ABSTRACT
Alzheimer’s disease (AD) is a progressive neuro-degenerative disease characterized by dementia. MicroRNAs (miRNAs) are involved in many diseases, including AD. MiR-132-3p has been identified to be downregulated in AD. In this study, we explored the effects of miR-132-3p on neuron apoptosis and impairments of learning and memory abilities. Aβ1-42-stimulated SH-SY5Y cells were used as in vitro models of AD. An AD-like homocysteine (Hcy) rat model was established to evaluate the effects of miR-132-3p on AD pathogenesis in vivo. RIP, RNA pull down and luciferase reporter assays were conducted to investigate the relationship between miR-132-3p and its downstream target genes. The viability and apoptosis of SH-SY5Y cells were measured by CCK-8 and TUNEL assays. The rat spatial learning and memory abilities were accessed using Morris water maze test. Results indicated that miR-132-3p was downregulated in SH-SY5Y cells after Aβ1-42 treatment and promoted cell apoptosis. Mechanistically, miR-132-3p targeted heterogeneous nuclear ribonucleoprotein U (HNRNPU). HNRNPU acted as an RNA binding protein (RBP) to regulate the mRNA stability of β-site amyloid precursor protein cleaving enzyme 1 (BACE1). Overexpression of HNRNPU or BACE1 reversed the effects of miR-132-3p overexpression on the viability and apoptosis of Aβ1-42-treated SH-SY5Y cells. In vivo experiments revealed the downregulation of miR-132-3p in the hippocampus of Hcy-treated rats. MiR-132-3p suppressed levels of apoptotic genes in hippocampus and reduced impairments of learning and memory abilities in Hcy-treated rats. In conclusion, miR-132-3p reduces apoptosis of SH-SY5Y cells and alleviates impairments of learning and memory abilities in AD rats by modulating the HNRNPU/BACE1 axis.
KEYWORDS:
Introduction
Alzheimer’s disease (AD), a prevalent form of dementia, is a neuro-degenerative disease caused by the degenerative confusion of the central nervous system [Citation1,Citation2]. AD usually occurs in older adults with memory loss and behavioral changes [Citation3,Citation4]. Early diagnosis and treatment of AD require further improvement [Citation5,Citation6]. It is imperative to search novel biomarkers and therapeutic agents for AD.
MicroRNAs (miRNAs) are endogenous small non-coding RNAs at a length of 18–25 nucleotides, which are able to regulate gene expression at the post-transcriptional level by binding with 3ʹ-untranslated region (UTR) of mRNAs [Citation7–9]. In recent years, miRNAs have received increasing attention and have been documented to act as important regulators in AD. For example, miR-204-3p is found to be downregulated in the hippocampus and plasma of APP/PS1 mice and ameliorates memory and synaptic deficits in APP/PS1 mice by downregulating Nox4 [Citation10]. MiR-34 c is upregulated in serum of patients with amnestic mild cognitive impairment and regulates synaptic and memory deficits by downregulating SYT1 via the ROS-JNK-p53 pathway [Citation11]. MiR-181a is highly expressed in the hippocampus of an AD mouse model and its expression is negatively correlated with plasticity-related protein levels. MiR-181 inhibition alleviates memory deficits by modulating AMPA receptors [Citation12].
Previous investigations have revealed the aberrant expression of miR-132-3p in many neurodegenerative diseases [Citation13,Citation14]. MiR-132-3p is dysregulated in central nervous system (CNS) disorders [Citation15]. MiR-132-3p overexpression exerts protective effect on an AD model in vitro, which can be reversed by accumulation of BACE1-AS [Citation16]. The aberrant expression of miR-132-3p is identified in the Parkinson’s disease, and miR-132-3p binds to the 3ʹUTR of SNCA mRNA, which is critically implicated in the pathophysiology of Parkinson’s disease [Citation17]. However, the downstream molecular mechanism of miR-132-3p on neuron apoptosis and impairments of learning and memory abilities in AD remains unknown.
In our study, the in vitro (Aβ1-42-stimulated SH-SY5Y cells) and in vivo models (Hcy-treated rats) of AD were established. The aim of this study was to explore the underlying mechanism of miR-132-3p in AD. This study may provide a promising therapeutic target for AD treatment.
