ABSTRACT
Dysfunctional histone deacetylases (HDACs) elicit unrestrained fibrosis and damage to organs. With regard to the link between HDACs and fibrosis, this research is practiced to decipher the concrete mechanism of HDAC3 in hyperuricemia (HN)-induced renal interstitial fibrosis (RIF) from microRNA-19b-3p/splicing factor 3b subunit 3 (miR-19b-3p/SF3B3) axis.
The HN model was established on rats to induce RIF by oral administration of adenine and potassium oxalate. HN rats were injected with miR-19b-3p- or HDAC3-related vectors to figure out their effects on RIF through detecting 24-h urine protein, uric acid (UA), blood urea nitrogen (BUN) and serum creatinine (Scr) contents and α-smooth muscle actin (α-SMA), transforming growth factor β1 (TGF-β1) and fibronectin (FN) contents in renal tissues and observing pathological damages and RIF index of renal tissues. HDAC3, miR-19b-3p and SF3B3 expression in renal tissues were tested, along with their interactions.
Elevated HDAC3 and SF3B3 and reduced miR-19b-3p were displayed in renal tissues of HN rats. Suppressed HDAC3 or promoted miR-19b-3p relieved HN-induced RIF, as reflected by their inhibitory effects on 24 h urine protein, UA, BUN, Scr, α-SMA, TGF-β1, and FN contents and RIF index and their ameliorated effects on pathological damages of renal tissues. HDAC3 bound to the promoter of miR-19b-3p to regulate SF3B3. MiR-19b-3p depletion abrogated down-regulated HDAC3-induced effects on HN-induced RIF.
It is delineated that depressed HDAC3 relives HN-induced RIF through restoring miR-19b-3p and knocking down SF3B3, replenishing the references for RIF curing.
Introduction
Hyperuricemia is a threat for the occurrence of chronic kidney diseases (CKD), contributing to renal fibrosis [Citation1]. Severely, renal fibrosis is the pathway guiding to end-stage renal disease [Citation2]. Renal fibrosis is mainly specified into renal interstitial fibrosis (RIF) and glomerular sclerosis [Citation3]. Specifically, RIF, referring to myofibroblast activation and extracellular matrix protein reservoir, is often stimulated by inflammatory responses and epithelial–mesenchymal transition (EMT) [Citation4]. RIF clinically results in renal tubule atrophy and chronic and progressive renal dysfunction, and renal replacement is the standard option for RIF patients [Citation5]. Given the insufficiency of drug administration for RIF, new but effective drug administrations are at wanting.
Histone deacetylases (HDACs) are defined to act with and deacetylate non-histone proteins in the regulation of cellular functions, and their inhibitions have the probability to treat acute kidney injury [Citation6]. For instance, HDAC8 knockdown is reported to obstruct EMT and renal fibrosis [Citation7] and class I HDAC inhibitor (FK228) suppresses renal fibroblast activation and proliferation and attenuates RIF [Citation8]. Focused on HDAC3, its deranged expression is a required part in renal damages in CKD, while its repression has the therapeutic prospect for CKD [Citation9]. Relevantly, HDACs are believed to collaborate with microRNAs (miRNAs) to cooperatively regulate the relevant target genes in chronic diseases [Citation10]. miR-19b-3p is connected with kidney function when patient receiving coronary angiography [Citation11]. miR-19b-3p has also been revealed to involve in chronic renal failure with its promoting effects on renal tubular epithelial cell proliferation and attenuated effects on renal fibrosis and EMT [Citation12]. In addition, miR-19b has the potential to be an effective biomarker in focal segmental glomerulosclerosis [Citation13]. Splicing factor 3b subunit 3 (SF3B3), an identified haplotype, is threatening on the onset of diabetic nephropathy [Citation14]. Moreover, it is evident that SF3B3 is attributable to the tumorigenesis of renal cancer [Citation15]. Combing through the involvements of HDAC3, miR-19b-3p and SF3B3 in renal diseases, it is assumed that they may function in combination in RIF. In this work, we aim to unearth the mechanism of HDAC3/miR-19b-3p/SF3B3 axis in hyperuricemic nephropathy (HN)-induced RIF.
Methods and materials
Ethics statement
This study was approved by the Laboratory Animal Ethics Committee of Hainan General Hospital, and strictly in compatibility with the guidelines of Hainan General Hospital. Every effort has been made to minimize animal suffering.
Rat HN modeling and treatment
Male Sprague Dawley rats (8 weeks old; 250 g; Jackson Laboratory, Maine, USA) were reared with a 12 h light/dark cycle. Rats were orally administered with adenine (0.1 g/kg) and potassium oxalate (1.5 g/kg, Macklin, Shanghai, China) every day to establish a HN rat model for a total of 3 weeks [Citation1]. Then, rats were injected with 100 μL HDAC3 low-expression vector negative control (sh-NC), HDAC3 low-expression vector (sh-HDAC3), miR-19b-3p mimic NC, miR-19b-3p mimic, or sh-HDAC3 and miR-19b-3p-inhibitor. The injection was performed twice a week, for 6 weeks [Citation16].
