ABSTRACT
This study was to explore the function of circular progastricsin (circ-PGC) in NSCLC. The histological morphology of tumor tissues was observed by hematoxylin and eosin (HE) staining. The expression of circ-PGC, miR-532-3p and forkhead box R2 (FOXR2) mRNA was measured by real-time quantitative polymerase chain reaction (RT-qPCR). The protein level of FOXR2 was checked by western blot. In functional analyses, cell viability, colony formation, cell apoptosis, cell migration and cell invasion were investigated using cell counting kit-8 (CCK-8) assay, colony formation assay, flow cytometry assay, wound healing assay and transwell assay. Besides, glycolysis metabolism was assessed by the levels of glucose consumption, lactate production and adenosine triphosphate (ATP) production. The predicted relationship between miR-532-3p and circ-PGC and FOXR2 was verified by dual-luciferase reporter assay and RNA immunoprecipitation (RIP) assay. The results showed that circ-PGC and FOXR2 were upregulated in NSCLC tissues and cells. Circ-PGC knockdown or FOXR2 knockdown inhibited NSCLC cell viability, colony formation, cell migration, invasion and glycolysis metabolism, and FOXR2 overexpression rescued these inhibitory effects caused by circ-PGC knockdown. MiR-532-3p harbored the same binding site with circ-PGC and FOXR2, and circ-PGC positively regulated FOXR2 expression by targeting miR-532-3p. The expression of β-catenin and c-Myc was decreased in cells after circ-PGC knockdown but recovered with miR-532-3p inhibition or FOXR2 overexpression. Circ-PGC downregulation also inhibited tumor growth in vivo. In conclusion, circ-PGC positively regulated FOXR2 expression by competitively binding to miR-532-3p, thereby promoting the development of NSCLC, and the Wnt/β-catenin signaling pathway might be activated by the circ-PGC/miR-532-3p/FOXR2 network.
Introduction
Lung cancer remains the key cause of cancer-related death worldwide. Non-small cell lung cancer (NSCLC) accounts for 85% of all lung cancer cases, including squamous cell carcinoma, adenocarcinoma and large cell carcinoma [Citation1]. With the improvement of overall survival, a therapeutic plateau has reached with a response rate of about 20% and a median survival of 8–10 months [Citation2,Citation3]. Nevertheless, the status of NSCLC therapy seems unsatisfactory. Studies on the molecular genetics of NSCLC have found that abnormal regulation occurs in oncogenes during the progression of NSCLC, which encode signaling proteins essential for cell proliferation and survival [Citation4,Citation5]. Exploring novel oncogenes or tumor suppressors in NSCLC may be helpful in understanding the pathogenesis of this cancer and improve therapeutic strategy.
The evolutionary studies on non-coding RNAs (ncRNAs) provide numerous oncogenic drivers and tumor suppressors, such as microRNAs (miRNAs), long ncRNAs (lncRNAs) and circular RNA (circRNAs) [Citation6]. CircRNA, a type of ncRNAs, is characterized by its stable closed-loop structure [Citation7]. An increasing number of circRNAs are discovered due to the development of high-throughput sequencing technology, and numerous circRNAs are recognized to be aberrantly expressed in tumor tissues compared to non-tumor tissues [Citation8,Citation9]. Progressive research has demonstrated that circRNAs play important functions in NSCLC. For example, circFARSA expression was increased in NSCLC cancerous tissues, and circFARSA overexpression promoted cell migration and invasion in NSCLC A459 cells [Citation10]. F-circEA-2a was also an oncogenic circRNA with stimulative effects on NSCLC cell migration and invasion [Citation11]. Circular progastricsin (circ-PGC), also known as hsa_circ_0076305, was originally shown to be differently expressed in gastric cancer [Citation12]. However, the function of circ-PGC remains not to be fully explored in NSCLC.
Forkhead box (FOX) proteins are a class of transcriptional regulators that are characterized by the forkhead DNA binding domains [Citation13]. Studies found that the deregulation of FOX proteins was commonly involved in human health and diseases, even in tumorigenesis and cancer progression [Citation13,Citation14]. Forkhead box R2 (FOXR2), a member of FOX proteins, is defined as an oncogene in multiple cancers, such as hepatocellular carcinoma, colorectal cancer and NSCLC [Citation15–17]. These findings highlighted the emerging role of FOXR2 as a direct and indirect target for therapeutic intervention, as well as biomarkers for therapeutic responses. Nevertheless, the mechanisms of FOXR2 action in cancer development are complex and not fully expounded.
The circRNA-miRNA-mRNA regulatory network is commonly demonstrated to be involved in cancer progression [Citation18,Citation19], which provides new insights to understand cancer pathogenesis. Whether FOXR2 is involved in circ-PGC-mediated regulatory networks in NSCLC is a mystery, but it is worth exploring.
In this study, we measured the expression levels of circ-PGC and FOXR2 in NSCLC tumor tissues and cells and conducted functional experiments to confirm the oncogenic functions of circ-PGC and FOXR2 in NSCLC. In addition, the circ-PGC/miR-532-3p/FOXR2 axis was constructed to provide a novel mechanism of circ-PGC and FOXR2 function in NSCLC.
Materials and methods
Clinical tissues
Tumor tissues and adjacent normal tissues were collected from NSCLC patients (n = 33) who were diagnosed and underwent surgery at The First College of Clinical Medical Science, China Three Gorges University. All patients had provided the written informed consent and approved the use of these tissues. Fresh tissues were immediately frozen in liquid nitrogen and stored in a − 80°C refrigerator. This study obtained the approval of the Ethics Committee of The First College of Clinical Medical Science, China Three Gorges University. The association of circ-PGC expression and clinicopathological characteristics of NSCLC patients is shown in .
