ABSTRACT
Previously our results showed miR-222-3p was significantly downregulated in retinoic acid-induced neural tube defect (NTD) mouse model through transcriptome. Down-regulation of miR-222-3p may be a causative biomarker in NTDs. In this study, RNA was extracted from mouse embryos at E8.5, E9.5 and E10.5, and the expression level of miR-222-3p was measured by quantitative real-time PCR analysis. The preliminary mechanism of miR-222-3p in NTDs involved in cell proliferation, apoptosis and migration was investigated in mouse HT-22 cell line. The expression of miR-222-3p was significantly decreased at E8.5, E9.5 and E10.5 developed in mouse embryos which were consistent with our transcriptome sequencing. Suppression of miR-222-3p in HT-22 cells resulted in the inhibition of cell proliferation and migration, cell cycle and apoptosis. Moreover, DNA damage transcript 4 (Ddit4) was identified as a direct and functional target of miR-222-3p. miR-222-3p is negatively regulated by Ddit4. The mutation of binding site of Ddit4 3ʹUTR abrogated the responsiveness of luciferase reporters to miR-222-3p and showed that Ddit4 expression partially attenuated the function of miR-222-3p. We preliminatively confirmed that low expression of miR-222-3p has reduced the expression of β-catenin, TCF4 and other related genes in the Wnt/β-catenin signaling pathway.
Collectively, these results demonstrated that miR-222-3p regulates the Wnt/β-catenin signaling pathway through Ddit4 inhibition in HT-22 cells, resulted in cell proliferation and apoptosis imbalance, and thus led to neural tube defects.
1. Introduction
Neural tube defects (NTDs) are the most common (second in occurrence after congenital heart disease) and serious birth defects of central nervous system [Citation1], due to failure of complete neural tube closure during embryonic development [Citation2]. World-wide, it has been estimated that approximately 300,000 NTDs cases are reported each year [Citation3]. According to the World Health Organization estimate, the incidence rate of birth defects in China is about 5.6%, which is close to the average level of middle-income countries in the world [Citation4]. Li et al. has reported that prevalence of NTDs tends to be higher in northern China (18.7 per 10,000 births) than in southern China (9.7 per 10,000 births) [Citation5]. The clinical manifestations of NTDs have two main sub-groups (open and closed defects) NTDs including anencephaly, spina bifida and myelomeningocele [Citation5–8]. NTDs are multifactorial disorders induced both by genetic and environmental factors [Citation9–11]. Despite many etiological studies in humans [Citation11–15], pathogenesis of NTDs remains poorly understood.
Table 1. List of primers used
Retinoic acid (RA) is a vitamin A metabolite and nutritionally derived small molecule involved in anterior-posterior and neural tube patterning [Citation16,Citation17]. Excess or insufficient RA levels can lead to neural tube abnormalities which have been extensively investigated in the developmental toxicity with retinoids in animal models, as well as in humans after taking RA in multivitamin preparations during pregnancy [Citation18–21]. During early embryogenesis (E7-E10), excessive RA exposure can lead to NTDs, including anencephaly, exencephaly and spina bifida [Citation16,Citation22].
MicroRNAs (miRNAs) are non-coding RNAs, composed of approximately 22 nucleotides which regulate gene expression by complementarily binding to the 3ʹUTR sequence of their target mRNA and thus lead to mRNA degradation or translational repression. MiRNAs are widely involved in crucial life processes such as cell cycle, embryonic development, cell growth and differentiation, apoptosis, stress response, metabolism and morphogenesis [Citation23–26]. It has been confirmed that miRNAs play key role in neurobiology of neuronal lineage, synapses and neurogenesis, and have significant effects on differentiation of neuronal cells [Citation27]. An increasing number of studies have implied that miRNAs could be a biomarker for NTDs, but still the underlying mechanism involved is not clear. Recently, several miRNAs have been reported to be associated with NTDs. miR-129-1, miR-129, miR-9, miR-197, miR-124 and miR-27 have been shown to influence neural tube formation [Citation28–33]. Our previous transcriptome sequencing showed that miR-222-3p has markedly down-regulated in NTDs mice. The expression of miR-222-3p has been reported to be up-regulated in human breast cancer [Citation34], thyroid cancer [Citation35] and glial cancers [Citation36]. However, miR-222 is elevated in Human TK-6 and DKO cells under folate deficiency [Citation37,Citation38]. The function of miR-222-3p in neural tube closure and formation of defects (NTDs), as well as its possible target genes, were still unknown.
Further review of relevant literature showed that the closure of the neural tube requires coordination of various cellular mechanisms, including apical constriction, cell proliferation, and apoptosis [Citation39]. Recent studies have shown that NTDs are mainly a result of excessive apoptosis of neural crest cells or ventral cells of neural tube [Citation40,Citation41]. Moreover, abnormal cell proliferation leads to NTDs in the process of neuronal furrow formation [Citation42]. Previous results of our research group have shown that apoptosis is one of the main causes of NTDs [Citation43]. Yang et al. [Citation44] evaluated that miR-222 has blocked cell cycle by inhibiting the expression of p27 gene and proliferation of hepatoma cells. Terasawa et al. [Citation45] studied that low expression of miR-222-3p inhibited cell growth, migration, and promoted apoptosis. The above research indicated that miR-222-3p may affect the proliferation, apoptosis and cycle of cells, thus affecting the normal cell function. However, the effect of miR-222-3p on neural tube defects remained largely unknown. Therefore, we speculated that miR-222-3p may cause NTDs by affecting cell proliferation and apoptosis.