Specimen collection
At 6 weeks post treatment, rats were fasted for 12 h and euthanized with pentobarbital sodium anesthesia. Blood samples were taken and centrifuged (3000 rpm), followed by preservation at −80°C for biochemical detection. The resected renal tissues were weighed and divided into two parts: one part was fixed with 4% paraformaldehyde for histopathological detection, and the other part was stored at −80°C for further reverse transcription quantitative polymerase chain reaction (RT-qPCR) and Western blot assay [Citation17].
Biochemical detection
With a standard commercial kit and a biochemical analyzer (AU5800; Beckman Coulter Commercial Enterprise Co., Ltd., Shanghai, China), 24 h urine protein, serum uric acid (UA), blood urea nitrogen (BUN) and serum creatinine (Scr) levels in serum were measured [Citation18].
Hematoxylin-eosin (H&E) staining
Renal tissues were fixed in 4% paraformaldehyde for 48 h and embedded in paraffin, followed by sectioning and staining with hematoxylin and eosin. Histopathological analysis was carried out under an optical microscope.
Masson staining
The degree of renal fibrosis was observed with Masson’s trichrome reagent staining. The renal tissue sections were soaked in potassium dichromate overnight and dyed with hematoxylin and Ponceaux. Then, the sections were then stained with phospho-molybdic acid and aniline blue and sealed with neutral gum. The blue fiber region (positive area) was measured under a microscope (400×) within randomly opted 5 non-overlapping fields of view (large blood vessels and glomeruli were supposed to be avoided). RIF index (%) = (positive area/total tubulointerstitial area) × 100% [Citation19].
Immunohistochemistry
Dewaxed in xylene, the tissue sections were dehydrated with ethanol and incubated with antigen retrieval solution at 97°C. Subsequently, the sections were immersed in 3% H2O2 deionized and blocked with goat serum, followed by treatment with anti-α-smooth muscle actin (α-SMA, 1:200, Invitrogen, CA, USA), anti-transforming growth factor β1 (TGF-β1, 1:200; R&D Systems, MN, USA) and anti-fibronectin (FN, 1:100; Millipore Sigma, MO, USA). Next, the sections were supplemented with immunoglobulin G (IgG; 1:5000; Abcam, MA, USA) and incubated with horseradish peroxidase (HRP) labeled streptavidin. Processed through diaminobenzidine (DAB) and hematoxylin staining, the sections were then dehydrated and sealed with neutral gum. A microscope (Nikon, Tokyo, Japan) was utilized to capture and count the positive signals [Citation20].
Transferase-mediated deoxyuridine triphosphate-biotin nick end labeling (TUNEL) staining
TUNEL staining was completed by an in situ cell death detection kit (Roche, Shanghai, China). Renal tissue sections were treated with 0.1% Triton X-100 (Servicebio, Hubei, China), blocked with 3% H2O2, and placed in 10% trypsin solution and 90% marking solution. Apoptotic cells were stained in dark brown by DAB (Servicebio) and observed under a microscope (Olympus, Tokyo, Japan) [Citation21].
RT-qPCR
Total RNA was extracted from renal tissues by Trizol kit (Invitrogen). Reverse transcription was finished by HiScript QRT SuperMix (Vazyme, Nanjing, China) or miRNA 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China). Real-time quantitative PCR was evaluated by ChamQ® SYBR® qPCR Master Mix (Vazyme, Nanjing, China) and LightCycler 480II real-time PCR system (Roche). The primers of HDAC3, miR-19b-3p and SF3B3 are presented as follows: HDAC3: forward: ACCGTGGCGTATTTCTACGAC; reverse: CCTGGTAAGGCTTGAAGACGA; miR-19b-3p: RT: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCTCAGTTTT; forward: TCGGCAGGTGTGCAAATCCATGC; reverse: CTCAACTGGTGTCGTGGA; SF3B3: forward: ACTGTTGCCATAACCAAGACG; reverse: ATTTCCTGTTGTTTGGTTCCAGAG; U6: forward: GCTTCGGCAGCACATATACTAAAAT; reverse: CGCTTCACGAATTTGCGTGTCAT; GAPDH: forward:ATGGTGAAGGTCGGTGTG; reverse: AACTTGCCGTGGGTAGAG. Gene expression was calculated by 2−ΔΔCt [Citation22].
Western blot assay
The total protein was extracted from renal tissues through radio-immunoprecipitation assay lysis buffer with protease inhibitors and separated by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis. After that, protein samples were transferred to a polyvinylidene fluoride membrane, followed by blockade with 5% skimmed milk and probing with anti-HDAC3 (1:1000; Abcam), SF3B3 (1:1000; Proteintech, IL, USA), glyceraldehyde-3-phosphate dehydrogenase (1:1000, Abcam) and HRP-conjugated secondary antibody. The protein membrane was finally detected by an ECL Detection system (Amersham Pharmacia Biotech, Little Chalfont, UK) [Citation23].