Table 1. Association of circ-PGC expression with clinicopathological characteristics of patients with NSCLC
Hematoxylin and eosin (HE) staining
The architecture and pathological characteristics of tumor tissues and normal tissues were distinguished by HE staining using the H&E Staining Kit (Abcam, Cambridge, MA, USA). In brief, fresh tissues were fixed with 4% paraformaldehyde. Then, the fixed tissues were dehydrated by gradient levels of ethanol. After hyalinization with xylene, the tissues were embedded in paraffin overnight and sliced into 4-μm-thick sections, followed by the staining using HE for histological investigation.
Cell lines
NSCLC cell lines, including H460 and PC9, and human bronchial epithelial cells (BEAS-2B) were purchased from EK-Bioscience (Shanghai, China). According to the instructions, H460 and PC9 cells were cultured in RPMI1640 (EK-Bioscience) containing 10% fetal bovine serum (FBS; EK-Bioscience), and BEAS-2B cells were cultured in DMEM (EK-Bioscience) containing 10% FBS. All cells were maintained in a 37°C incubator supplemented with 5% CO2.
Real-time quantitative polymerase chain reaction (RT-qPCR)
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was utilized to extract total RNA, and the quality of RNA was tested using Nanodrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA). Next, the complementary DNA (cDNA) was synthesized from RNA using the riboSCRIPT Reverse Transcription Kit (Ribobio, Guangzhou, China). Afterward, the SYBR Green Real-Time PCR Master Mix (Thermo Fisher Scientific) was used to perform amplification reaction. β-actin or U6 was set as the internal parameter, and the relative expression was obtained using the 2−∆∆Ct method. The primer sequences were listed as below:
Circ-PGC forward, 5ʹ-CTAGACTTGCTGCCTCGACA-3ʹ and reverse, 5ʹ-TCTCACCAAAGTAGGCAGCTAA-3ʹ; PGC forward, 5ʹ-TCTGGTCTAGGTCCACTGCT-3ʹ and reverse, 5ʹ-CCAGAGGCTTCTCTGGTAGC-3ʹ; FOXR2 forward, 5ʹ-GGTTGGTGCCAAGTCTGTCT-3ʹ and reverse, 5ʹ-AACAACAGCCACTCTCCTCG-3ʹ; β-actin forward, 5ʹ-TCCCTGGAGAAGAGCTACGA-3ʹ and reverse, 5ʹ-AGCACTGTGTTGGCGTACAG-3ʹ; miR-532-3p forward, 5ʹ-CCCTCCCACACCCAAGG-3ʹ and reverse, 5ʹ-CCCAGTAGTCGTTCAGTCCA-3ʹ; U6 forward, 5ʹ-CTCGCTTCGGCAGCACA-3ʹ and reverse, 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ.
Subcellular location
RNA from cytoplasmic fraction and nuclear fraction was separately isolated using the PARIS kit (Invitrogen) as the protocol suggested. The enrichment of circ-PGC in cytoplasmic fraction and nuclear fraction was determined by RT-qPCR, using GAPDH or U6 as the internal reference in cytoplasmic fraction or nuclear fraction.
Western blot
Protein extraction was performed using the RIPA lysis buffer (Beyotime, Shanghai, China), and the equal protein (20 μg) was then separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and subsequently transferred onto polyvinylidene fluoride (PVDF) membranes (Bio-Rad, Hercules, CA, USA). After incubated with 10% bovine serum albumin (BSA), the membranes were incubated with the primary antibodies at 4°C overnight, including anti-FOXR2 (ab151476; Abcam, Cambridge, MA, USA), anti-Hexokinase 2 (HK-2; ab227198; Abcam), anti-matrix metalloproteinase 9 (MMP9; ab76003; Abcam), anti-E-cadherin (E-cad; ab40772; Abcam), anti-β-catenin (ab16051; Abcam), anti-c-Myc (ab32072; Abcam) and anti-β-actin (ab8227; Abcam). Next, the membranes were incubated with the goat anti-rabbit secondary antibody (ab201758; Abcam) at room temperature for 2 h. The protein bands were finally visualized using the enhanced chemiluminescent (ECL) Detection Reagents (Beyotime) and quantified using Image J software (NIH, Bethesda, MA, USA).
RNase R treatment
Total RNA (10 μg) from H460 and PC9 cells was incubated with 40 U RNase R (Epicenter, Madison, WI, USA) at 37 °C for 1 h. RNA transcripts were then precipitated with ethanol and resuspended in RNase-free H2O for RT-qPCR.
Cell transfection
Small interference RNA (siRNA)-mediated circ-PGC knockdown (si-circ-PGC#1 and #2) and FOXR2 knockdown (si-FOXR2) were obtained from Genepharma (Shanghai, China), with si-NC as a negative control. FOXR2 overexpression plasmid (pcDNA-FOXR2) was constructed by Genepharma, with pcDNA plasmid as a control. For miR-532-3p overexpression or inhibition, miR-532-3p mimics (miR-532-3p) or miR-532-3p inhibitors (anti-miR-532-3p) and matched negative control (miR-NC or anti-miR-NC) were purchased from Ribobio (Guangzhou, China). H460 and PC9 cells were transfected with these oligonucleotides or plasmids for functional analysis using the Lipofectamine 3000 transfection kit (Invitrogen) according to the manufacturer’s instruction.