In this study, we used a retinoic acid (RA)-induced mouse NTDs model to correlate the spatial and temporal expression pattern of miR-222-3p with NTDs. Surprisingly, miR-222-3p was differentially expressed in NTDs group as compared with the control group. We then investigated the role of miR-222-3p on cell proliferation, apoptosis and migration in HT-22 cell line. To explore the underlying mechanism of miR-222-3p in NTDs, we hope to explore a promising target in HT-22 cells. We identified potential target genes of miR-222-3p using bioinformatics algorithms Targetscan, PicTar, miRanda, PITA and miRDB. All resulted in one prediction that Ddit4 was an important target gene of miR-222-3p and is also an oxidative stress gene [Citation46]. We identified Ddit4 as a target gene of miR-222-3p and demonstrated that it can partially mediate the function of miR-222-3p in HT-22 cells. Furthermore, we preliminarily confirmed that low expression of miR-222-3p inhibited the Wnt/β-catenin signaling pathways. Taken together, our results demonstrated a link between miR-222-3p expression andNTDs, which enhanced our understanding regarding abnormal epigenetic modification in NTDs.
2. Materials and methods
2.1 Animals
SPF C57BL/6 mice (8–12 weeks, 19–25 g) were purchased from the Experimental Animal Center of Shanxi Medical University (Taiyuan, China). Mature male and female mice were mated overnight and the vaginal plug was detected in the morning. Pregnant mice were randomly divided into control and NTDs group. At 7.5 day of pregnancy (E7.5), NTDs model group mice were gavaged with all-trans retinoic acid (ATRA) (Sigma, USA, dissolved in sesame oil) at a dose of 28 mg/kg, and the control group was treated with the same dose of sesame oil. At E8.5, E9.5 and E10.5, cervical dislocation was used to euthanize pregnant mice, and embryonic neural tissues were collected according to previous research method (Mukhopadhyay P, Brock G, Appana S, Webb C, Greene RM, Pisano MM. MicroRNA gene expression signatures in the developing neural tube. Birth Defects Res A. 2011;91:744–62.).
2.2 Cell culture and transfection
The mouse clonal hippocampal neuronal cell line HT-22 cells (Jennio Biotech Co, Guangzhou, China) were cultured in DMEM high glucose medium (Hyclone, Logan, UT) supplemented with 10% fetal bovine serum (FBS, Gibco, USA) and 1% penicillin/streptomycin (Sigma, USA). All cells were maintained in a humidified incubator with 5% CO2 and 37 °C atmospheres. 5 × 105 cells/ml was plated in antibiotic-free medium for 24 h before transfections. miR-222-3p mimics (mim-222), miR-222-3p inhibitor (anti-222), miR mimics negative control (mim-NC), miR inhibitor negative control (anti-NC) and siRNA-Ddit4 plasmid were designed and synthesized by GenePharma Co (Shanghai, China). When the HT22 cells become 75–85% confluent, cells were transiently transfected with 100 nM miRNA or siRNA using Lipofectamine RNAiMax (Santa Cruz Biotechnology, USA), according to the manufacturer’s instructions. After 6 hours of transfection, the culture medium was replaced with fresh medium containing 10% FBS. The transfections were performed for 24 or 48 h, and then harvested for further analysis. The sequences of the small molecules are as follows:
mim-222: AGCUACAUCUGGCUACUGGGUCU;
ACCCAGUAGCCAGAUGUAGCUUU.
anti-222: AGACCCAGUAGCCAGAUGUAGCU;
mim-NC: CAGUACUUUUGUGUAGUACAA.
siRNA-Ddit4:UGAGAGUCAUCAAGAAGAATT;
UUCUUCUUGAUGACUCUGATT.
anti-NC:UUCUCCGAACGUGUCACGUTT;
ACGUGACACGUUCGGAGAATT.
2.3 RNA extraction and qRT-PCR
Total RNAs from E8.5, E9.5 and E10.5 embryonic brain tissues were extracted using TRIzol reagent (Invitrogen, Carlsbad, CA), the quality and concentration of total RNA were measured by SoftMax Pro7.1 (NanoDrop, Thermo, US). For mRNA detection, reverse-transcribed complementary DNA was synthesized using the RevertAidlst cDNA Synth Kit (Thermo, USA). Quantitative PCR was performed on a Real-Time PCR platform (ABI Stepone Real-Time PCR system, USA) with SYBR Premix EX Taq (Takara, Japan). The procedure was as follows: (95°C, 10 min)×1 cycle; (95°C, 15 s; 60°C, 1 min)×40 cycles. The results were normalized to β-actin expression. For miRNA detection, the reverse transcribed cDNA was synthesized with the Hairpin-it miRNAs qRT-PCR and quantification kit qRT-PCR detection of miR-222-3p (GenePharma, China), and normalized by U6 small nuclear RNA. The data was analyzed using 2−ΔΔCt method and all qPCR reactions were performed in triplicate. Primer sequences were shown in Additional file 1: Table S1.
2.4 Cell proliferation assay
For the cell proliferation assay, the Cell Counting Kit-8 (CCK-8; Dojindo) was used to determine the viability of cells. After 24 h of transfection, HT22 cells at a density of 1 × 103 cells/well were plated in 96-well microplates and incubated in normal culture conditions. The cell viability assay was performed every 24 h for 96 h by adding 10 μl of CCK-8 reagent to each well, incubated at 37°C for an additional 1 h and then measured the absorbance/optical density (OD) at a wavelength of 450 nm using a Bio-Rad imarkTM microplate absorbance reader (Bio-Rad Laboratories, Hercules, CA, USA).
2.5 Cell cycle analysis
The cell cycle was analyzed by flow cytometry using a cell cycle detection kit (KeyGEN, Suzhou, China). Transfected cells were seeded in 6-well plates and incubated for 48 h. Cells were washed twice with phosphate-buffered saline (PBS), trypsinized and then centrifuged at 1000 × g for 5 min. The harvested cells were re-washed twice with PBS, fixed in 70% cold ethanol at 4°C for 24 h and stained with 50 μg/ml propidium iodide (PI) for 30 min. Cell cycle distribution was analyzed by Coulter Epics XL flow cytometry (BD Biosciences, San Jose, CA, USA) and cells in distinct cell cycle phases were determined using ModFit LT3.2 (Verity Software House).