Chromatin immunoprecipitation (ChIP)
Rat renal tissues were cross-linked with 3.7% formaldehyde and sonicated to obtain genomic DNA (200–400 bp). ChIP analysis was practiced on a ChIP analysis kit (Millipore, MA, USA) and HDAC3 antibody (Abcam), with rabbit IgG as a NC [Citation24].
RNA immunoprecipitation (RIP)
AGO2 RIP was initiated in HEK293T cells with EZ Magna RIP kit (Millipore). Cells were lysed by RIP lysis buffer (Millipore) with protease and RNase inhibitors and added with anti-AGO2 (1:50) and IgG (1:100, both from Abcam). Subsequently, the cells were incubated with proteinase K. The enrichment of miR-19b-3p and SF3B3 was determined by RT-qPCR [Citation25].
Dual luciferase reporter gene assay
The targeting relationship between miR-19b-3p and SF3B3 was verified by the RNA22 website. After restriction of endonuclease digestion, the target fragment was inserted into the pMIR-reporter plasmid by T4 DNA ligase. Plasmids (SF3B3-WT, and SF3B3-MUT) were transected into HEK293T cells with miR-19b-3p mimic and mimic NC. The luciferase activity was measured by a dual luciferase reporter reagent assay kit luminometer (LB960, Berthold Technologies, Germany) [Citation26].
Statistical analysis
All data, expressed as the mean ± standard deviation, were evaluated using SPSS 21.0 (IBM, NY, USA). The t-test was used for comparison between two groups. At P < 0.05, it was of statistical significance.
Results
HN induces RIF
HN model was established by oral administration with adenine and potassium oxalate. The biochemical indicators in serum (24 h urine protein, Scr, BUN, and UA) were tested to verify the success of the modeling. It was demonstrated that these indices were all increased, proving that the modeling was successful ()).
Figure 1. HN induces RIF. (a). 24 h urine protein, Scr, BUN and UA contents in serum of HN rats; (b). H&E staining analyzed pathological damages of renal tissues of HN rats; (c). Masson staining analyzed the degree of fibrosis and RIF index of HN rats; (d). α-SMA, TGF-β1 and FN contents in renal tissues of HN rats; (e). TUNEL staining detected renal cell apoptosis of HN rats. Data were expressed as mean ± standard deviation. The t-test was used for comparison between two groups. * P < 0.05 compared with the normal group.
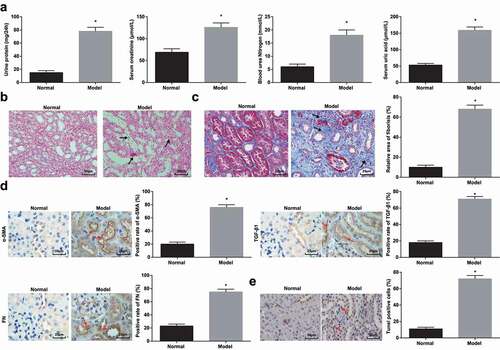
Histopathological detection was processed on rat renal tissues. H&E staining pictured that clearly demarcated renal medulla and renal cortex, clear glomerular capillary structure, normal tubular epithelial cell structure, and neatly and regularly arranged brush borders without inflammation were observed in normal rats. In HN-induced RIF rats, it could be recognized with renal tubular epithelial cell edema in the renal cortex, renal tubular epithelial cell necrosis, karyopyknosis and fragmentation, and renal tubular distension. It was also observed that tubular cavity was severely expanded, epithelial cells were flattened, shed, and vacuolated, and renal tubules were atrophied, accompanied by fibrosis and edema in the renal interstitial and infiltration of inflammatory cells such as lymphocytes and monocytes ()).
Subsequently, Masson staining was adopted to observe the degree of renal fibrosis. In normal rats, tissue structure was normal, and a small amount of collagen fibers were positively stained in the renal interstitial. In HN-induced RIF rats, blue collagen fibers were bundled and reticularly distributed, renal tubules were dilated with a small amount were contracted, and RIF area was widened ()).
Activated interstitial fibroblasts (also called myofibroblasts) played a key role in the occurrence and development of renal fibrosis, which was characterized by α-SMA expression [Citation1]. TGF-β was widely regarded as a key cytokine to promote renal fibrosis [Citation27], while FN was a key component of interstitial matrix [Citation1]. Immunohistochemistry manifested that these three actors were mainly located in tubulointerstitium, and α-SMA, TGF-β1, and FN positive cells increased in HN-induced RIF rats ()).