Cell counting kit-8 (CCK-8) assay
A CCK-8 kit (Dojindo, Tokyo, Japan) was adapted to check cell viability. Briefly, H460 and PC9 cells (5 × 103 cells/well) were planted into 96-well plates and incubated for 48 h. Cells in each well were treated with CCK-8 solution for another 4 h. The absorbance was measured at 450 nm using a Multiskan Ascent reader (Thermo Fisher Scientific) to obtain relative cell viability.
Colony formation assay
H460 and PC9 cells were plated into a 6-well plate at a density of 300 cells/well and placed into a 37°C incubator supplemented with 5% CO2. After colony growth for two weeks, the colony surface was washed with phosphate-buffered saline (PBS; Beyotime), fixed with 4% paraformaldehyde (Beyotime), stained with 1% crystal violet (Beyotime) and then photographed. The number of colonies was counted under a microscope (Olympus, Tokyo, Japan).
Flow cytometry assay
Flow cytometry assay was performed to monitor cell apoptosis using the Annexin V- FITC apoptosis detection kit (BD Biosciences, San Jose, CA, USA). In brief, H460 and PC9 cells were collected at 48 h post-transfection, then washed with pre-cooled PBS (Beyotime) and incubated with the Annexin V-FITC binding buffer, followed by the staining with FITC-Annexin V and Propidium Iodide (PI) based on the instruction. The apoptotic cells were analyzed by FACS cytometer (BD Biosciences) and Flowjo software (Tree Star Corp, San Carlos, CA, USA).
Wound healing assay
Wound healing assay was implemented for cell migration analysis. In brief, H460 and PC9 cells were plated in 6-well plates (5 × 105 cells/well) and incubated for 24 h until a cell monolayer was formed. Then, a 10 μL pipette tip was utilized to create a mimic wound on the cell monolayer. Cells were washed with PBS, and wound healing was observed after 24 h under a reversed microscope (40×; Olympus). The migrated rate was analyzed using Image J software.
Transwell assay
Transwell assay was performed to monitor cell invasion. In brief, transwell chambers (Corning Costar, Corning, NY, USA) were coated with Matrigel (Corning Costar) at 4°C overnight. A total of 4 × 104 cells in serum-free culture medium were added into the top of chambers, and meanwhile, the bottom of chambers was filled with RPMI1640 medium containing 10% FBS. Cells were incubated in a 37°C incubator supplemented with 5% CO2 for 24 h, and then cells in the lower surface were fixed with 4% paraformaldehyde (Beyotime) and stained with 1% crystal violet (Beyotime). Five randomly selected areas were used to observe cell invasion under a microscope (100×; Olympus).
Glycolysis metabolism analysis
Glycolysis metabolism was analyzed according to the levels of glucose consumption, lactate production and adenosine triphosphate (ATP) production. The levels of glucose consumption, lactate production and ATP production were evaluated using the Glucose Assay kit (Abcam), Lactate Assay kit (Abcam) and ATP Assay kit (Abcam), respectively. All experimental procedures were performed following the protocol from each kit.
Target prediction
The relationship between circ-PGC and miR-532-3p was predicted by Circular RNA interactome (https://circinteractome.nia.nih.gov/). The relationship between FOXR2 and miR-532-3p was predicted by Target Scan Human7.2 (http://www.targetscan.org/vert_72).
Dual-luciferase reporter assay
Dual-luciferase reporter assay was conducted to verify the predicted relationship between miR-532-3p and circ-PGC or FOXR2. According to the predicted binding site between miR-532-3p and circ-PGC or FOXR2 3ʹUTR, mutations at miR-532-3p binding site in circ-PGC or FOXR2 3ʹUTR was designed by Genepharma. Then, the wild-type sequence fragment and mutant sequence fragment of circ-PGC or FOXR2 3ʹUTR were cloned into pmirGLO plasmid (Promega, Madison, WI, USA) to generate fusion report plasmid, naming as wt-circ-PGC, mut-circ-PGC, wt-FOXR2 3ʹUTR or mut-FOXR2 3ʹUTR. For dual-luciferase reporter assay, H460 and PC9 cells were transfected with miR-532-3p or miR-NC together with wt-circ-PGC, mut-circ-PGC, wt-FOXR2 3ʹUTR or mut-FOXR2 3ʹUTR and then cultured for 48 h. The luciferase activity in these cells was examined using the Dual-Luciferase Reporter Assay System (Promega).
RNA immunoprecipitation (RIP)
RIP assay was conducted to further verify the relationship between miR-532-3p and circ-PGC or FOXR2 using a Magna RIP Kit (Millipore Corp, Billerica, MA, USA). In brief, H460 and PC9 cells were lysed in 100 μL RIP lysis buffer containing a proteinase inhibitor and RNase inhibitor. Cell lysates were then diluted with 900 μL RIP buffer and incubated for 3 h with magnetic beads combined with Argonaute 2 (Ago2) antibody (Millipore Corp) or Immunoglobulin G (IgG) antibody (Millipore Corp). Beads were then washed six times with RIP wash buffer to elute immunoprecipitate. RNA was isolated using TRIzol reagent (Invitrogen) for RT-qPCR detection.