Materials and methods
Rat model
All procedures were approved by the ethics committee of Xinjiang Military General Hospital. Fifty adult male Sprague-Dawley rats (220–250 g) were supplied by Vital River Co. Ltd. (Beijing, China). All rats were kept in cages accessible to food and water under a light/dark cycle of 12/12 h, in a 55 ± 15% humidity, and a constant room temperature (20–24°C). These rats were randomly divided into the following 5 groups: sham, Hcy, Hcy+AAV1-NC and Hcy+AAV1-miR-132-3p, Hcy+AAV1-miR-132-3p+AAV-BACE1 (n = 10 per group). Adeno associated virus (AAV) particles (serotype: AAV1; titer: 1.0 × 109) (Vigene Biosciences, shanghai, China) were injected into the vena caudalis of rats. Homocysteine (Hcy) was dissolved in saline with 0.9% NaCl to 400 μg/mL. Rats were treated with Hcy (400 μg/kg) by tail vein injection or normal saline (Control) from 9:00 am to 2:00 pm each day for 14 consecutive days [Citation18,Citation19]. Rats in the sham group were injected with the same dose of saline.
Cell culture
Human neuroblastoma SH-SY5Y cells provided by Shanghai Institute for Cell Research (shanghai, China) were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, USA) with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (Sigma-Aldrich, USA) in a humidified atmosphere with 5% CO2 at 37°C. SH-SY5Y cells were treated with Aβ1-42 oligomers (0, 2.5, 5, 10 μM, Sigma-Aldrich) to mimic the in vitro model of AD. For some assays, cells were treated with actinomycin D (100 μg/mL) followed by detection of the stability of BACE1.
Transfection
MiR-132-3p mimics were used to overexpress miR-132-3p, with NC mimics as the negative control. The short hairpin RNAs targeting HNRNPU (sh-HNRNPU) were utilized to knockdown HNRNPU, with sh-NC as the negative control. The coding region of HNRNPU or BACE1 was constructed into pcDNA3.1 plasmid to overexpress HNRNPU or BACE1, and empty pcDNA3.1 was the negative control. All plasmids were purchased from GeneChem (Shanghai, China) and were transfected into SH-SY5Y cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA).
Cell viability assay
SH-SY5Y cells after indicated transfection were plated in 96-well plates. Then, Aβ1-42 at different dozes (0, 2.5, 5, 10 μL) was used to treat SH-SY5Y cells for 24 h. Each well was added with 3-(4,5)-dimethylthiahiazo (-z-y1)-3,5-di- phenytetrazoliumromide (MTT) solution (10 μL, 5 mg/mL) for 4 h of incubation. After removing culture medium, 100 μL of DMSO was used for crystal dissolution. A microplate reader (Bio-Tek Instruments, USA) was used to examine optical density at absorbance of 490 nm.
Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay
Cell apoptosis was assessed with the In Situ Cell Death Detection kit (Roche Applied Science, Germany). Paraformaldehyde (4%) was used to fix the SH-SY5Y cells and 0.2% Triton X-100 was used for permeabilization. Next, the TUNEL reaction mixture (50 μL) was used to dye the cells which were then counterstained with 4ʹ,6-diamidino-2-phenylindole (DAPI). The TUNEL staining positive cells were observed under the fluorescence microscope (Leica, Wetzlar, Germany).
Reverse transcription quantitative polymerase chain reaction (RT-qPCR) assay
Total RNAs were isolated from SH-SY5Y cells or hippocampus by TRIzol reagent (Invitrogen). Mice in each group were anesthetized with sodium pentobarbital, and brain sections were dissected and homogenized in TRIzol reagent (15,596,018, Thermo Fisher, Shanghai, China) on ice for RNA extraction. Subsequently, the collected RNAs were reverse transcribed into cDNA with a PrimeScript® miRNA cDNA Synthesis Kit (Takara, Dalian, China). The RT-qPCR was performed using SYBR® Premix Ex TaqTM II (Takara) on an Applied Biosystems 7500 Real-Time PCR System. GAPDH and U6 served as endogenous controls. The relative expression level of genes was assessed with the 2−ΔΔCt method. The primer sequences used for PCR were shown in .