TUNEL staining detected apoptosis in renal tissues. Observed by a light microscope, cells with brownish yellow nucleus or cytoplasm were apoptotic cells. In normal rats, renal cells with normal morphology without obvious apoptotic cells were manifested. A large number of cells in renal tissues of HN rats showed a typical crescent-shaped chromatin margination, nuclear condensation, and a large number of positive signals for apoptotic cells ()).
The above results indicated that a rat model of RIF induced by HN was successfully established.
Suppressed HDAC3 relieves HN-induced RIF
A study has pointed out that abnormal HDAC3 was a key factor involved in kidney injury [Citation9], and its expression was raised in the kidneys of diabetic nephropathy mice [Citation28]. In this research, HDAC3 expression was tested to find that (,b)) it was up-regulated in HN-induced RIF rats.
Figure 2. Suppressed HDAC3 relieves HN-induced RIF. (a/b). RT-qPCR and Western blot assay detected HDAC3 expression of HN rats; (c/d). RT-qPCR and Western blot assay detected HDAC3 expression of HN rats after down-regulation of HDAC3; (e). 24 h urine protein, Scr, BUN and UA in serum of HN rats after down-regulation of HDAC3; (f). H&E staining analyzed pathological damages of renal tissues of HN rats after down-regulation of HDAC3; (g). Masson staining analyzed the degree of fibrosis and RIF index of HN rats after down-regulation of HDAC3; (h). α-SMA, TGF-β1 and FN contents in renal tissues of HN rats after down-regulation of HDAC3; (i). TUNEL staining detected renal cell apoptosis of HN rats after down-regulation of HDAC3. Data were expressed as mean ± standard deviation. The t-test was used for comparison between two groups. # P < 0.05 compared with the normal group; * P < 0.05 compared with the sh-NC group.
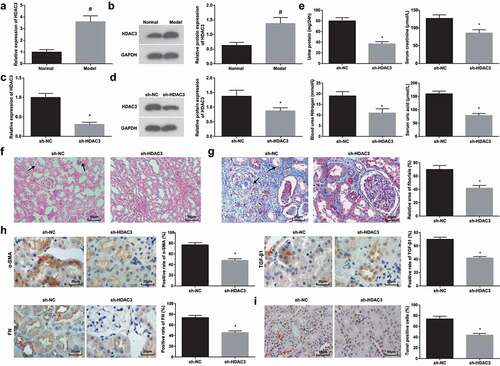
HDAC3 was knocked down to further define its effects on HN-induced RIF rats. RT-qPCR and Western blot assay confirmed that HDAC3 expression in renal tissues was decreased by sh-HDAC3 (,d)). Moreover, followed by HDAC3 knockdown, except for decreases in 24 h urinary protein, UA, BUN, Scr, α-SMA, TGF-β1, and FN contents, it could be also noticed that the structure of renal tubular epithelial cells was relatively normal, and the brush borders of renal tubulars were arranged regularly without inflammation, the positive area and apoptotic cells were reduced (-i)).
HDAC3 binds to the promoter of miR-19b-3p
Studies have evidenced that miR-19b-3p was down-regulated in chronic renal failure [Citation29] and chronic kidney disease [Citation11]. In this work, miR-19b-3p expression was manifested to down-regulate in HN-induced RIF rats ()), which was consistent with the miR-19b-3p manifestation in kidney disease. Moreover, it was speculated that there was a certain correlation between HDAC3 and miR-19b-3p in RIF.
Figure 3. HDAC3 binds to the promoter of miR-19b-3p. (a). RT-qPCR detected miR-19b-3p expression in renal tissues of HN rats; (b). ChIP detected HDAC3 recruitment to the miR-19b-3p promoter; (c). RT-qPCR detected miR-19b-3p expression after down-regulation of HDAC3. Data were expressed as mean ± standard deviation. The t-test was used for comparison between two groups. # P < 0.05 compared with the normal group; * P < 0.05 compared with the sh-NC group.
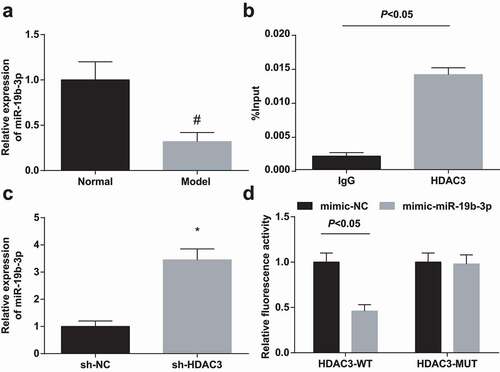
It has been reported that HDAC3 could bind to the miRNA promoter to regulate miRNA expression. For instance, HDAC3 could be bound to the miR-17-92 cluster promoter and the host Mirg of most Dlk1-Dio3 miRNAs [Citation24]. To investigate whether HDAC3 could directly inhibit miR-19b-3p expression, ChIP was conducted to examine whether HDAC3 could bind to the miR-19b-3p promoter. The results were recognizable by the fact that HDAC3 was bound to the miR-19b-3p promoter to inhibit miR-19b-3p expression ()). RT-qPCR highlighted that miR-19b-3p expression was increased after inhibiting HDAC3 ()).