Xenograft experiments
A total of 10 nude mice (female, 6 weeks old, 18–21 g) were purchased from Charles River Labs (Beijing, China) and then maintained at a pathogen-free room. Short hairpin RNA (shRNA) targeting circ-PGC (sh-circ-PGC) for circ-PGC downregulation and its negative control (sh-NC) were synthesized and cloned into lentiviral vector by HanBio (Shanghai, China). After obtaining lentivirus particles (1 × 108PFU/mL) containing sh-circ-PGC or sh-NC, H460 cells were infected with sh-circ-PGC-introduced or sh-NC-introduced lentivirus particles and then subcutaneously inoculated into nude mice, respectively. The mice were classified into two groups (sh-circ-PGC group and sh-NC group, n = 5 per group). During tumor growth, tumor volume (length×width2 × 0.5) was measured once a week for 5 weeks. Then, mice underwent anesthesia were killed to excise tumor tissues. Tumor tissues were collected for the analysis of circ-PGC, miR-532-3p and FOXR2 expression. This study was implemented with the approval of the Animal Care and Use Committee of The First College of Clinical Medical Science, China Three Gorges University.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 7 (GraphPad Prism, La Jolla, CA, USA). Measured data following normal distribution were used for the comparisons between groups using Student’s t-test, while the comparisons among multiple groups were performed using analysis of variance (ANOVA). The data were shown as mean ± standard deviation (SD). The curve of overall survival was analzyed by Kaplan–Meier plot and log-rank test. The correlation between the expression of two parameters was analyzed using the Pearson R value analysis. P < 0.05 was considered to be statistically significant.
Results
Circ-PGC and FOXR2 were significantly upregulated in NSCLC tumor tissues and cell lines
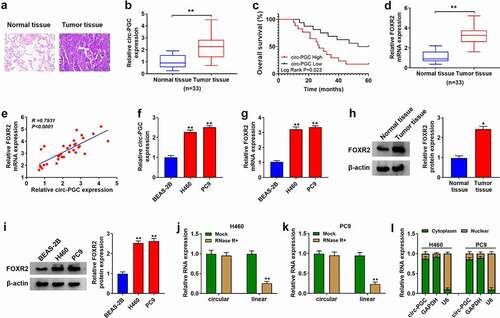
At first, we investigated the expression levels of circ-PGC and FOXR2 in NSCLC tumor tissues and cells. To distinguish tumor tissues and adjacent normal tissues, the result from HE staining led us to know that the number of cell nucleus was increased, and the atypia and split images were also increased in NSCLC tumor tissues compared to normal tissues (). Then, the data from RT-qPCR showed that the expression of circ-PGC was significantly elevated in tumor tissues (n = 33) compared to normal tissues (n = 33) (), and high circ-PGC expression was associated with poor overall survival of patients within 5 years (). The expression of FOXR2 mRNA was also enhanced in tumor tissues (n = 33) (), and FOXR2 mRNA level was positively correlated with circ-PGC expression in these tumor tissues (). Meanwhile, the data from RT-qPCR also showed that the expression of circ-PGC and FOXR2 mRNA was notably enhanced in H460 and PC9 cells compared with that in BEAS-2B cells (). The expression of FOXR2 protein was also enhanced in tumor tissues and cell lines relative to normal tissues and non-cancer cell line, respectively (). Moreover, we found RNase R treatment led to a significant decrease of linear PGC mRNA expression but scarcely affected circ-PGC expression in H460 and PC9 cells (), suggesting that circ-PGC was resistant to RNase R. Additionally, we characterized that circ-PGC was mainly distributed in the cytoplasm of H460 and PC9 cells but not nucleus (). These data hinted that circ-PGC and FOXR2 were involved in NSCLC development.
Figure 1. The expression of circ-PGC and FOXR2 was elevated in NSCLC tissues and cells
Circ-PGC knockdown inhibited the malignant phenotypes of H460 and PC9 cells
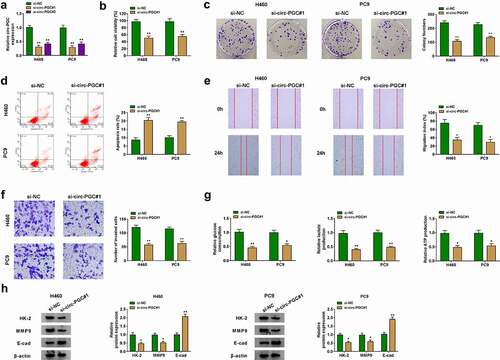
Si-circ-PGC-dependent suppression of circ-PGC expression was utilized to determine the function of circ-PGC in NSCLC cells. Firstly, we noticed that the expression of circ-PGC was notably decreased in H460 and PC9 cells after the transfection of si-circ-PGC#1 or si-circ-PGC#2, and si-circ-PGC#1 transfection led to a greater decrease of circ-PGC expression (). Herein, si-circ-PGC#1 was used in the subsequent assays. In function, we found si-circ-PGC#1 transfection not only weakened cell viability but also suppressed the ability of colony formation through CCK-8 and colony formation assays (). From flow cytometry assay, we found that circ-PGC knockdown remarkably promoted cell apoptosis (). Cell migration and invasion were assessed using wound healing assay and transwell assay, respectively. The data observed that the capacities of migration and invasion were significantly blocked in H460 and PC9 cells transfected with si-circ-PGC#1 relative to si-NC (). In addition, we also monitored the glycolysis metabolism affected by circ-PGC knockdown. The data showed that circ-PGC knockdown resulted in a remarkable decrease of glucose consumption, lactate production and ATP production (). Additionally, the expression of glycolysis-related marker (HK-2) and migration/invasion-related markers (MMP9 and E-cad) was detected by western blot, and the result presented that the expression of HK-2 and MMP9 was declined, while the expression of E-cad was enhanced in H460 and PC9 cells after circ-PGC knockdown (), suggesting that circ-PGC knockdown inhibited glycolysis and migration/invasion in H460 and PC9 cells. These data provided evidence that circ-PGC knockdown inhibited the malignant phenotypes of NSCLC cells.