Table 1. The primer sequences used for PCR
Western blot
Brains were dissected on ice. Hippocampus of rats were obtained, and sonication was applied to homogenize the rat hippocampus in lysis buffer. Subsequently, after centrifuging the homogenates for ten min at 12,000 g, the supernatant was obtained and maintained at −80°C. Proteins from SH-SY5Y cells or hippocampus of rats were extracted by RIPA regent with 100 mg/mL phenylmethyl sulfonyl fluoride and Halt™ Protease Inhibitor Cocktail (#78,430, Thermo Fisher, Shanghai, China), and quantified by the bicinchoninic acid (BCA) protein quantitative kit. Later, the isolation of proteins was performed with 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis. The isolated proteins were transferred onto the polyvinylidene fluoride membranes. Then, the membranes were blocked with 5% defatted milk and incubated with primary antibodies, including anti-Bax (#ab182734, 1/1000), anti-Bcl-2 (#ab32124, 1/1000), anti-Caspase-3 (#ab32351, 1/5000), anti-Cleaved Caspase-3 (#ab2302, 1/500), anti-APP-1 (#ab241616, 1/2000), anti-BACE1 (#ab108394, 1/1000), anti-beta Amyloid 1–42 (#ab180956, 1/1000), anti-HNRNPU (#PA5-109,827, 1/100, Thermo Fisher) at 4°C overnight, and next incubated with horse radish peroxidase conjugated IgG (#ab205718, 1/2000) at room temperature for 1 h. Except Anti-HNRNPU (Thermo Fisher), all the other antibodies were purchased from the Abcam company. The enhanced chemiluminescence detection system (Millipore) was used to examine the protein bands.
Luciferase reporter assay
Luciferase reporter assay was performed according to a previous study [Citation20]. The wild type sequences of HNRNPU 3ʹUTR and the mutated sequences were inserted into pmirGLO vectors for the construction of HNRNPU-WT and HNRNPU-Mut reporters, which were co-transfected with NC mimics or miR-132-3p mimics into SH-SY5Y cells with Lipofectamine 2000. Two days later, a Dual-Luciferase Assay System (Promega, Madison, WI, USA) was used for luciferase activity examination. Relative luciferase activity was defined as the ratio of firefly luciferase activity to renilla luciferase activity.
RNA pull‐down
Biotinylated BACE1 sense (Bio-BACE1 sense) and BACE1 antisense (Bio-BACE1 antisense) probes were supplied by Sangon (Shanghai, China). SH-SY5Y cells were lysed by cell lysis buffer. The probes, cell lysate and Dynabeads M-280 Streptavidin (Invitrogen) were all mixed, and then the eluted proteins were tested by western blot analysis.
RNA immunoprecipitation (RIP)
The EZ‐Magna RIP Kit (Millipore, Billerica, MA) was utilized for RIP assay. After lysing SH-SY5Y cells with RIPA lysis buffer, cell extracts were mixed with RIPA buffer added with magnetic beads conjugated with human anti‐HNRNPU (Abcam). Purified RNA was detected with RT‐qPCR analysis.
Morris water maze (MWM) test
The MWM test was conducted to examine spatial learning and memory abilities. Rats were allowed to swim freely for 1 minute in the pool without the platform. Rats were trained to look for the hidden platform within 1 minute in each trail. If they failed, the rats will be guided to stand on platform for 30 s. The training was done for consecutive 3 days (3 trails/day). The evaluation of spatial memory was performed on day 46. After removing the platform, the escape latency and the time spend in the target quadrant were documented with a Noldus video tracking system (Noldus Information Technology, Holland).
Statistical analysis
The experiment results were presented as the mean values ± standard deviation and analyzed with SPSS 19.0 (SPSS, Chicago, IL). The two-group difference evaluation was assessed by student’s t-test. The one-way analysis of variance followed by Dunnett’s and Tukey’s post-hoc tests as well as two-way analysis of variance were used for difference evaluation among multiple groups. p < 0.05 had statistical significance.