2.6 Cell apoptosis assay
The apoptosis of transfected cells was measured using Annexin V/Fluorescein isothiocyanate (FITC) apoptosis detection kit (KeyGEN, Suzhou, China). After 48 hours of transfection, the cells were digested with 0.25% trypsin without ethylenediaminetetraacetic acid (EDTA) and centrifuged at 1000 × g for 5 min. A mixture of 500 µL binding buffer, 5 µL PI and 5 µL Annexin V/FITC was added to the cells, and incubated for 15 minutes. The apoptotic rate was detected on flow cytometry (BD Biosciences, San Jose, CA, USA). All experiments were performed in triplicate for each group.
2.7 Acridine Orange/Ethidium Bromide staining (AO/EB)
Acridine Orange/Ethidium Bromide staining (AO/EB) (Solarbio, Beijing, China) was used to detect the cell apoptosis according to the manufacturer’s instructions. Transfected cells were seeded in 24-well plates and incubated for 24 h. The cells were then washed thrice with 1x PBS and stained with 10 μg/ml Hoechst and 10 μg/ml Acridine Orange/Ethidium Bromide (AO/EB) for 15 min in a humidified chamber. The cells were washed again with 1x PBS and visualized by fluorescence inverted microscope (Nikon, Tokyo, Japan).
2.8 Cell migration assays
Transfected cells were seeded in 6-well plates, cultured for 16–18 h, and serum-starved overnight. A perpendicular line was drawn at the center of the cells monolayer with the help of p10 pipette tip to create the wound. The cells were washed thrice with PBS and incubated in serum-free medium at 37°C and 5% CO2. The wound closure was photographed at specified time intervals (0, 24 h) by an inverted microscope with a microscope digital camera. Wound healing area was calculated using ImageJ with the following formula: Wound healing rate = (initial scratch width – final scratch width)/initial scratch width × 100%. These experiments were performed in triplicate.
2.9 Western blotting
After 72 h of transfection, the cells were lysed with RIPA lysis buffer (Solarbio, Beijing, China) containing 10 mM PMSF (Solarbio, Beijing, China). The concentration of protein was measured with BCA Protein Assay kit (Thermo Fisher Scientific, USA). The samples were subjected to SDS-PAGE and then electro-transferred into a PVDF membrane. The membrane was blocked from nonspecific first antibody in 5% skim milk with Tris-buffered saline containing 0.1% Tween 20 solution (TBST) at room temperature for 1 h. Subsequently, the membrane was incubated with primary antibodies (Rabbit anti- C-myc, β-catenin, GAPDH, PCNA, Cleaved Caspase-3, Cycin-D1, 1:3000, Abcam, United Kingdom; Mouse anti-TCF4, Ddit4, 1:3000, Abcam, United Kingdom) at 4°C overnight, followed by TBST washing and incubation with the corresponding secondary HRP-conjugated antibody (HRP-labeled goat anti-rabbit IgG and anti-mouse IgG, 1:10,000, ZXGB-BIO, Beijing, China). After washing with TBST, targeted proteins on the membrane were detected by using ECL chemiluminescent detection system (minibio, Shangxi, China). The blots were scanned and detected on the Gene Gnome Western blot imaging system (Bio-RAD, US). The band density was measured by ImageJ software.
2.10 Luciferase reporter gene assay
The target-binding site of miR-222-3p on Ddit4 was predicted by TargetScan (http://www.targetscan.org/). The sequence fragments of Ddit4 wild-type (Ddit4-WT) and mutant (Ddit4-MUT) were synthesized and cloned into GP-miRGLO luciferase vector (GenePharma, China). After transfection with miRNA mimics for 24 hours, GP-miRGLO-Ddit4-Wt/Mut (130 ng per well in 96-well plate) along with GP-miRGLO-control (130 ng per well) were transfected into the cells. After 24 h of incubation, the cells were harvested and lysed with passive lysis buffer (Promega, US). Luciferase activity was measured by a dual-luciferase reporter system (Promega, US). The luminescence intensity of Firefly luciferase was normalized to that of Renilla luciferase. All experiments were performed five times for each group.
2.11 Statistical analysis
All reactions were performed at least three times and each independent experiment was carried out in triplicate for each condition according to the manufacturer’s instructions. Statistical analysis was performed using SPSS 17.0 software (SPSS, Chicago, IL, USA). The data were shown as mean ± SD. The Student’s t-test was used to analyze the difference between two samples. *p < 0.05, **p < 0.01 or ***p < 0.001 was considered statistically significant.
3. Results
3.1 Down-regulation of miR-222-3p in NTDs mouse
In this study, NTDs mouse models were induced with 28 mg/kg dose of RA on 7.5 days of gestation, the failure of closure at the level of the forebrain at this stage leads to significant anencephaly [Citation47,Citation48]. As shown in , significant differences were shown in embryonic morphology between RA treatment group and control group on E10.5. The embryos of the normal group were intact with obvious outlines, clear sections and full brain. RA treated embryos showed a typical anencephalic phenotype with blurred outlines, small size and unclear body segments. Our previous sRNA sequencing in those brain tissues showed that the expression level of miR-222-3p was significantly decreased in the RA treated group on E8.5, E9.5 and E10.5 compared with the control group [Citation49] (). Subsequently, we confirmed that miR-222-3p expression was down-regulated in RA treatment group on E8.5, E9.5 and E10.5 compared with the control group by qRT-PCR (). Results were consistent with Transcriptome sequencing. These results suggested that miR-222-3p may play a key role in the pathogenesis of NTDs.