Figure 2. Circ-PGC knockdown suppressed H460 and PC9 cell malignant phenotypes
FOXR2 knockdown attenuated the malignant phenotypes of H460 and PC9 cells
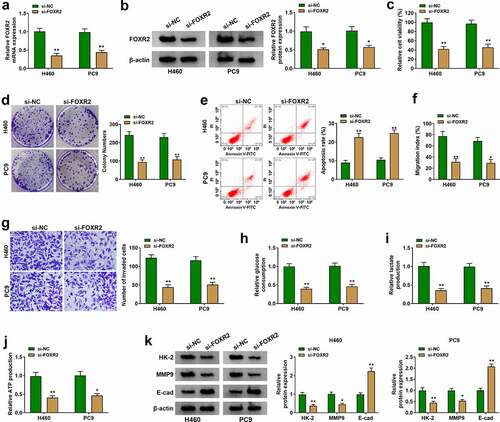
We next investigated the function of FOXR2 in NSCLC cells. At first, the data from RT-qPCR and western blot showed that the expression of FOXR2 was pronouncedly reduced in H460 and PC9 cells after the transfection of si-FOXR2 (). In function, si-FOXR2 transfection triggered a significant decrease of cell viability and colony formation ability (), while si-FOXR2 transfection notably induced cell apoptosis (). Besides, FOXR2 knockdown notably weakened the capacities of cell migration and cell invasion (). In addition, the levels of glucose consumption, lactate production and ATP production were significantly blocked in H460 and PC9 cells transfected with si-FOXR2 (). Western blot assay showed that the expression levels of HK-2 and MMP9 were decreased, while the expression level of E-cad was elevated in H460 and PC9 cells transfected with si-FOXR2 (). These data presented that FOXR2 knockdown attenuated the malignant phenotypes of H460 and PC9 cells.
Figure 3. FOXR2 knockdown inhibited H460 and PC9 cell malignant phenotypes
FOXR2 overexpression rescued the inhibitory malignant phenotypes of NSCLC cells caused by circ-PGC knockdown
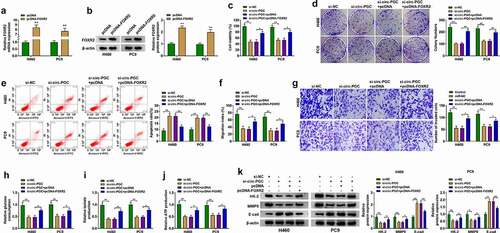
To investigate the association between circ-PGC and FOXR2, pcDNA-FOXR2 or pcDNA empty vector was transfected with si-circ-PGC into H460 and PC9 cells to test whether FOXR2 overexpression could rescue circ-PGC knockdown-resulted effects. The efficiency of pcDNA-FOXR2 was checked, and we found that the expression of FOXR2 at both mRNA and protein levels was remarkably elevated in H460 and PC9 cells after the transfection of pcDNA-FOXR2 compared to pcDNA (). Circ-PGC knockdown-resulted suppression of cell viability and colony formation was recovered by FOXR2 reintroduction (). Unsurprisingly, circ-PGC knockdown-induced cell apoptosis was largely suppressed by FOXR2 reintroduction (). Besides, cell migration and cell invasion were blocked in H460 and PC9 cells transfected with si-circ-PGC but partly promoted in cells transfected with si-circ-PGC+pcDNA-FOXR2 (). Moreover, the levels of glucose consumption, lactate production and ATP production were also recovered by the reintroduction of FOXR2 (). As expected, the expression of HK-2 and MMP9 weakened in si-circ-PGC-transfected cells was pronouncedly restored in cells transfected with si-circ-PGC+pcDNA-FOXR2, while the expression of E-cad promoted in si-circ-PGC-transfected cells was largely impaired in cells transfected with si-circ-PGC+pcDNA-FOXR2 (). These data indicated that FOXR2 overexpression could rescue the effects caused by circ-PGC knockdown, thereby recovering the malignant phenotypes of NSCLC cells.
Figure 4. FOXR2 overexpression rescued the inhibitory effects on H460 and PC9 cell malignant phenotypes caused by circ-PGC knockdown
MiR-532-3p was a target of circ-PGC, and FOXR2 was a target of miR-532-3p
Bioinformatics analysis (Circular RNA interactome and Target Scan Human7.2) predicted that miR-532-3p was a potential target of circ-PGC, and FOXR2 was a potential target of miR-532-3p. As shown in , miR-532-3p harbored the same binding site within circ-PGC fragment and FOXR2 3ʹUTR fragment. Then, dual-luciferase reporter assay was performed to verify the predicted relationship using wild-type and mutant-type reporter plasmids. The data showed that the luciferase activity in H460 and PC9 cells transfected with miR-532-3p and wt-circ-PGC was significantly lessened compared with that in cells transfected with miR-NC and wt-circ-PGC, while the luciferase activity was hardly changed in cells transfected with miR-532-3p and mut-circ-PGC relative to miR-NC (). Similarly, the data also showed that miR-532-3p along with wt-FOXR2 3ʹUTR transfection but not mut-FOXR2 3ʹUTR transfection markedly reduced the luciferase activity in H460 and PC9 cells (). Meanwhile, RIP assay was performed to further verify the relationship between miR-532-3p and circ-PGC or FOXR2. The results displayed that both miR-532-3p and circ-PGC could be largely detected in the anti-Ago2 group compared to anti-IgG in RIP assay (), and both miR-532-3p and FOXR2 were enriched in the anti-Ago2 RIP group relative to anti-IgG (). The evidence strongly supported that miR-532-3p was a target of circ-PGC, and miR-532-3p directly bound to FOXR2. In addition, we examined the expression levels of miR-532-3p in NSCLC tissues and cells. As shown in , the expression of miR-532-3p was markedly declined in NSCLC tumor tissues compared to normal tissues, also in H460 and PC9 cells compared to BEAS-2B cells through RT-qPCR analysis. Importantly, miR-532-3p expression was negatively correlated with circ-PGC expression in tumor tissues, and FOXR2 mRNA expression was negatively correlated with miR-532-3p expression in tumor tissues ().