Elevated miR-19b-3p attenuates HN-induced RIF
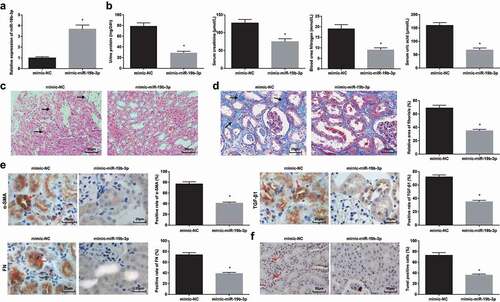
For further comprehension of the effects of miR-19b-3p on RIF, miR-19b-3p up-regulation was implemented in HN-induced RIF rats()). Notably, the phenomena (biochemical indicators, pathological damages in renal tissues, fibrosis-related parameters, and cell apoptosis) were improved in HN rats treated with miR-19b-3p mimic (-f)).
Figure 4. Elevated miR-19b-3p attenuates HN-induced RIF. (a). RT-qPCR detected miR-19b-3p expression after up-regulation of miR-19b-3p in HN rats; (b). 24 h urine protein, Scr, BUN and UA contents in serum of HN rats after up-regulation of miR-19b-3p; (c). H&E staining analyzed pathological damages of renal tissues of HN rats after up-regulation of miR-19b-3p; (d). Masson staining analyzed the degree of fibrosis and RIF index of HN rats after up-regulation of miR-19b-3p; (e). α-SMA, TGF-β1 and FN contents in renal tissues of HN rats after up-regulation of miR-19b-3p; F. TUNEL staining detected renal cell apoptosis of HNl rats after up-regulation of miR-19b-3p. Data were expressed as mean ± standard deviation. The t-test was used for comparison between two groups. * P < 0.05 compared with the mimic-NC group.
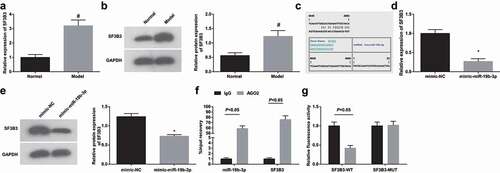
miR-19b-3p negatively regulates SF3B3
SF3B3 has been suggested to increase the risk of diabetic nephropathy [Citation14], and it was up-regulated in clinical renal cell carcinoma specimens [Citation15]. In this work, the effects of SF3B3 on RIF were interpreted first from detecting its expression. It was displayed that HN-induced RIF rats expressed high SF3B3 expression in renal tissues (,b)). Through the bioinformatics website RNA22, it was revealed miR-19b-3p bound to the 3ʹ-untranslated region of SF3B3 ()). In addition, Western blot assay depicted that restoring miR-19b-3p suppressed SF3B3 expression in HN-induced RIF rat renal tissues ()). RIP determined the enrichment of miR-19b-3p and SF3B3 in the immunoprecipitation complex, with IgG as an NC. The results outlined that miR-19b-3p and SF3B3 were preferentially enriched in miRNA riboprotein complexes containing AGO2. Dual-luciferase reporter gene assay validated that the luciferase activity of the 3’UTR in SF3B3-WT was significantly lowered by miR-19b-3p mimic (,f)), showing that miR-19b-3p and SF3B3 had a targeting relationship.
Figure 5. miR-19b-3p negatively regulates SF3B3. (a/b). RT-qPCR and Western blot assay detected SF3B3 expression in renal tissues of HN rats; (c). Bioinformatics website predicted the binding site of miR-19b-3p and SF3B3; (d). Western blot assay detected SF3B3 expression after up-regulation of miR-19b-3p; (e). RIP verified the binding relations of miR-19b-3p/SF3B3 and AGO2; (f). Dual luciferase reporter gene assay verified the regulatory relationship between miR-19b-3p and SF3B3. Data were expressed as mean ± standard deviation. The t-test was used for comparison between two groups. # P < 0.05 compared with the normal group; * P < 0.05 compared with the mimic-NC group.
MiR-19b-3p depletion abrogates down-regulated HDAC3-induced effects on HN-induced RIF
To verify whether HDAC3 regulated RIF through miR-19b-3p/SF3B3 axis, SF3B3 expression was detected after inhibiting HDAC3. It was mirrored that silencing HDAC3 inhibited SF3B3 expression in the renal tissues of HN-induced RIF rats (,b)).