Results
MiR-132-3p promoted cell viability and reduced cell apoptosis in Aβ1-42-treated SH-SY5Y cells
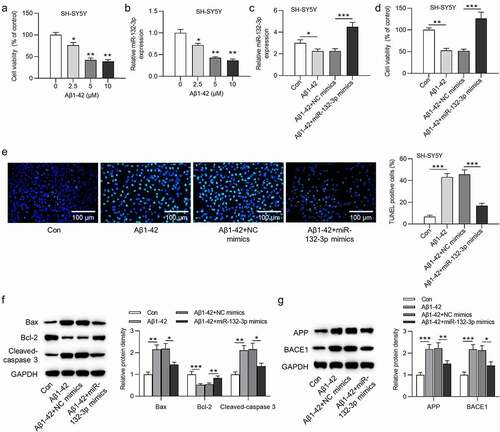
The viability of SH-SY5Y cells was dose-dependently decreased by Aβ1-42 (F(3,8) = 211.187, P < 0.001; )). Reduced expression level of miR-132-3p was observed in SH-SY5Y cells after treatment of Aβ1-42 in a dose-dependent manner (F(3,8) = 98.315, P < 0.001; )). Additionally, the overexpression efficiency of miR-132-3p was verified in ) (F(3,8) = 52.363, P < 0.001). MiR-132-3p mimics reversed the suppressive effect of Aβ1-42 on SH-SY5Y cell viability (F(3,8) = 44.088, P < 0.001; )). Based on results of TUNEL assay, the increased cell apoptosis rate induced by Aβ1-42 was counteracted by overexpressing miR-132-3p (F(3,8) = 401.339, P < 0.001; )). Based on results of western blot analysis, the cell apoptosis related proteins (Bax, Bcl-2, cleaved-caspase 3) and AD related proteins (APP, BACE1) were measured. MiR-132-3p overexpression neutralized the Aβ1-42-induced promotive effects on Bax (F(3,8) = 20.050, P < 0.001), cleaved-caspase 3 (F(3,8) = 22.153, P < 0.001), APP (F(3,8) = 44.153, P < 0.001) and BACE1 (F(3,8) = 34.377, P < 0.001) expression, as well as the inhibitive effect of Aβ1-42 on Bcl-2 (F(3,8) = 42.485, P < 0.001) expression ()).
Figure 1. MiR-132-3p was lowly expressed, and reduced cell apoptosis in SH-SY5Y cells after the treatment of Aβ1-42
MiR-132-3p negatively regulated HNRNPU in SH-SY5Y cells after the treatment of Aβ1-42
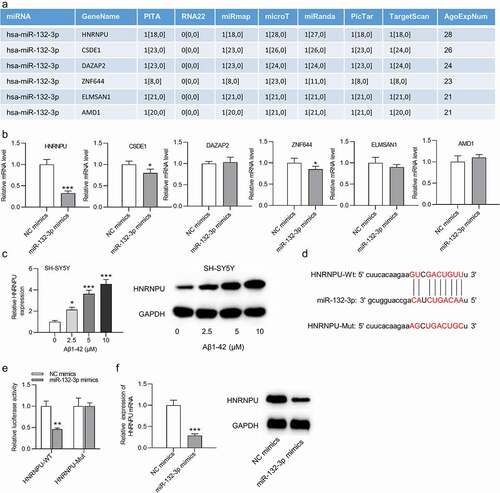
Six mRNAs were predicted to be targeted by miR-132-3p under conditions of CLIP-Data ≥5, pan-Cancer ≥2, program Num ≥ 1, programs: microT, miRanda, miRmap, PITA, PicTar, TargetScan based on the starBase website (http://starbase.sysu.edu.cn/index.php) ()). The expression of these mRNAs was detected after transfection of miR-132-3p mimics, and we discovered that HNRNPU showed the most significant downregulation by overexpressed miR-132-3p (t(4) = 22.523, P < 0.001; )). The HNRNPU mRNA and protein levels were elevated in SH-SY5Y cells after the treatment of Aβ1-42 in a dose-dependent manner (F(3,8) = 319.238, P < 0.001; )). The binding sequences between miR-132-3p and HNRNPU were shown in ). Luciferase reporter assay indicated that overexpressed miR-132-3p reduced the relative luciferase activities of wild-type HNRNPU reporters (t(4) = 7.065, P = 0.002), and no significant change (t(4) = 0.426, P = 0.692) was observed in HNRNPU-Mut group ()). Moreover, the expression of HNRNPU at the mRNA and protein levels was reduced after upregulating miR-132-3p (t(4) = 23.302, P < 0.001; )).