Figure 5. MiR-532-3p was a target of circ-PGC, and miR-532-3p bound to FOXR2
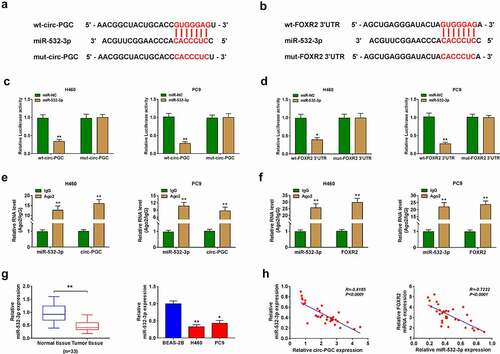
Circ-PGC positively regulated FOXR2 expression by targeting miR-532-3p
Furthermore, we found that the expression of FOXR2 was prominently decreased in H460 and PC9 cells transfected with si-circ-PGC, while its expression was largely recovered in H460 and PC9 cells transfected with si-circ-PGC+anti-532-3p (), suggesting that circ-PGC could positively regulate FOXR2 expression by targeting miR-532-3p.
Figure 6. Circ-PGC positively regulated FOXR2 expression by targeting miR-532-3p
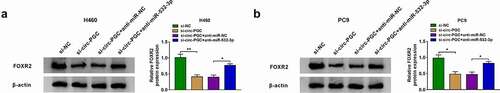
Wnt/β-catenin signaling pathway might be involved in the circ-PGC/miR-532-3p/FOXR2 regulatory network
Herein, the expression of β-catenin and c-Myc protein was remarkably reduced in H460 and PC9 cells transfected with si-circ-PGC, while their expression was largely recovered in H460 and PC9 cells transfected with si-circ-PGC+anti-miR-532-3p or si-circ-PGC+pcDNA-FOXR2 (). The changes of β-catenin and c-Myc expression in these transfected cells suggested that Wnt/β-catenin signaling pathway might be regulated by the circ-PGC/miR-532-3p/FOXR2 axis in NSCLC development.
Figure 7. Wnt/β-catenin signaling pathway might be involved in the circ-PGC/miR-532-3p/FOXR2 regulatory network
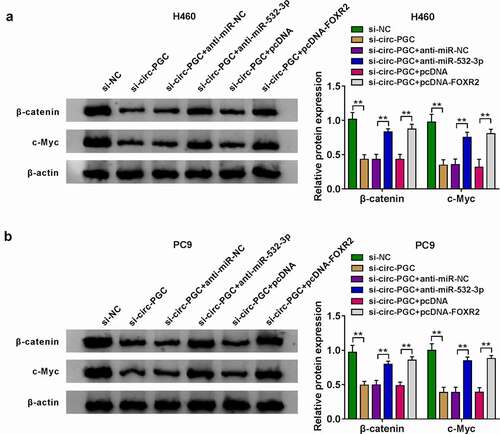
Circ-PGC downregulation blocked tumor growth in vivo
Xenograft model was constructed in nude mice to expound the role of circ-PGC in vivo. As a result, circ-PGC downregulation strikingly restrained the tumor size (). As shown in , tumor volume and tumor weight were significantly blocked in the sh-circ-PGC group compared with that in the sh-NC group. The data indicated that circ-PGC downregulation blocked tumor growth in vivo. In addition, the expression of circ-PGC and the expression of FOXR2 at both mRNA and protein levels were significantly lower in tumor tissues from the sh-circ-PGC group, while the expression of miR-532-3p was notably higher in tumor tissues from the sh-circ-PGC group compared with that from the sh-NC group (), suggesting that circ-PGC affected tumor growth in vivo by targeting the miR-532-3p/FOXR2 axis. Moreover, we found that the protein levels of MMP9, c-Myc and β-catenin were strikingly decreased, while the protein level of E-cad was strikingly elevated in tumor tissues from the sh-circ-PGC group (), suggesting that circ-PGC knockdown blocked the activation of Wnt/β-catenin signaling in vivo.
Figure 8. Circ-PGC inhibited tumor growth in vivo.
Discussion
NSCLC has long been a focus of public concern due to its high morbidity and mortality. In recent years, molecular targeted therapy has emerged gradually and is considered as a promising method to improve the therapeutic effects of NSCLC [Citation20]. With the advance of RNA sequencing technology, a growing number of circRNAs are identified to be differently expressed in cancers. Accumulating evidence discloses that specific circRNAs are ideal biomarkers for cancer diagnosis and therapy, and they act as oncogenes or tumor suppressor genes in function [Citation10,Citation21,Citation22]. In our study, we illustrated that circ-PGC functioned as an oncogene in NSCLC to deteriorate the development of NSCLC.