Figure 1. miR-222-3p expression was decreased in RA-treated mouse embryos. (a) Normal and RA-treated mouse embryos at E10.5. The red arrows indicate growth retardation in the forebrain and the posterior brain. (b) qRT-PCR was used to detect the mRNA level of miR-222-3p in normal and malformed brain tissue at E8.5-E10.5. (c) miR-222-3p miRNA-Seq results
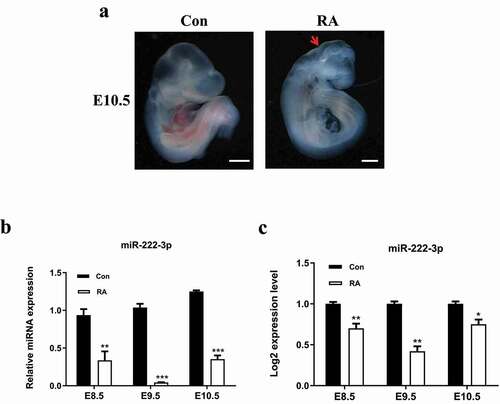
3.2 miR-222-3p down-regulation inhibits the proliferation of HT-22 cells in vitro
To investigate the biological function of miR-222-3p in NTDs, we selected mouse hippocampal neuronal cell line (HT-22 cells) . According to the results (), the expression of miR-222-3p in NTDs mice was decreased. Therefore, subsequent studies were performed on cell function by transfecting anti-222 to evaluate expression of miR-222-3p. Various types of miR with GFP were successfully transfected into HT-22 cells and the transfected efficacy is shown in . qRT-PCR showed that the expression level of miR-222-3p was significantly decreased compared with the anti-NC (miR inhibitor negative control) and the control groups (con, no transfection group) (). CCK-8 assay showed that low expression of miR-222-3p suppressed the viability of HT-22 cells (). Western blotting confirmed that knockdown of miR-222-3p significantly inhibited the expression level of proliferating cell nuclear antigen (PCNA), a marker of proliferation ().
Figure 2. Inhibition of miR-222-3p affects the proliferation of HT-22 cells. (a) Transfection efficiency was detected by flow cytometry. (b) The mRNA level of miR-222-3p was analyzed by qRT-PCR in con (blank control group), anti-NC and anti-222 groups. The U6 was used as a control. (c) The proliferation of con, anti-NC and anti-222 groups cells were measured by CCK8. (d) The PCNA protein level in con, anti-NC and anti-222 groups were evaluated via western blot and the expression of β-tubulin was used for confirming equal loading
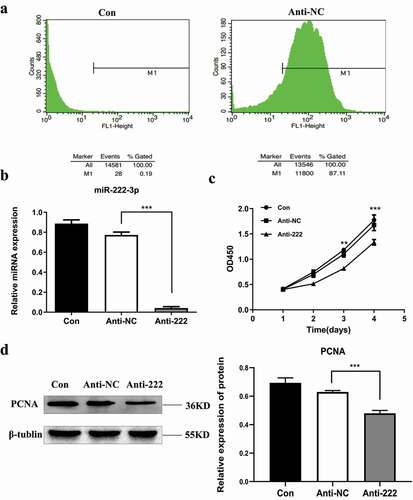
3.3. Knockdown of miR-222-3p promotes HT-22 cells apoptosis in vitro
To further investigate the mechanism of miR-222-3p on HT-22 cell proliferation, cell cycle and apoptosis, flow cytometry was used to show that the percentage of early apoptotic and late apoptotic cells were increased (the second quadrant represents late apoptotic cells, and the fourth quadrant represents early apoptotic cells) in the anti-222 transfected group compared with anti-NC and blank groups (). The expression of apoptosis-related proteins showed that cleaved caspase-3 was dramatically elevated in HT-22 cells post transfection with anti-222 (). AO/EB staining () further confirmed that most cells in the control and anti-NC group have green fluorescence representing normal, viable and non-apoptotic cells with intact structure. However, anti-222 transfected cells showed orange and red fluorescence which increased significantly, representing apoptotic cells with condensed or fragmented chromatin. These results indicated that knockdown of miR-222-3p significantly promoted apoptosis in HT-22 cells. Flow cytometry assay showed that knockdown of miR-222-3p significantly increased the proportion of cells in the G1 phase, while the proportion of cells in the S phase were decreased between two groups (). These results indicated that low expression of miR-222-3p affects cell proliferation by promoting apoptosis and cell cycle distribution of HT-22 cells.
Figure 3. Inhibition of miR-222-3p affects the Cell function of HT-22 cells. (a) The cell apoptosis in con, anti-NC and anti-222 groups were analyzed by flow cytometry. (b) The Cleaved Caspase-3 protein level in con, anti-NC and anti-222 groups were evaluated via western blot and the expression of β-tubulin was used for confirming equal loading. (c) Fluorescence results of AO-EB double staining HT-22 cell nuclei. The arrows indicate apoptotic cells. (d) The cell cycle distribution in con, anti-NC and anti-222 groups was analyzed by flow cytometry. (e) The migration ability in con, anti-NC and anti-222 groups was analyzed by cell scratch experiment
3.4. Knockdown of miR-222-3p promotes migration of HT-22 cells in vitro
In view of the facts that occurrence of NTDs is related to neuronal migration, we further evaluated the effect of miR-222-3p on migration of HT-22 cells. The results showed that knockdown of miR-222-3p significantly inhibited migration of HT-22 cells ().
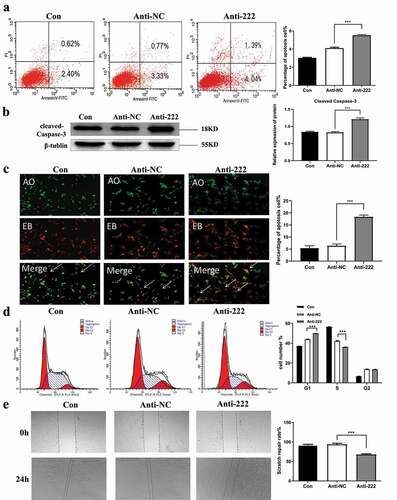
3.5. Ddit4 is a direct target of miR-222-3p in HT-22 cells
Real-time qPCR showed that mRNA expression of Ddit4 was up-regulated in RA treated group compared with the normal group (), which was consistent with the sequencing. In order to determine whether miR-222-3p endogenously regulates Ddit4, miR-222-3p mimic (mim-222) and anti-222 were transfected into the cells to observe the expression of Ddit4. showed that the expression of miR-222-3p in HT22 cells could be well increased after transfection with mim-222. RT-qPCR showed that the mRNA expression level of Ddit4 was significantly increased after transfection with anti-222 (), while it was significantly decreased after transfection with mim-222 (). Similarly, WB showed that Ddit4 protein level was significantly increased in HT-22 cells after transfection with anti-222 while significantly decreased after transfection with mim-222 (, f). Luciferase activity assay was used for validation of interaction between miR-222-3p and target gene Ddit4. In the empty vector group without Ddit4 expression (Ddit4-PC), mir-222 overexpression had no effect on Luc/R-LuC compared with miRNA simulated negative control. However, in the wild-type vector group (Ddit4-WT) with normal expression of Ddit4, the overexpression of miR-222-3p significantly decreased the luciferase activity of HT-22 cells transfected with GP-mirglo-DDIT4-WT. The deletion mutation of Ddit4 binding site (Ddit4-MUT) eliminated the effect of miR-222-3p on luciferase activity (). At the same time, the seed sequence, mutation sequence and complementary sequence diagram of Mir-222 and DDIT4 3ʹUTR are also shown in .
