in four cell lines used in the previous assays was observed using qRT-PCR. The results displayed that the expression of miR-532-3p in ccRCC cell lines was lower than that in human renal tubular epithelial cell line HKC ()). Then miR-532-3p was overexpressed in the Caki-2 cell line, and qRT-PCR was performed to verify that miR-532-3p was successfully overexpressed ()). The expression changes of TROAP in cells were assessed, and the results exhibited that overexpressed miR-532-3p in Caki-2 cells dramatically reduced TROAP mRNA and protein levels (, )). Finally, the dual-luciferase reporter gene assay revealed that miR-mimic did not affect the TROAP-Mut group significantly, but it could reduce the luciferase activity of the TROAP-Wt group, which substantiated that miR-532-3p and TROAP had a targeted binding relationship ()). In sum, miR-532-3p could directly bind to TROAP mRNA to inhibit the expression of TROAP.
3.4. MiR-532-3p modulates cell viability, migration, and invasion by TROAP in ccRCC
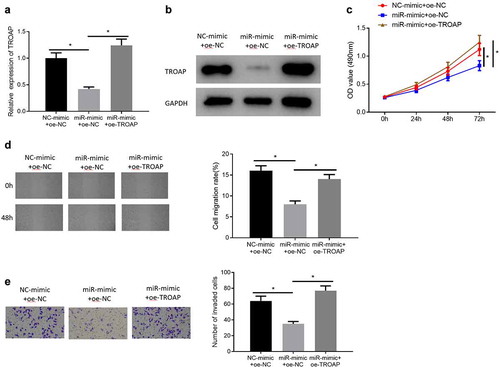
In order to further confirm that miR-532-3p could restrain cell proliferative rate, migratory and invasive abilities of ccRCC by targeting TROAP, miR-532-3p was firstly overexpressed and then miR-532-3p and TROAP were overexpressed simultaneously in Caki-2 cells (NC-mimic+oe-NC group, miR-mimic+oe-NC group, miR-mimic+oe-TROAP group). The expression of TROAP in each group was detected by qRT-PCR and western blot. The results indicated that compared with the miR-532-3p overexpression group, the TROAP expression level was higher in the miR-532-3p and TROAP simultaneously overexpressed group (, )). Then the changes in the viability of the three groups of transfected cells were observed. The results manifested that miR-532-3p could suppress the viability of cancer cells, which could be reversed by TROAP ()). Similarly, the results of wound healing and cell invasion assays also clearly pointed out that miR-532-3p could inhibit the migration and invasion of Caki-2 cells, while overexpression of TRAP could reverse the inhibition effect of miR-532-3p on the migration and invasion of Caki-2 cells (, )). Through the above assays, it could be determined that TROAP could reverse the inhibition effects of miR-532-3p on cell viability, migration and invasion of ccRCC.
Figure 4. MiR-532-3p inhibits cell viability, migration and invasion of ccRCC by targeting TROAP (a): The results of qRT-PCR of TROAP mRNA expression in cells after various treatments; (b): The results of western blot of TROAP protein expression in cells after various treatments; (c): Changes in cell proliferative ability after various treatments were measured by MTT assay; (d): Cell migratory ability after various treatments was detected via wound healing assay (×40); (e): Cell invasion ability after various treatments was assessed through cell invasion assay (×100); * p < 0.05
4. Discussion
As is well known, many genes are abnormally expressed in cancer and play an important regulatory role in the occurrence and progression of cancer [Citation23,Citation24]. Therefore, in this study, in order to discover the related genes that regulate ccRCC cells, DEmRNAs in ccRCC were screened out through bioinformatics analysis. By consulting related studies, it is found that TROAP is up-regulated in a variety of cancers except ccRCC, including liver cancer, breast cancer, gastric cancer, etc., and it can promote the progression of these cancers [Citation10,Citation15,Citation22]. Thus, TROAP was selected as the research object, followed by qRT-PCR and western blot to detect the expression of TROAP in human renal tubular epithelial cell line and ccRCC cell lines. It was confirmed that TROAP was up-regulated in ccRCC. Afterward, the effect of TROAP on ccRCC was observed through cell biology functional experiments, which confirmed that TROAP could promote cell viability, migration and invasion of ccRCC. These studies fully elucidated that TROAP as an oncogene exacerbated malignant behaviors of cancer cells.