Circ-PGC was previously shown to be downregulated in gastric cancer tissues, suggesting that circ-PGC was involved in gastric cancer progression [Citation12]. However, a recent study exposed that the expression of circ-PGC was elevated in NSCLC tissues and cells, and downregulated circ-PGC inhibited cisplatin resistance and thus blocked NSCLC progression [Citation23]. The results indicated that circ-PGC had different expression patterns and functions in different cancers. In our study, we found that the expression of circ-PGC was promoted in NSCLC tissues and cells. Functionally, circ-PGC knockdown suppressed NSCLC cell malignant behaviors, including proliferation, migration and invasion. Intense energy metabolism is an emblematical feature of cancer progression, of which glycolysis is a representative process [Citation24]. Progressive glycolysis manifested as increased glucose consumption, increased lactic acid production, and overexpression of HK-2 [Citation25]. Herein, circ-PGC knockdown was shown to lessen the levels of glucose consumption, lactate and ATP production, leading to inhibitory glycolysis metabolism. On the whole, our findings provide the functions of circ-PGC not only on cell proliferation and migration/invasion but also on energy metabolism in NSCLC.
Regarding the role of FOXR2 in NSCLC, previous studies led us to know that FOXR2 deficiency inhibited cell proliferation and invasion in vitro as well as tumor growth and metastasis [Citation17]. Moreover, the carcinogenic effects of FOXR2 were also determined in other cancers. For example, enhanced expression of FOXR2 resulted in aggressive cell malignant phenotypes, including proliferation, angiogenesis, migration and epithelial-mesenchymal transition (EMT) in ovarian cancer, colorectal cancer and prostate cancer [Citation16,Citation26,Citation27]. Consistent with these consequences, our data showed that FOXR2 was highly expressed in NSCLC tissues and cells, and FOXR2 knockdown repressed NSCLC cell proliferation, migration, invasion and glycolysis metabolism.
Further experiments monitored that FOXR2 overexpression rescued the inhibitory effects of circ-PGC knockdown on NSCLC cell malignant phenotypes. Considering the vital role of circ-PGC and FOXR2 in NSCLC, we hypothesized a correlation between circ-PGC and FOXR2. Luckily and interestingly, we found that miR-532-3p harbored the same binding site with circ-PGC and FOXR2 3ʹUTR, and circ-PGC positively regulated the expression of FOXR2 by targeting miR-532-3p, which was accordant with the competing endogenous RNA (ceRNA) mechanism that circRNA functioned as a molecular sponge of target miRNA to regulate the expression of downstream mRNA [Citation28,Citation29]. It was documented that low expression of miR-532-3p was detected in NSCLC tissues and cells, and miR-532-3p restoration impaired NSCLC cell proliferation, migration and invasion [Citation30,Citation31]. Herein, circ-PGC acted as miR-532-3p sponge to suppress miR-532-3p expression and decoy the tumor suppressor role of miR-532-3p.
Moreover, our data showed that deregulated circ-PGC affected the expression of β-catenin and c-Myc, which were vital proteins in the Wnt/β-catenin signaling pathway. The activation of Wnt/β-catenin signaling pathway is closely associated with tumorigenesis and progression [Citation32]. Previous studies recorded that FOXR2 knockdown blocked NSCLC progression by reducing the levels of β-catenin, cyclinD1 and c-Myc in the Wnt/β-catenin signaling pathway [Citation17]. Combined with our data, FOXR2-activated Wnt/β-catenin signaling pathway might be mediated by the circ-PGC/miR-532-3p/FOXR2 axis.
In conclusion, we found that the expression of circ-PGC and FOXR2 was elevated in NSCLC tissues and cells. Circ-PGC increased the expression of FOXR2 by targeting miR-532-3p, thus promoting the development of NSCLC, which might be attributed to the activation of the Wnt/β-catenin signaling pathway. The evidence from our study strongly supports the carcinogenic effect of circ-PGC, which may be a targeted strategy for NSCLC treatment.
Highlights
Circ-PGC and FOXR2 are highly expressed in NSCLC tumor tissues and cells.
Circ-PGC knockdown or FOXR2 knockdown inhibits NSCLC cell malignant phenotypes.
Circ-PGC positively regulates FOXR2 expression by targeting miR-853-3p.
Wnt/β-catenin signaling pathway may be involved in the circ-PGC/miR-532-3p/FOXR2 regulatory network.
Circ-PGC downregulation inhibits tumor growth in vivo.
Ethics approval and consent to participate
The present study was approved by the ethical review committee of The First College of Clinical Medical Science, China Three Gorges University. Written informed consent was obtained from all enrolled patients.
Consent for publication
Patients agree to participate in this work
Acknowledgments
Not applicable
Disclosure statement
No potential conflict of interest was reported by the author(s).
Availability of data and materials
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
Additional information
Funding
References
- Saijo N. Problems involved in the clinical trials for non-small cell lung carcinoma. Cancer Treat Rev. 2012;38(3):194–202.
- Group N M-A C. Chemotherapy in addition to supportive care improves survival in advanced non-small-cell lung cancer: a systematic review and meta-analysis of individual patient data from 16 randomized controlled trials. J Clin Oncol. 2008;26(28):4617–4625.
- Kumarakulasinghe NB, van Zanwijk N, Soo RA. Molecular targeted therapy in the treatment of advanced stage non-small cell lung cancer (NSCLC). Respirology. 2015;20(3):370–378.
- Moravcikova E, Krepela E, Donnenberg VS, et al. BOK displays cell death-independent tumor suppressor activity in non-small-cell lung carcinoma. Int J Cancer. 2017;141(10):2050–2061.