Figure 4. Ddit4 is a direct target of miR-222-3p in HT-22 cells. (a) qRT-PCR was used to detect the mRNA level of Ddit4 gene in normal and malformed brain tissue at E8.5-E10.5. (b) The mRNA level of Ddit4 gene was analyzed by qRT-PCR in con, anti-NC and anti-222 groups. The β-actin gene was used as a control. (c) The mRNA level of miR-222-3p was analyzed by qRT-PCR in con, mim-NC and mim-222 groups. The U6 was used as a control. (d) The mRNA level of Ddit4 gene was analyzed by qRT-PCR in con, mim-NC and mim-222 groups. The β-actin gene was used as a control. (e) The Ddit4 protein level in con, anti-NC and anti-222 and mim-222 groups was evaluated via western blot and the expression of β-tubulin was used for confirming equal loading. (f) Histogram analysis of graph E. (g) The fluorescence values of Ddit4 PC, Ddit4 WT and Ddit4 MUT combined with miR-222-3p mimis or NC were determined by double luciferase assay. (i) The seed sequence, mutated sequence, and complementary sequence of mir-222 and ddit4 3` UTR
These results strongly suggested that miR-222-3p regulates the expression of Ddit4 by directly binding to the Ddit4 3ʹUTR sequence in HT-22 cells. These results indicated that Ddit4 is a potential target gene of miR-222-3p and the expression of Ddit4 was regulated by miR-222-3p.
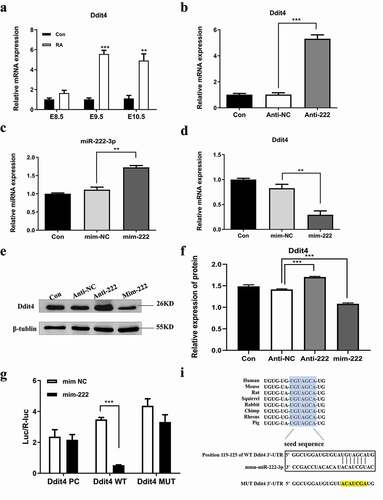
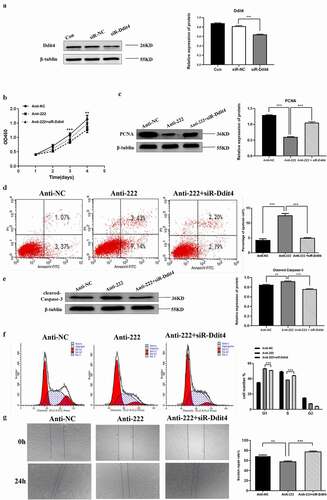
3.6. Ddit4 reverses the effect of miR-222-3p on HT-22 cells
Previous studies have shown that Ddit4 is associated with cell apoptosis and cell proliferation [Citation46]. Subsequently, Ddit4 was knocked down by a small interfering RNA (siRNA). As shown in , a significant reduction in Ddit4 was achieved in siR-Ddit4 compared with siRNA control (siR-NC). The CCK8 assay showed that cell proliferation capacity was reduced by miR-222 inhibitor, which was reversed by co-transfection of siR-Ddit4 (). Identically, PCNA was significantly decreased in HT-22 cells after transfected with miR-222 inhibitor and expression was reversed after Ddit4 was silenced (). Similarly, knockdown of Ddit4 protected anti-222-induced apoptosis in HT-22 cells (, e)). In addition, down-regulation of Ddit4 ablated anti-222-mediated cell cycle arrest in HT-22 cells (). Besides, low expression of Ddit4 also weakened the inhibitory effect of miR-222 on migration of HT-22 cells (). These results indicated that down-regulation of miR-222-3p in HT-22 cells affects proliferation, apoptosis and migration of HT-22 cells by binding and promoting expression of the target gene Ddit4 of miR-222-3p.