Figure 2. MiR-132-3p negatively regulated HNRNPU in SH-SY5Y cells
Knockdown of RBP HNRNPU reduced the stability of mRNA BACE1 in SH-SY5Y cells
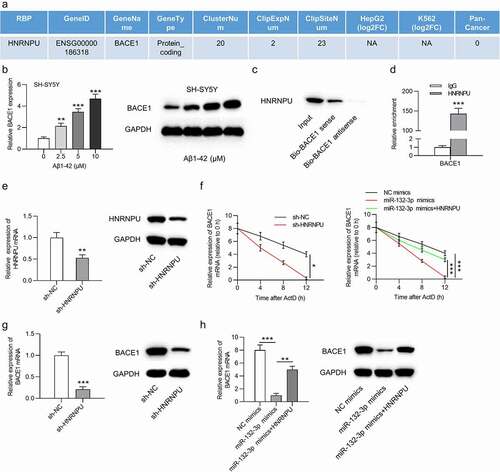
HNRNPU has been identified as an RNA binding protein (RBP) to promote RNA stability. Based on the starBase website, HNRNPU protein was found to bind with BACE1 mRNA ()). The BACE1 mRNA expression and protein levels were elevated in SH-SY5Y cells after the treatment of Aβ1-42 in a dose-dependent manner (F(3,8) = 659.496, P < 0.001; )). The critical role of BACE1 in AD progression has been documented in the previous studies [Citation21–23]. The interaction between HNRNPU and BACE1 was verified by RNA pull down assay followed by western blot analysis and RIP assay. The results of RNA pull down assay revealed the abundant enrichment of HNRNPU in the complex pulled by Bio-BACE1 sense (t(4) = 15.783, P < 0.001; )). RIP assay indicated that BACE1 was enriched in the anti-HNRNPU group, while no enrichment of BACE1 was found in the anti-IgG group ()). HNRNPU expression at the mRNA and protein levels was significantly downregulated in SH-SY5Y cells after transfection of sh-HNRNPU (t(4) = 8.133, P < 0.001; )). Furthermore, the stability of BACE1 mRNA was reduced by silenced HNRNPU (F = 3.107, P = 0.029), and the decreased stability of BACE1 mRNA caused by miR-132-3p mimics (F = 7.465, P < 0.001) was reversed by overexpressing HNRNPU (F = 8.543, P < 0.001) in Aβ1-42-stimulated SH-SY5Y cells after treatment of actinomycin D ()). HNRNPU inhibition decreased the mRNA and protein expression of BACE1 (t(4) = 22.934, P < 0.001), and HNRNPU upregulation rescued (F(2,6) = 83.469, P < 0.001) the inhibited BACE1 mRNA and protein expression caused by overexpressed miR-132-3p ()).
Figure 3. HNRNPU regulated BACE1 expression by acting as an RBP
MiR-132-3p affects cell phenotype by regulating expression of HNRNPU or BACE1 in SH-SY5Y cells
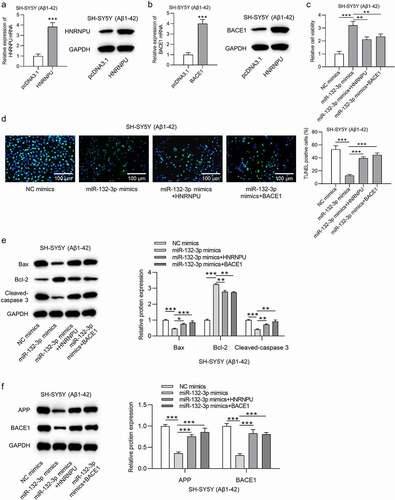
The overexpression efficiency of HNRNPU (t(4) = 17.753, P < 0.001) and BACE1 (t(4) = 13.992, P < 0.001) was verified in ). The promoted cell viability induced by miR-132-3p upregulation was counteracted by overexpressed HNRNPU or BACE1 (F(3,8) = 64.023, P < 0.001; )). Overexpression of HNRNPU or BACE1 rescued the increased apoptosis rate of SH-SY5Y cells caused by miR-132-3p (F(3,8) = 47.076, P < 0.001; )). Upregulation of HNRNPU or BACE1 neutralized the suppressive influence induced by miR-132-3p overexpression on Bax (F(3,8) = 43.809, P < 0.001), cleaved-caspase 3 (F(3,8) = 29.781, P < 0.001), APP (F(3,8) = 42.300, P < 0.001) and BACE1 (F(3,8) = 45.596, P < 0.001) expression, as well as rescued the positive effects of miR-132-3p mimics on Bcl-2 (F(3,8) = 214.125, P < 0.001) expression in SH-SY5Y cells after Aβ1-42 treatment ()).