- Alaee M, Nool K, Pasdar M. Plakoglobin restores tumor suppressor activity of p53(R175H) mutant by sequestering the oncogenic potential of beta-catenin. Cancer Sci. 2018;109(6):1876–1888.
- Ghafouri-Fard S, Shoorei H, Branicki W, et al. Non-coding RNA profile in lung cancer. Exp Mol Pathol. 2020;114:104411.
- Ebbesen KK, Hansen TB, Kjems J. Insights into circular RNA biology. RNA Biol. 2017;14(8):1035–1045.
- Lopez-Jimenez E, Rojas AM, Andres-Leon E. RNA sequencing and prediction tools for circular RNAs analysis. Adv Exp Med Biol. 2018;1087:17–33.
- Wang C, Tan S, Liu WR, et al. RNA-Seq profiling of circular RNA in human lung adenocarcinoma and squamous cell carcinoma. Mol Cancer. 2019;18(1):134.
- Hang D, Zhou J, Qin N, et al. A novel plasma circular RNA circFARSA is a potential biomarker for non-small cell lung cancer. Cancer Med. 2018;7(6):2783–2791.
- Tan S, Sun D, Pu W, et al. Circular RNA F-circEA-2a derived from EML4-ALK fusion gene promotes cell migration and invasion in non-small cell lung cancer. Mol Cancer. 2018;17(1):138.
- Dang Y, Ouyang X, Zhang F, et al. Circular RNAs expression profiles in human gastric cancer. Sci Rep. 2017;7(1):9060.
- Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7(11):847–859.
- Benayoun BA, Caburet S, Veitia RA. Forkhead transcription factors: key players in health and disease. Trends Genet. 2011;27(6):224–232.
- Wang X, He B, Gao Y, et al. FOXR2 contributes to cell proliferation and malignancy in human hepatocellular carcinoma. Tumour Biol. 2016;37(8):10459–10467.
- Lu SQ, Qiu Y, Dai WJ, et al. FOXR2 promotes the proliferation, invasion, and epithelial-mesenchymal transition in human colorectal cancer cells. Oncol Res. 2017;25(5):681–689.
- Wang XH, Cui YX, Wang ZM, et al. Down-regulation of FOXR2 inhibits non-small cell lung cancer cell proliferation and invasion through the Wnt/beta-catenin signaling pathway. Biochem Biophys Res Commun. 2018;500(2):229–235.
- Liang ZZ, Guo C, Zou MM, et al. circRNA-miRNA-mRNA regulatory network in human lung cancer: an update. Cancer Cell Int. 2020;20(1):173.
- Liang L, Zhang L, Zhang J, et al. Identification of circRNA-miRNA-mRNA networks for exploring the fundamental mechanism in lung adenocarcinoma. Onco Targets Ther. 2020;13:2945–2955.
- Molina JR, Yang P, Cassivi SD, et al. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83(5):584–594.
- Liu W, Ma W, Yuan Y, et al. Circular RNA hsa_circRNA_103809 promotes lung cancer progression via facilitating ZNF121-dependent MYC expression by sequestering miR-4302. Biochem Biophys Res Commun. 2018;500(4):846–851.
- Lei B, Tian Z, Fan W, et al. Circular RNA: a novel biomarker and therapeutic target for human cancers. Int J Med Sci. 2019;16(2):292–301.
- Dong Y, Xu T, Zhong S, et al. Circ_0076305 regulates cisplatin resistance of non-small cell lung cancer via positively modulating STAT3 by sponging miR-296-5p. Life Sci. 2019;239:116984.
- Abbaszadeh Z, Cesmeli S, Biray Avci C. Crucial players in glycolysis: cancer progress. Gene. 2020;726:144158.
- Zhou L, Li M, Yu X, et al. Repression of Hexokinases II-Mediated glycolysis contributes to piperlongumine-induced tumor suppression in non-small cell lung cancer cells. Int J Biol Sci. 2019;15(4):826–837.
- Li B, Huang W, Cao N, et al. Forkhead-box R2 promotes metastasis and growth by stimulating angiogenesis and activating hedgehog signaling pathway in ovarian cancer. J Cell Biochem. 2018;119(9):7780–7789.
- Xu W, Chang J, Liu G, et al. Knockdown of FOXR2 suppresses the tumorigenesis, growth and metastasis of prostate cancer. Biomed Pharmacother. 2017;87:471–475.
- Zhang S, Cheng J, Quan C, et al. circCELSR1 (hsa_circ_0063809) contributes to Paclitaxel resistance of ovarian cancer cells by regulating FOXR2 expression via miR-1252. Mol Ther Nucleic Acids. 2020;19:718–730.
- Tian X, Zhang L, Jiao Y, et al. CircABCB10 promotes nonsmall cell lung cancer cell proliferation and migration by regulating the miR-1252/FOXR2 axis. J Cell Biochem. 2019;120(3):3765–3772.
- Jiang W, Zheng L, Yan Q, et al. MiR-532-3p inhibits metastasis and proliferation of non-small cell lung cancer by targeting FOXP3. J BUON. 2019;24(6):2287–2293.
- Liu D, Liu H, Jiang Z, et al. Long non-coding RNA DARS-AS1 promotes tumorigenesis of non-small cell lung cancer via targeting miR-532-3p. Minerva Med. 2021;112(3):408–409.
- Li YF, Zhang J, Yu L. Circular RNAs regulate cancer onset and progression via Wnt/beta-Catenin signaling pathway. Yonsei Med J. 2019;60(12):1117–1128.