Figure 5. Ddit4 reverses the effect of miR-222-3p on HT-22 cells function. (a) The Ddit4 protein level in Con, siR-NC and siR-Ddit4 groups was evaluated via western blot and the expression of β-tubulin was used for confirming equal loading.(b)The proliferation of anti-NC, anti-222 and anti-222 + siR-Ddit4 groups cells were measured by CCK8. (c) The PCNA protein level in anti-NC, anti-222 and anti-222 + siR-Ddit4 groups was evaluated via western blot and the expression of β-tubulin was used for confirming equal loading. (d) The cell apoptosis in anti-NC, anti-222 and anti-222 + siR-Ddit4 was analyzed by flow cytometry. (e) The Cleaved Caspase-3 protein level in anti-NC, anti-222 and anti-222 + siR-Ddit4groups was evaluated via western blot and the expression of β-tubulin was used for confirming equal loading. (f)The cell cycle distribution in anti-NC, anti-222 and anti-222 + siR-Ddit4 groups was analyzed by flow cytometry. (g) The migration ability in anti-NC, anti-222 and anti-222 + siR-Ddit4 groups was analyzed by cell scratch experiment
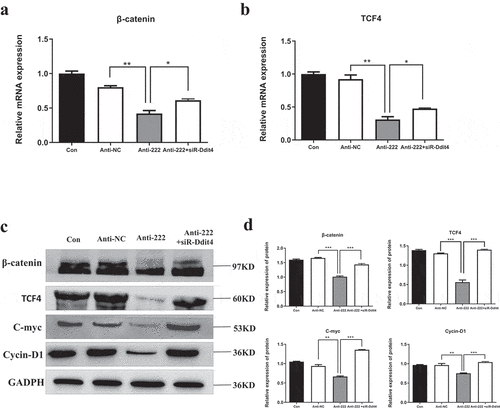
3.7 miR-222-3p is involved in regulation of Wnt signaling pathway
Ddit4 regulatesvWnt/β-catenin signaling pathway and affects the development of the dorsal-ventral axis of the embryo [Citation50,Citation51]. The present study confirmed that Ddit4 was a direct target of miR-222-3p. Wnt signaling pathway and its downstream indicators were investigated at mRNA and protein levels (). RT-qPCR results showed that expression of β-catenin and TCF4 was significantly decreased in HT-22 cells after transfected with anti-222 (), and was greatly reversed by co-transfection of siR-Ddit4 (). WB showed that the protein levels of β-catenin, TCF4, c-Myc and cyclin D1 were significantly decreased in HT-22 cells after transfected with anti-222 compared with anti-NC (). This effect was enormously reversed by co-transfection of siR-Ddit4 (). These results suggested that down-regulation of miR-222-3p could restrain Wnt signaling pathway through Ddit4 inhibition.
Figure 6. Inhibition of miR-222-3p regulates Wnt signaling pathway. (a) The mRNA level of β-catenin gene was analyzed by qRT-PCR in con, anti-NC, anti-222 and anti-222+ siR-Ddit4 groups. The β-actin gene was used as a control. (b) The mRNA level of TCF4 gene was analyzed by qRT-PCR in con, anti-NC, anti-222 and anti-222+ siR-Ddit4 groups. The β-actin gene was used as a control. (c) The β-catenin, TCF4, C-myc, Cycin D1 protein level in con, anti-NC, anti-222 and anti-222+ siR-Ddit4 groups was evaluated via western blot and the expression of GADPH was used for confirming equal loading
4. Discussion
In the present study, we demonstrated that miR-222-3p was significantly down-regulated in NTDs mouse embryonic brain tissues. At the same time, an important link was observed between miR-222-3p and Ddit4 that we found miR-222-3p is negatively correlated with Ddit4 in HT-22 cells. Previous studies have demonstrated that aberrant expression of Ddit4 is associated with the occurrence progression and recurrence of various cancers, such as gastric and ovarian cancer, but not with neural tube malformation [Citation52–54]. Collectively, these data supported the present study that Ddit4 is a direct target gene of miR‑222‑3p in NTDs.
Ddit4 (DNA damage-inducing transcript 4) belongs to the DDIT (DNA Damage Inducible Transcript) or GADD (Growth Arrest DNA Damage) protein family. This family member is involved in both physiological and pathological processes such as growth, development, DNA damage repair, apoptosis, inflammatory response, stress and tumor formation [Citation55–57]. The Ddit4 proteins in human and mouse are conserved at its C-terminus, and both contain RTP801-C protein domain with a sequence identity of 61%, and its homologous genes scylla and charybde are involved in the formation of the head of fruit fly [Citation58]. During mouse embryonic development, Ddit4 is specifically expressed in the ectodermal axis and participates in the development and migration of cerebral cortical neurons during early embryonic development [Citation22]. Studies have shown that Ddit4 gene is associated with a variety of neurodegenerative diseases, and its overexpression promotes apoptosis and inhibits cell proliferation [Citation59,Citation60].
Therefore, it may be a new breakthrough to understand the pathogenesis of NTDs through the study of miR-222-3p targeting Ddit4. Slawny and O’Shea [Citation61] showed that Ddit4 regulates the Wnt/β-catenin signaling pathway, the imbalance of which leads to abnormal neural embryonic formation during embryonic development, and thus cause abnormal closure of neural tube [Citation62]. To investigate whether Ddit4 was implicated in miR-222-mediated NTDs progression, HT-22 cells were transfected with anti-NC, anti-222 and co-transfection of anti-222 and siR-Ddit4 simultaneously. The decreased expression of β-catenin, TCF4, c-Myc and Cyclin D1 in HT-22 cells transfected with miR-222 inhibitor was reversed by co-transfection of siR-Ddit4, suggesting that Ddit4 was involved in the regulation of Wnt/β-catenin pathway. Therefore, the present study has concluded an association between miR-222-3p and Wnt/β-catenin pathway, and the results suggested that knockdown of miR-222-3p suppressed the expression of Wnt signaling-related genes. Furthermore, the present study indicated that miR-222-3p inhibited HT-22 cell progression by targeting Ddit4 and further regulates the Wnt/β-catenin signaling pathway.
5. Conclusion
In conclusion, this study suggested a crucial role of miR-222-3p in the occurrence of NTDs, indicating a potential clinical value of miR-222-3p as an effective biomarker for diagnosis and prognosis of NTDs. In addition, our study demonstrated that miR-222-3p exerted its role by targeting Ddit4 through Wnt/β-catenin signaling pathway inhibition. Therefore, miR-222-3p may be a novel therapeutic target for the treatment of NTDs.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Zhu J, Li X, Wang Y, et al. Prevalence of neural tube defect pregnancies in China and the impact of gestational age of the births from 2006 to 2008: a hospital-based study[J]. Journal of Maternal-Fetal and Neonatal Medicine. 2012;25(9):1730–1734.
- Greene NDE, Stanier P, Copp AJ. Genetics of human neural tube defects[J]. Hum Mol Genet. 2009;18(R2):R113–R129.
- Zaganjor I, Sekkarie A, Tsang BL, et al. Describing the Prevalence of Neural Tube Defects Worldwide: a Systematic Literature Review[J]. PLOS ONE. 2016;11(4):e0151586.