Figure 4. MiR-132-3p affects cell phenotype by targeting HNRNPU/BACE1
MiR-132-3p alleviated neuron apoptosis and impairments of learning and memory abilities in Hcy-treated rats
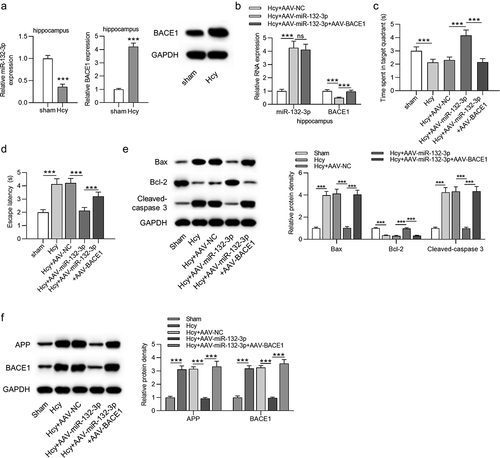
Then, the AD rat model was established by injecting Hcy into Sprague-Dawley rats. Expression of miR-132-3p was downregulated in hippocampus of Hcy-treated rats, while BACE1 expression at the mRNA (t(18) = 23.206, P < 0.001) and protein (t(18) = 40.937, P < 0.001) levels was increased in Hcy-treated hippocampus samples ()). The miR-132-3p overexpression efficiency in hippocampus of Hcy-treated rats was verified, and injection of AAV-BACE1 elevated the levels of BACE1(F(2,27) = 78.504, P < 0.001) but had no influence on miR-132-3p expression ()). Results from MWM tests revealed that rats in the Hcy+AAV-miR-132-3p group spent more time in the target quadrant and had shorter escape latency, compared to rats in the Hcy+ AAV-NC group, and the injection of AAV-BACE1 reversed the effect of miR-132-3p on the learning (F(4,45) = 58.766, P < 0.001) and memory abilities of rats (F(4,45) = 117.531, P < 0.001) ()). Additionally, the levels of apoptosis-related proteins, Bax (F(4,45) = 313.240, P < 0.001), cleaved-caspase 3 (F(4,45) = 412.267, P < 0.001), and AD-related proteins APP (F(4,45) = 194.280, P < 0.001), BACE1 (F(4,45) = 336.562, P < 0.001) were reduced, and Bcl-2 expression (F(4,45) = 560.246, P < 0.001) was increased after upregulating miR-132-3p in Hcy-treated rats, while the injection of AAV-BACE1 reversed the effect caused by miR-132-3p overexpression ()).
Figure 5. MiR-132-3p alleviated neuron apoptosis and impairments of learning and memory abilities in Hcy-treated rats
Discussion
The cognitive impairment and memory decline are typical features of AD, characterized with neuron apoptosis, beta-amyloid (Aβ) plaque accumulation and neurofibrillary tangle formation [Citation24,Citation25]. Aβ, the cleavage product of amyloid precursor protein (APP), can induce neuron apoptosis, and the deposition of Aβ is one of the major pathological changes spotted in AD brain [Citation26,Citation27]. In this study, Aβ1-42 was used to stimulate SH-SY5Y cells for the establishment of the in vitro model of AD. The aberrant expression of miR-132-3p has been identified in various neurodegenerative diseases. However, whether miR-132-3p influences neuron apoptosis and learning and memory abilities in AD is unknown. In this study, miR-132-3p was expressed at a low level in Aβ1-42-stimulated SH-SY5Y cells and improved SH-SY5Y cell apoptosis.
Accumulating evidence has identified that miRNAs bind with the 3ʹ-UTR of mRNAs to regulate the occurrence and progression of diseases [Citation28,Citation29]. In the present study, miR-132-3p was verified to negatively regulate HNRNPU in SH-SY5Y cells after the treatment of Aβ1-42. HNRNPU is an RBP associated with cell self-renewal and survival and can combine with RNAs to regulate their stability. Studies have also reported that HNRNPU is involved in the progression of neurodegenerative diseases. For example, variants of HNRNPU are reported to result in early onset epilepsy and severe intellectual disability [Citation30]. hnRNPU was documented to show significant alterations in Amyotrophic lateral sclerosis [Citation31]. Moreover, HNRNPU was predicted by the starBase website as an RBP for BACE1. BACE1 has been confirmed to be a key factor in AD. MiR-361-3p targets BACE1 to reduce β-amyloid accumulation and ameliorate cognitive deficits in AD [Citation21]. Aβ generation at pathological levels disrupts synaptic functions. Silencing of BACE1 reduces Aβ levels and Aβ-mediated synaptic toxicity, which provides promising targets for AD treatment [Citation22]. In Alzheimer transgenic mice, MMP13 inhibition regulates BACE1 to reverse cognitive decline [Citation23]. It was revealed in our study that HNRNPU acted as an RBP to regulate the mRNA stability of BACE1. Overexpression of HNRNPU or BACE1 reversed miR-132-3p-mediated effects on SH-SY5Y cell viability and apoptosis after Aβ1-42 treatment. Moreover, miR‑132‑3p [Citation32] and BACE1 [Citation33] have been reported to be associated with angiogenesis that is involved in neuronal apoptosis protection. Suppression of the function of Beclin 1 interactome can impair autophagy and promote AD pathology [Citation34]. MiR-132-3p can also repress autophagy and detoxification of reactive oxygen species [Citation35].