Figure 6. miR-19b-3p depletion abrogates down-regulated HDAC3-induced effects on HN-induced RIF. (a/b). RT-qPCR and Western blot assay detected SF3B3 expression after down-regulation of HDAC3 in HN rats; (c/d). RT-qPCR and Western blot assay detected SF3B3 expression after down-regulation of HDAC3 and miR-19b-3p in HN rats; (e). 24 h urine protein, Scr, BUN and UA contents after down-regulation of HDAC3 and miR-19b-3p in HN rats; (f). H&E staining analyzed pathological damages of renal tissues after down-regulation of HDAC3 and miR-19b-3p in HN rats; (g). Masson staining analyzed the degree of fibrosis and RIF index after down-regulation of HDAC3 and miR-19b-3p in HN rats; (h). α-SMA, TGF-β1 and FN contents in renal tissues after down-regulation of HDAC3 and miR-19b-3p in HN rats; (i). TUNEL staining detected renal cell apoptosis after down-regulation of HDAC3 and miR-19b-3p in HN rats. Data were expressed as mean ± standard deviation. The t-test was used for comparison between two groups. & P < 0.05 compared with the sh-NC group; * P < 0.05 compared with the sh-HDAC3 group.
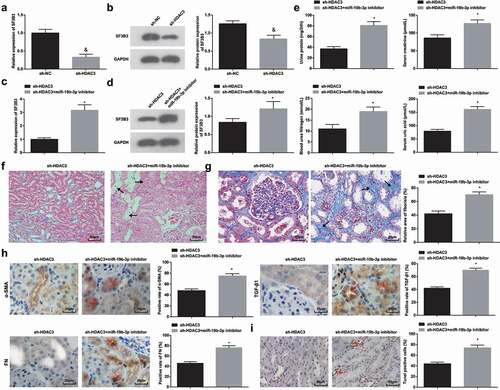
The effect of HDAC3-mediated miR-19b-3p on RIF was deciphered. miR-19b-3p downregulation mitigated the effect of knocking out HDAC3 on the renal tissues of HN-induced RIF rats, as evidenced by downregulated SF3B3, deteriorated RIF, a large number of blue collagen fibers in the renal interstitial, raised α-SMA, TGF-β1, FN, 24 h urine protein, Scr, BUN, and UA contents and enhanced apoptosis (-i)).
Discussion
RIF is the last trackway of progressive and chronic renal diseases [Citation30]. HDAC3, miR-19b-3p, and SF3B3 have been independently documented to position in renal diseases, but their loop feedback in RIF has not been clarified comprehensively. Given that, this work is triggered to figure out their reciprocals in HN-induced RIF. To the end, our research has worked out that HDAC3 down-regulation promotes miR-19b-3p to depress SF3B3, thereby attenuating HN-induced RIF.
To begin with, HN rats are modeled to induce RIF. In these HN-induced RIF rats, the performance of HDAC3 is disclosed. First, we have observed that HDAC3 manifests a reduction in its expression. Then, down-regulation assays of HDAC3 have stratified that silencing HDAC3 attenuates HN-induced RIF, as testified by decreased 24 h urine protein, UA, BUN, and Scr contents in serum, inhibited α-SMA, TGF-β1, and FN levels in renal tissues, together with ameliorated pathological damages and reduced RIF index of renal tissues. Consistent with the discoveries in this work, a late research has discussed that HDAC3 expression goes toward an elevation in CKD mice, while HDAC inhibitor (trichostatin A) attenuates pathological abnormalities and depresses BUN and α-SMA contents in the kidneys [Citation9]. Besides that, HDAC suppression by trichostatin A has the capacity to restrict α-SMA and FN expression, and diminishes renal fibrosis via inhibiting the activation of JNK/Notch-2 pathway in renal tubulointerstitial fibrosis [Citation31]. Complementary to the results, another literature has declared that HDAC3 expression is accumulated in the kidneys of diabetic mellitus mice, and eliminating HDAC3 prevents FN expression going uprising [Citation28]. Impressively, HDAC3 silencing by largazole is regarded to partially reduce α-SMA expression, and relieves liver fibrosis and angiogenesis [Citation32]. Similarly, HDAC depletion facilitates to impair RIF and reduce interstitial α-SMA and FN expression in renal ischemia-reperfusion injury [Citation33]. Pharmacologically, knocking down HDAC by trichostatin A impedes the process of glomerulosclerosis, renal inflammation, and tubulointerstitial fibrosis [Citation34].
Moreover, it has been discovered that the mediation of HDAC3 on miR-19b-3p. Briefly, HDAC3 binds to the promoter of miR-19b-3p to inhibit miR-19b-3p expression. In this work, miR-19b-3p has been found to down-regulate in HN-induced RIF, while its restoration is conducive to relieving RIF by decreasing biochemical indicators and fibrosis-related parameters, suppressing RIF index, and improving pathological damages. At present, nearly no study has reported the specific binding connection between HDAC3 and miR-19b-3p, which requires further investigations. In an existing research, reduced miR-19b-3p has witnessed the progression of chronic renal failure, and elevating miR-19b-3p is contributory to renal tubular epithelial cell proliferation and renal fibrosis amelioration [Citation12]. Also, an observation in a previous study has identified that miR-19b is down-regulated in focal segmental glomerulosclerosis [Citation13]. In a similar fashion, miR-19b up-regulation with adeno-associated virus serotype 2 vector promotes liver function recovery and hampers hepatic fibrosis (HF), which is proved by decreased α-SMA and TGF-β receptor II contents [Citation35]. In addition to that, a reduction can be seen in miR-19b expression in HF while miR-19b elevation attenuates fibrosis in livers [Citation36]. Commonly speaking, our study findings are compatible with the manifestation and function of miR-19b-3p in renal disease or fibrosis-related diseases.