- Ministry of Health of the People’s Republic of China. Report on prevention and treatment of birth defects in China[J]. China Practical medicine 2012; 19(20): 3–5
- Li X, Zhu J, Wang Y, et al. Geographic and urban–rural disparities in the total prevalence of neural tube defects and their subtypes during 2006–2008 in China: a study using the hospital-based birth defects surveillance system[J]. BMC Public Health. 2013;13(1): 161-161. https://doi.org/10.1186/1471-2458-13-161.
- Copp AJ. Greene N D E. Genetics and development of neural tube defects[J]. J Pathol. 2010;220(2):217–230.
- Nicholas DE. Greene, Andrew J Copp. Neural Tube Defects[J]. Annual Review of Neuroscience. 2014;37(1):221–242.
- Liu W, Wang K, Lv X, et al. Up-regulation of RNA binding proteins contributes to folate deficiency-induced neural crest cells dysfunction[J]. Int J Biol Sci. 2020;16(1):85–98.
- Yi K, Han D, Hengxing Z, et al. Epidemiology of worldwide spinal cord injury: a literature review[J]. J Neurorestoratol. 2017;6:1–9.
- Li X, Zhou W, Li X, et al. SOX19b regulates the premature neuronal differentiation of neural stem cells through EZH2-mediated histone methylation in neural tube development of zebrafish[J]. Stem Cell Res Ther. 2019;10(1):389.
- Wallingford JB, Niswander LA, Shaw GM, et al. The continuing challenge of understanding, preventing, and treating neural tube defects[J]. Science. 2013;339(6123): 1222002-1222002. https://doi.org/10.1126/science.1222002.
- Blom HJ, Shaw GM, Den Heijer M, et al. Neural tube defects and folate: case far from closed [J]. Nat Rev Neurosci. 2006;7(9):724–731.
- Murdoch JN, Damrau C, Paudyal A, et al. Genetic interactions between planar cell polarity genes cause diverse neural tube defects in mice. Dis. Model. 2014;7(17): 1153–1163
- Wilde JJ, Petersen JR, Niswander L. Genetic, Epigenetic, and Environmental Contributions to Neural Tube Closure[J]. Annu Rev Genet. 2014;48(1):583–611.
- Wang L, Ren A, Tian T, et al. Whole-Exome Sequencing Identifies Damaging de novo Variants in Anencephalic Cases[J]. Front Neurosci. 2019;13(29):1285.
- Pennimpede T, Cameron DA, Maclean GA, et al. The role of CYP26 enzymes in defining appropriate retinoic acid exposure during embryogenesis[J]. Birth Defects Research Part A Clinical & Molecular Teratology. 2010;88(10):883–894.
- Piersma AH, Pennings J, Tonk ECM. An adverse outcome pathway framework for neural tube and axial defects mediated by modulation of retinoic acid homeostasis[J]. Reprod Toxicol. 2015;56:15.
- Lammer E, Kamp HG, Hisgen V, et al. Development of a flow-through system for the fish embryo toxicity test (FET) with the zebrafish (Danio rerio)[J]. Toxicol in Vitro. 2009;23(7):0–1442.
- Lammer EJ, Chen DT, Hoar RM, et al. Retinoic acid embryopathy[j]. N Engl J Med. 1985;313(14):837–841.
- Geelen JA Hypervitaminosis A induced teratogenesis[J]. CRC Crit Rev Toxicol. 1979;6(4):351–375
- Rosa F. Teratogenicity of isotretinoin[J]. Lancet. 1983;322(8348):513.
- Gongal PA, March LD, Holly VL, et al. Hmx4 regulates Sonic hedgehog signaling through control of retinoic acid synthesis during forebrain patterning[J]. Dev Biol. 2011;355(1):55–64.
- Chen H, Yang Q, Chen K, et al. Integrated microRNA and transcriptome profiling reveals a miRNA-mediated regulatory network of embryo abortion under calcium deficiency in peanut (Arachis hypogaeaL.)[J]. BMC Genomics. 2019;20(1): 5770
- Yakovlev AF. The Role of miRNA in Differentiation, Cell Proliferation, and Pathogenesis of Poultry Diseases[J]. Russian Journal of Developmental Biology. 2019;50(3):102–112
- Majzoub RE, Fayyad-kazan M, and Dine ANE, et al. A thiosemicarbazone derivative induces triple negative breast cancer cell apoptosis: possible role of miRNA-125a-5p and miRNA-181a-5p[J]. Genes Genomics. 2019;41(12):1431-1443.;().
- Ravichandran S, Ragupathy R, Edwards T, et al. MicroRNA-guided regulation of heat stress response in wheat[J]. BMC Genomics. 2019;20(1):s12864-019-5799-6.
- Fineberg SK, Kosik KS, Davidson BL. MicroRNAs Potentiate Neural Development[J]. Neuron. 2009;64(3):303–309.
- Wang F, Xu C, Reece EA, et al. Protein kinase C-alpha suppresses autophagy and induces neural tube defects via miR-129-2 in diabetic pregnancy[J]. Nat Commun. 2017;8:15182.
- Wang LL, Zhang Z, Li Q, et al. Ethanol exposure induces differential microRNA and target gene expression and teratogenic effects which can be suppressed by folic acid supplementation[J]. Hum Reprod. 2009;24(3):562.
- Cao X, Pfaff SL, Gage FH. A functional study of miR-124 in the developing neural tube[J]. Genes Dev. 2007;21(5):531–536.
- Maller Schulman R, Liang B, Stahlhut XC, et al. The let-7 microRNA target gene, Mlin41/Trim71 is required for mouse embryonic survival and neural tube closure[J]. Cell Cycle. 2008;7(24):3935–3942.
- Debernardi S, Skoulakis S, Molloy G, et al. MicroRNA miR-181a correlates with morphological sub-class of acute myeloid leukaemia and the expression of its target genes in global genome-wide analysis[J]. Leukemia. 2007;21(5):116–119.
- Tsuchiya, Y. MicroRNA Regulates the Expression of Human Cytochrome P450 1B1[J]. Cancer Res. 2006;66(18):9090–9098.