Hcy, as a methionine cycle intermediate product, can induce hyperhomocysteinemia that is identified as an independent cause of AD [Citation36,Citation37]. Hcy was used to establish an AD rat model in our work. In vivo experiments indicated that miR-132-3p expression was downregulated in Hcy-treated rats. Furthermore, miR-132-3p alleviated SH-SY5Y cell apoptosis and mitigated impairments of learning and memory abilities of Hcy-treated rats.
In conclusion, we confirmed that miR-132-3p alleviates SH-SY5Y cell apoptosis and ameliorates impairments of learning and memory abilities in an AD rat model by regulating the expression of HNRNPU and BACE1, which may shed new light on AD treatment.
Acknowledgments
We thank all participants for their contributions.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Lane CA, Hardy J, Schott JM. Alzheimer’s disease. Eur J Neurol. 2018 Jan;25(1):59–70.
- Alzheimer's Association. 2016 Alzheimer’s disease facts and figures. Alzheimers Dement. 2016 Apr;12(4):459–509.
- Reitz C. Genetic diagnosis and prognosis of Alzheimer’s disease: challenges and opportunities. Expert Rev Mol Diagn. 2015 Mar;15(3):339–348.
- Mucke L. Neuroscience: alzheimer’s disease. Nature. 2009 Oct 15;461(7266):895–897.
- Mantzavinos V, Alexiou A. Biomarkers for Alzheimer’s Disease Diagnosis. Curr Alzheimer Res. 2017;14(11):1149–1154.
- Mangialasche F, Solomon A, Winblad B, et al. Alzheimer’s disease: clinical trials and drug development. Lancet Neurol. 2010 Jul;9(7):702–716.
- Liu B, Li J, Cairns MJ. Identifying miRNAs, targets and functions. Brief Bioinform. 2014 Jan;15(1):1–19.
- Tafrihi M, Hasheminasab E. MiRNAs: biology, biogenesis, their web-based tools, and databases. Microrna. 2019;8(1):4–27.
- Donato L, Bramanti P, Scimone C, et al. miRNAexpression profile of retinal pigment epithelial cells under oxidative stress conditions. FEBS Open Bio. 2018 Feb;8(2):219–233.
- Tao W, Yu L, Shu S, et al. miR-204-3p/Nox4 mediates memory deficits in a mouse model of Alzheimer’s disease. Mol Ther. 2020 Jan;29(1):396-408.
- Shi Z, Zhang K, Zhou H, et al. Increased miR-34c mediates synaptic deficits by targeting synaptotagmin 1 through ROS-JNK-p53 pathway in Alzheimer’s disease. Aging Cell. 2020 Mar;19(3):e13125.
- Rodriguez-Ortiz CJ, Prieto GA, Martini AC, et al. miR-181a negatively modulates synaptic plasticity in hippocampal cultures and its inhibition rescues memory deficits in a mouse model of Alzheimer’s disease. Aging Cell. 2020 Mar;19(3):e13118.
- Lau P, Bossers K, Janky R, et al. Alteration of the microRNA network during the progression of Alzheimer’s disease. EMBO Mol Med. 2013 Oct;5(10):1613–1634.
- Juźwik CA, Zhang SSD, Zhang Y, et al. microRNA dysregulation in neurodegenerative diseases: a systematic review. Prog Neurobiol. 2019 Nov;182:101664.
- Van Den Berg MMJ, Krauskopf J, Ramaekers JG, et al. Circulating microRNAs as potential biomarkers for psychiatric and neurodegenerative disorders. Prog Neurobiol. 2020;185:101732.
- Ge Y, Song X, Liu J, et al. The combined therapy of berberine treatment with lncRNA BACE1-AS depletion attenuates Aβ(25-35) induced neuronal injury through regulating the expression of miR-132-3p in neuronal cells. Neurochem Res. 2020 Apr;45(4):741–751.