Next, the mechanism of miR-19b-3p targeting SF3B3 has been translated. Also, SF3B3 expression presents a decline in HN-induced RIF rats. No existing literature has announced the finding about miR-19b-3p targeting SF3B3, which is expected to be validated in proceeding explorations. As for the presentation of SF3B3, there are studies mentioning that it is up-regulated in clear cell renal cell carcinoma tissues and breast cancer [Citation15,Citation37], which are echoed with SF3B3 expression in this work.
In short, it is explained in this work that restrained HDAC3 improves HN-induced RIF with the facilitation of restored miR-19b-3p and depleted SF3B3, widening the horizon to the mechanism and management of RIF. For the promising application of HDAC3/miR-19b-3p/SF3B3 axis-oriented targets in RIF, more logical researches are required for further confirmation of the results of this work.
Acknowledgments
We would like to acknowledge the reviewers for their helpful comments on this paper.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Liu N, Wang L, Yang T, et al. EGF receptor inhibition alleviates hyperuricemic nephropathy. J Am Soc Nephrol. 2015;26(11):2716–2729.
- Wang JL, Chen CW, Tsai MR, et al. Antifibrotic role of PGC-1alpha-siRNA against TGF-beta1-induced renal interstitial fibrosis. Exp Cell Res. 2018;370(1):160–167.
- Ji X, Cao J, Zhang L, et al. Kaempferol protects renal fibrosis through activating the BMP-7-Smad1/5 signaling pathway. Biol Pharm Bull. 2020;43(3):533–539.
- Xiao H, Shen HY, Liu W, et al. Adenosine A2A receptor: a target for regulating renal interstitial fibrosis in obstructive nephropathy. PLoS One. 2013;8(4):e60173.
- Ruan GP, Xu F, Li ZA, et al. Induced autologous stem cell transplantation for treatment of rabbit renal interstitial fibrosis. PLoS One. 2013;8(12):e83507.
- Hyndman KA. Histone deacetylases in kidney physiology and acute kidney injury. Semin Nephrol. 2020;40(2):138–147.
- Zhang Y, Zou J, Tolbert E, et al. Identification of histone deacetylase 8 as a novel therapeutic target for renal fibrosis. FASEB J. 2020;34(6):7295–7310.
- Yang M, Chen G, Zhang X, et al. Inhibition of class I HDACs attenuates renal interstitial fibrosis in a murine model. Pharmacol Res. 2019;142:192–204.
- Lin W, Zhang Q, Liu L, et al. Klotho restoration via acetylation of peroxisome proliferation-activated receptor gamma reduces the progression of chronic kidney disease. Kidney Int. 2017;92(3):669–679.
- Swierczynski S, Klieser E, Illig R, et al. Histone deacetylation meets miRNA: epigenetics and post-transcriptional regulation in cancer and chronic diseases. Expert Opin Biol Ther. 2015;15(5):651–664.
- Muendlein A, Geiger K, Leiherer A, et al. Evaluation of the associations between circulating microRNAs and kidney function in coronary angiography patients. Am J Physiol Renal Physiol. 2020;318(2):F315–F321.
- Chen W, Zhou ZQ, Ren YQ, et al. Effects of long non-coding RNA LINC00667 on renal tubular epithelial cell proliferation, apoptosis and renal fibrosis via the miR-19b-3p/LINC00667/CTGF signaling pathway in chronic renal failure. Cell Signal. 2019;54:102–114.
- Xiao B, Wang LN, Li W, et al. Plasma microRNA panel is a novel biomarker for focal segmental glomerulosclerosis and associated with podocyte apoptosis. Cell Death Dis. 2018;9(5):533.
- Liao LN, Chen CC, Wu FY, et al. Identified single-nucleotide polymorphisms and haplotypes at 16q22.1 increase diabetic nephropathy risk in Han Chinese population. BMC Genet. 2014;15:113.
- Chen K, Xiao H, Zeng J, et al. Alternative splicing of EZH2 pre-mRNA by SF3B3 contributes to the tumorigenic potential of renal cancer. Clin Cancer Res. 2017;23(13):3428–3441.
- Ding H, Xu Y, Jiang N. Upregulation of miR-101a suppresses chronic renal fibrosis by regulating KDM3A via blockade of the YAP-TGF-beta-Smad signaling pathway. Mol Ther Nucleic Acids. 2020;19:1276–1289.