Accumulating evidence exhibited that miRNAs can directly regulate gene expression by inhibiting the mRNA translation or reducing mRNA stability to stimulate degradation[Citation25]. Prior work demonstrated that TROAP can be regulated by miRNAs. For instance, miR-519d-3p can affect the proliferation and migration of colorectal cancer cells by targeting TROAP[Citation23]. Therefore, the upstream miRNAs that regulate TROAP were explored by bioinformatics analysis in this study, and finally two miRNAs (miR-532-3p; miR-138-5p) that had binding sites with TROAP were found, which were negatively correlated with TROAP. At last, miR-532-3p was selected, which was negatively correlated with the highest correlation coefficient of TROAP. MiR-532-3p is a popular miRNA in cancer research, which has been substantiated to play a pivotal tumor suppressor effect in different cancers by regulating mRNAs. For example, miR-532-3p restrains the NF-κB signaling pathway in prostate cancer cells by targeting TRAF1, TRAF2 and TRAF4, thereby inhibiting the osseous metastasis of prostate cancer[Citation26]. MiR-532-3p can reduce the expression of KIFC1 in liver cancer, thereby inhibiting the metastasis of liver cancer by activating the gankyrin/AKT/TWIST1 signaling pathway[Citation27]. However, the role of miR-532-3p in ccRCC has not yet to be fully defined. In this study, the expression of miR-532-3p in human renal tubular epithelial cell line and ccRCC cell lines was detected by qRT-PCR. It was determined that miR-532-3p was down-regulated in ccRCC cell lines. Moreover, it was confirmed by dual-luciferase reporter gene assay that miR-532-3p could directly target and regulate TROAP. Concurrently, it was also denoted in rescue experiment that TROAP reversed the inhibition effect of miR-532-3p on the viability, migration and invasion of ccRCC. This fully indicated that miR-532-3p could suppress the malignant progression of ccRCC by targeting TROAP.
Viewed in toto, this study found for the first time that TROAP was up-regulated in ccRCC, and could promote the viability, migration and invasion of cancer cells. At the same time, miR-532-3p was discovered that could target and inhibit TROAP. Furthermore, it was proved that TROAP could reverse the inhibition effect on viability, migration and invasion caused by miR-532-3p. Exploring interaction as well as the potential mechanism of miR-532-3p and TROAP in ccRCC may contribute to developing new avenues for the treatment of ccRCC.
Consent for publication
All authors consent to submit the manuscript for publication.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Authors’ contributions
Bin Gao contributed to the study design. Lijuan Wang conducted the literature search. Na Zhang acquired the data. Huancai Liu wrote the article. Dongli Sun performed data analysis. Bin Gao and Yifei Liu revised the article. Yifei Liu gave the final approval of the version to be submitted.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Song S, Long M, Yu G, et al. Urinary exosome miR-30c-5p as a biomarker of clear cell renal cell carcinoma that inhibits progression by targeting HSPA5. J Cell Mol Med. 2019;23(10):6755–6765.
- Fritz HKM, Lindgren D, Ljungberg B, et al. The miR(21/10b) ratio as a prognostic marker in clear cell renal cell carcinoma. Eur J Cancer. 2014;50:1758–1765.
- Gupta K, Miller JD, Li JZ, et al. Epidemiologic and socioeconomic burden of metastatic renal cell carcinoma (mRCC): a literature review. Cancer Treat Rev. 2008;34(3):193–205.
- Pires-Luis AS, Costa-Pinheiro P, Ferreira MJ, et al. Identification of clear cell renal cell carcinoma and oncocytoma using a three-gene promoter methylation panel. J Transl Med. 2017;15(1):149.
- Hsieh JJ, Purdue MP, Signoretti S, et al. Renal cell carcinoma. Nat Rev Dis Primers. 2017;3(1):17009.
- Nabi S, Kessler ER, Bernard B, et al. Renal cell carcinoma: a review of biology and pathophysiology. F1000Res. 2018;7:307.
- Keys KA, Daniali LN, Warner KJ, et al. Multivariate predictors of failure after flap coverage of pressure ulcers. Plast Reconstr Surg. 2010;125(6):1725–1734.