- Schulz J, Takousis P, Wohlers I, et al. Meta-analyses identify differentially expressed micrornas in Parkinson’s disease. Ann Neurol. 2019 Jun;85(6):835–851.
- Wei H, Zhang HL, Wang XC, et al. Direct activation of protein phosphatase 2A (PP2A) by tricyclic sulfonamides ameliorates Alzheimer’s disease pathogenesis in cell and animal models. Neurotherapeutics. 2020 Jul;17(3):1087–1103.
- Zhang CE, Tian Q, Wei W, et al. Homocysteine induces tau phosphorylation by inactivating protein phosphatase 2A in rat hippocampus. Neurobiol Aging. 2008 Nov;29(11):1654–1665.
- Donato L, Scimone C, Rinaldi C, et al. Stargardt phenotype associated with two ELOVL4 promoter variants and ELOVL4 downregulation: new possible perspective to etiopathogenesis? Invest Ophthalmol Vis Sci. 2018 Feb 1;59(2):843–857.
- Huang R, Hu Z, Cao Y, et al. MiR-652-3p inhibition enhances endothelial repair and reduces atherosclerosis by promoting Cyclin D2 expression. EBioMedicine. 2019 Feb;40:685–694.
- Yan R, Fan Q, Zhou J, et al. Inhibiting BACE1 to reverse synaptic dysfunctions in Alzheimer’s disease. Neurosci Biobehav Rev. 2016;65:326–340.
- Zhu BL, Long Y, Luo W, et al. MMP13 inhibition rescues cognitive decline in Alzheimer transgenic mice via BACE1 regulation. Brain. 2019 Jan 1;142(1):176–192.
- Bondi MW, Edmonds EC, Salmon DP. Alzheimer’s disease: past, present, and future. J Int Neuropsychol Soc. 2017 Oct;23(9–10):818–831.
- Serý O, Povová J, Míšek I, et al. Molecular mechanisms of neuropathological changes in Alzheimer’s disease: a review. Folia Neuropathol. 2013;51(1):1–9.
- Cai Z, Hussain MD, Yan LJ. Microglia, neuroinflammation, and beta-amyloid protein in Alzheimer’s disease. Int J Neurosci. 2014 May;124(5):307–321.
- Gouras GK, Olsson TT, Hansson O. β-Amyloid peptides and amyloid plaques in Alzheimer’s disease. Neurotherapeutics. 2015 Jan;12(1):3–11.
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009 Jan 23;136(2):215–233.
- Michlewski G, Cáceres JF. Post-transcriptional control of miRNA biogenesis. Rna. 2019 Jan;25(1):1–16.
- Bramswig NC, Lüdecke HJ, Hamdan FF, et al. Heterozygous HNRNPU variants cause early onset epilepsy and severe intellectual disability. Hum Genet. 2017 Jul;136(7):821–834.
- Bakkar N, Kovalik T, Lorenzini I, et al. Artificial intelligence in neurodegenerative disease research: use of IBM Watson to identify additional RNA-binding proteins altered in amyotrophic lateral sclerosis. Acta Neuropathol. 2018 Feb;135(2):227–247.
- Wang J, Zhang J, Ding X, et al. Differential microRNA expression profiles and bioinformatics analysis between young and aging spontaneously hypertensive rats. Int J Mol Med. 2018 Mar;41(3):1584–1594.
- Choi RJ, Roy A, Jung HJ, et al. BACE1 molecular docking and anti-Alzheimer’s disease activities of ginsenosides. J Ethnopharmacol. 2016 Aug 22;190:219–230.
- Salminen A, Kaarniranta K, Kauppinen A, et al. Impaired autophagy and APP processing in Alzheimer’s disease: the potential role of Beclin 1 interactome. Prog Neurobiol. 2013;106-107:33–54.
- Schimmel K, Stojanović SD, Huang CK, et al. Combined high-throughput library screening and next generation RNA sequencing uncover microRNAs controlling human cardiac fibroblast biology. J Mol Cell Cardiol. 2021;150:91–100.
- Liu Y, Zheng W, Pan Y, et al. Low expression of miR-186-5p regulates cell apoptosis by targeting toll-like receptor 3 in high glucose-induced cardiomyocytes. J Cell Biochem. 2019 Jun;120(6):9532–9538.
- Sun J, Wen S, Zhou J, et al. Association between malnutrition and hyperhomocysteine in Alzheimer’s disease patients and diet intervention of betaine. J Clin Lab Anal. 2017 Sep;31(5):e22090.