- Chen Y, Li C, Duan S, et al. Curcumin attenuates potassium oxonate-induced hyperuricemia and kidney inflammation in mice. Biomed Pharmacother. 2019;118:109195.
- Li L, Chen Y, Jiao D, et al. Protective effect of astaxanthin on ochratoxin A-induced kidney injury to mice by regulating oxidative stress-related NRF2/KEAP1 pathway. Molecules. 2020;25:6.
- Xie Y, Lan F, Zhao J, et al. Hirudin improves renal interstitial fibrosis by reducing renal tubule injury and inflammation in unilateral ureteral obstruction (UUO) mice. Int Immunopharmacol. 2020;81:106249.
- Zhou H, Qiu ZZ, Yu ZH, et al. Paeonol reverses promoting effect of the HOTAIR/miR-124/Notch1 axis on renal interstitial fibrosis in a rat model. J Cell Physiol. 2019;234(8):14351–14363.
- Chen L, Xiang E, Li C, et al. Umbilical cord-derived mesenchymal stem cells ameliorate nephrocyte injury and proteinuria in a diabetic nephropathy rat model. J Diabetes Res. 2020;2020:8035853.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408.
- Ji C, Zhang J, Zhu Y, et al. Exosomes derived from hucMSC attenuate renal fibrosis through CK1delta/beta-TRCP-mediated YAP degradation. Cell Death Dis. 2020;11(5):327.
- Wang Y, Frank DB, Morley MP, et al. HDAC3-dependent epigenetic pathway controls lung alveolar epithelial cell remodeling and spreading via miR-17-92 and TGF-beta signaling regulation. Dev Cell. 2016;36(3):303–315.
- Pan G, Mao A, Liu J, et al. Circular RNA hsa_circ_0061825 (circ-TFF1) contributes to breast cancer progression through targeting miR-326/TFF1 signalling. Cell Prolif. 2020;53(2):e12720.
- Li Y, Gao M, Xu LN, et al. MicroRNA-142-3p attenuates hepatic ischemia/reperfusion injury via targeting of myristoylated alanine-rich C-kinase substrate. Pharmacol Res. 2020;156:104783.
- Zhang L, Zhao S, Zhu Y. Long noncoding RNA growth arrest-specific transcript 5 alleviates renal fibrosis in diabetic nephropathy by downregulating matrix metalloproteinase 9 through recruitment of enhancer of zeste homolog 2. FASEB J. 2020;34(2):2703–2714.
- Shan Q, Zheng G, Zhu A, et al. Epigenetic modification of miR-10a regulates renal damage by targeting CREB1 in type 2 diabetes mellitus. Toxicol Appl Pharmacol. 2016;306:134–143.
- Ogawa Y, Maruko I, Koizumi H, et al. Quantification of choroidal vasculature by high-quality structure en face swept-source optical coherence tomography images in eyes with central serous chorioretinopathy. Retina. 2020;40(3):529–536.
- Yamaguchi I, Tchao BN, Burger ML, et al. Vascular endothelial cadherin modulates renal interstitial fibrosis. Nephron Exp Nephrol. 2012;120(1):e20–31.
- Tung CW, Hsu YC, Cai CJ, et al. Trichostatin A ameliorates renal tubulointerstitial fibrosis through modulation of the JNK-dependent Notch-2 signaling pathway. Sci Rep. 2017;7(1):14495.
- Liu Y, Wang Z, Wang J, et al. A histone deacetylase inhibitor, largazole, decreases liver fibrosis and angiogenesis by inhibiting transforming growth factor-beta and vascular endothelial growth factor signalling. Liver Int. 2013;33(4):504–515.
- Hyndman KA, Kasztan M, Mendoza LD, et al. Dynamic changes in histone deacetylases following kidney ischemia-reperfusion injury are critical for promoting proximal tubule proliferation. Am J Physiol Renal Physiol. 2019;316(5):F875–F888.
- Van Beneden K, Geers C, Pauwels M, et al. Comparison of trichostatin A and valproic acid treatment regimens in a mouse model of kidney fibrosis. Toxicol Appl Pharmacol. 2013;271(2):276–284.
- Brandon-Warner E, Benbow JH, Swet JH, et al. Adeno-associated virus serotype 2 vector-mediated reintroduction of microRNA-19b attenuates hepatic fibrosis. Hum Gene Ther. 2018;29(6):674–686.
- Ge S, Xie J, Liu F, et al. MicroRNA-19b reduces hepatic stellate cell proliferation by targeting GRB2 in hepatic fibrosis models in vivo and in vitro as part of the inhibitory effect of estradiol. J Cell Biochem. 2015;116(11):2455–2464.
- Gokmen-Polar Y, Neelamraju Y, Goswami CP, et al. Expression levels of SF3B3 correlate with prognosis and endocrine resistance in estrogen receptor-positive breast cancer. Mod Pathol. 2015;28(5):677–685.