- Chen W-X, Hu Q, Qiu M-T, et al. miR-221/222: promising biomarkers for breast cancer[J]. Tumor Biol. 2013;34(3):1361–1370.
- Takizawa T. The miR-221/222 cluster, miR-10b and miR-92a are highly upregulated in metastatic minimally invasive follicular thyroid carcinoma[J]. Int J Oncol. 2013;42(6):1858–1868.
- Pu P. miR-221/222 is the regulator of Cx43 expression in human glioblastoma cells[J]. Oncol Rep. 2012;27(5):1652.
- Lee D, Han S, Woo S, et al. Enhanced expression and purification of inositol 1,4,5-trisphosphate 3-kinase A through use of the pCold1-GST vector and a C-terminal hexahistidine tag in Escherichia coli[J]. Protein Expr Purif. 2014;97:72–80.
- Raanan G, Anna G, Tamar S, et al. Non-cell autonomous and non-catalytic activities of ATX in the developing brain[J]. Front Neurosci. 2015;9:53.
- Copp AJ, Stanier P, Greene ND. Neural tube defects: recent advances, unsolved questions, and controversies[J]. Lancet Neurol. 2013;12(8):799–810.
- Conaco C, Otto S, Han -J-J, et al. Reciprocal actions of REST and a microRNA promote neuronal identity[J]. Proc Natl Acad Sci USA. 2006;103(7):2422–2427.
- Chen W-X, HU Q, Qiu M-T, et al. MiR-221/222: promising biomarkers for breast cancer[J]. Tumour Biol. 2013;34(1):1361–1370.
- Kancherla V, Black RE. Historical perspective on folic acid and challenges in estimating global prevalence of neural tube defects[J]. Ann N Y Acad Sci. 2018;1414(1):20–30.
- Liu S, Guo Y, Yuan Q, et al. Melatonin prevents neural tube defects in the offspring of diabetic pregnancy[J]. J Pineal Res. 2015;59(4):508–517.
- Yang YF, Wang F, Xiao JJ, et al. MiR-222 overexpression promotes proliferation of human hepatocellular carcinoma HepG2 cells by downregulating p27.[J]. International Journal of Clinical & Experimental Medicine. 2014;7(4): 893–902
- Terasawa K, Ichimura A, Sato F, et al. Sustained activation of ERK1/2 by NGF induces microRNA-221 and 222 in PC12 cells[J]. FEBS J. 2009;276(12):3269–3276.
- Chen R, Wang B, Chen L, et al. DNA damage-inducible transcript 4 (DDIT4) mediates methamphetamine-induced autophagy and apoptosis through mTOR signaling pathway in cardiomyocytes[J]. Toxicol Appl Pharmacol. 2016;295:1–11.
- Zhang J, Li R, He Q, et al. All- trans -retinoic acid alters Smads expression in embryonic neural tissue of mice[J]. J Appl Toxicol. 2009;29(4):364–366.
- Yu J, Wang L, and Pei P, et al. Reduced H3K27me3 leads to abnormal Hox gene expression in neural tube defects[J]. Epigenetics Chromatin 2019;12:76.
- Juan Z, Lihong Y, and Juan Y, et al. Alteration of the microRNA expression profile and identification of miRNA/mRNA negative regulation pairs in neural tube defects[J]. Acta Biochim Biophys Sin (Shanghai) 2019 ;51 (7):7.
- Shoshani T, Faerman A, Mett I, et al. Identification of a Novel Hypoxia-Inducible Factor 1-Responsive Gene, RTP801, Involved in Apoptosis[J]. Mol Cell Biol. 2002;22(7):2283–2293.
- Qiang F, Xia Z, Ling L, et al. The Stress-Response Gene redd1 Regulates Dorsoventral Patterning by Antagonizing Wnt/β-catenin Activity in Zebrafish[J]. PLoS ONE. 2012;7(12):e52674.
- Pinto JA, Rolfo C, Raez LE, et al. In silico evaluation of DNA Damage Inducible Transcript 4 gene (DDIT4) as prognostic biomarker in several malignancies[J]. Sci Rep. 2017;7(1):1526.
- Feng D, Lina S Y C, et al. DDIT4 promotes gastric cancer proliferation and tumorigenesis through the p53 and MAPK pathways[J]. Cancer Commun. 2018;38(1):45.
- Fu X, Li Y, Alvero A, et al. MicroRNA-222-3p/GNAI2/AKT axis inhibits epithelial ovarian cancer cell growth and associates with good overall survival[J]. Oncotarget. 2016;7(49):80633.
- Brugarolas J. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex[J]. Genes Dev. 2004;18(23):2893–2904.
- Deyoung MP, Horak P, Sofer A, et al. Hypoxia regulates TSC1/2 mTOR signaling and tumor suppression through REDD1-mediated 14 3 3 shuttling[J]. Genes Dev. 2008;22(2):239–251.
- Miyazaki M, Esser KA. REDD2 is enriched in skeletal muscle and inhibits mTOR signaling in response to leucine and stretch[J]. AJP Cell Physiology. 2009;296(3):C583–92
- Scuderi A, Simin K, Kazuko SG, et al. scylla and charybde, homologues of the human apoptotic gene RTP801, are required for head involution in Drosophila[J]. Dev Biol. 2006;291(1):0–122.
- Shoshani T, Faerman A, Mett I, et al. Identification of a novel hypoxia-inducible factor 1-responsive gene, RTP801, Involved in Apoptosis[J]. Mol Cell Biol. 2002;22(7):2283–2293.
- Sofer A, Lei K, Johannessen CM, et al. Regulation of mTOR and cell growth in response to energy stress by REDD1[J]. Mol Cell Biol. 2005;25(14):5834–5845.
- Slawny NA, O’Shea KS. Dynamic changes in Wnt signaling are required for neuronal differentiation of mouse embryonic stem cells[J]. Molecular & Cellular Neuroence. 2011;48(3):205–216
- Bennett GD, An J, Craig JC, et al. Neurulation abnormalities secondary to altered gene expression in neural tube defect susceptible splotch embryos[J]. Teratology. 1998;57(1):17–29.