- Nie W, Ni D, Ma X, et al. miR122 promotes proliferation and invasion of clear cell renal cell carcinoma by suppressing forkhead box O3. Int J Oncol. 2019;54:559–571.
- Liu F, Chen Y, Chen B, et al. MiR-935 promotes clear cell renal cell carcinoma migration and invasion by targeting IREB2. Cancer Manag Res. 2019;11:10891–10900.
- Jing K, Mao Q, Ma P. Decreased expression of TROAP suppresses cellular proliferation, migration and invasion in gastric cancer. Mol Med Rep. 2018;18:3020–3026.
- Fukuda MN, Sato T, Nakayama J, et al. Trophinin and tastin, a novel cell adhesion molecule complex with potential involvement in embryo implantation. Genes Dev. 1995;9(10):1199–1210.
- Suzuki N, Zara J, Sato T, et al. A cytoplasmic protein, bystin, interacts with trophinin, tastin, and cytokeratin and may be involved in trophinin-mediated cell adhesion between trophoblast and endometrial epithelial cells. Proc Natl Acad Sci U S A. 1998;95(9):5027–5032.
- Fukuda MN, Nozawa S. Trophinin, tastin, and bystin: a complex mediating unique attachment between trophoblastic and endometrial epithelial cells at their respective apical cell membranes. Semin Reprod Endocrinol. 1999;17:229–234.
- Suzuki N, Nakayama J, Shih I-M, et al. Expression of trophinin, tastin, and bystin by trophoblast and endometrial cells in human placenta. Biol Reprod. 1999;60(3):621–627.
- Li K, Zhang R, Wei M, Zhao L, et al. TROAP promotes breast cancer proliferation and metastasis. Biomed Res Int. 2019;2019:6140951.
- Liu Z, Sun J, Liu B, et al. miRNA222 promotes liver cancer cell proliferation, migration and invasion and inhibits apoptosis by targeting BBC3. Int J Mol Med. 2018;42:141–148.
- Zhao J, Li D, Fang L. MiR-128-3p suppresses breast cancer cellular progression via targeting LIMK1. Biomed Pharmacother. 2019;115:108947.
- Xie Y, Chen L, Gao Y, et al. miR-363 suppresses the proliferation, migration and invasion of clear cell renal cell carcinoma by downregulating S1PR1. Cancer Cell Int. 2020;20:227.
- Pan W, Pang LJ, Cai HL, et al. MiR-1294 acts as a tumor suppressor in clear cell renal cell carcinoma through targeting HOXA6. Eur Rev Med Pharmacol Sci. 2019;23:3719–3725.
- Liu Y, Li Q, Dai Y, et al. miR-532-3p inhibits proliferation and promotes apoptosis of lymphoma cells by targeting beta-catenin. J Cancer. 2020;11:4762–4770.
- Han M, Yan H, Yang K, et al. Identification of biomarkers and construction of a microRNAmRNA regulatory network for clear cell renal cell carcinoma using integrated bioinformatics analysis. PLoS One. 2021;16:e0244394.
- Hu H, Xu L, Chen , et al. The upregulation of trophinin-associated protein (TROAP) predicts a poor prognosis in hepatocellular carcinoma. J Cancer. 2019;10:957–967.
- Ye X, Lv H. MicroRNA-519d-3p inhibits cell proliferation and migration by targeting TROAP in colorectal cancer. Biomed Pharmacother. 2018;105:879–886.
- Xia X, Wang J, Liu Y, et al. Lower cystic fibrosis transmembrane conductance regulator (CFTR) promotes the proliferation and migration of endometrial carcinoma. Med Sci Monit. 2017;23:966–974.
- Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. 2015;16(7):421–433.
- Wa Q, Zou C, Lin Z, et al. Ectopic expression of miR-532-3p suppresses bone metastasis of prostate cancer cells via inactivating NF-kappaB signaling. Mol Ther Oncolytics. 2020;17:267–277.
- Han J, Wang F, Lan Y, et al. KIFC1 regulated by miR-532-3p promotes epithelial-to-mesenchymal transition and metastasis of hepatocellular carcinoma via gankyrin/AKT signaling. Oncogene. 2019;38(3):406–420.
