ABSTRACT
Circular RNA derived from the SLC8A1 gene (circSLC8A1) has been implicated in the pathogenesis of several types of cancers. However, the role of circSLC8A1 in non-small cell lung cancer (NSCLC) remains unclear. In the present study, the expression levels of circSLC8A1 in NSCLC tissues and cell lines were determined by qRT-PCR analysis. Function-gain-assays were then carried out to further validate the role of circSLC8A1 in NSCLC in vitro. Online prediction software and the subsequent luciferase reporter assay were used to identify the target genes of circSLC8A1 and microRNA (miR)-106b-5p. CircSLC8A1 was found to be downregulated in NSCLC tissues and cell lines. Overexpression of circSLC8A1 significantly inhibited the proliferation and invasion of NSCLC cells. Further investigations shown that circSLC8A1 was able to bind to miR-106b-5p as well as inhibit the expression of miR-106b-5p in NSCLC cells. MiR-106b-5p mimics reversed the inhibitory effects of circSLC8A1 overexpression on cell proliferation and invasion. Furthermore, we found that forkhead box J3 (FOXJ3) to be a target gene of miR-106b-5p in NSCLC cells. Knockdown of FOXJ3 reversed the inhibitory effects of miR-106b-5p inhibitor on cell proliferation and invasion. Collectively, these findings indicate that circSLC8A1 exhibits anti-tumor activity in NSCLC, which might be mediated by the miR-106b-5p/FOXJ3 axis. The circSLC8A1/miR-106b-5p/FOXJ3 axis may thus represent a promising therapeutic target for the management of NSCLC.
1. Introduction
Lung cancer is one of the most common cancers which remains the leading cause of cancer-related deaths in China [Citation1,Citation2]. This cancer is categorized as small cell carcinoma (SCLC) or non-small cell lung cancer (NSCLC). NSCLC represents more than 85% of lung cancer cases, and published documents have validated various oncogenic driver genes to contribute to the development of targeted agents [Citation3]. Although continuous efforts have been devoted toward exploring the mechanism of pathogenesis of lung cancer, the therapeutic approaches for advanced NSCLC are poor, and till date, the overall five-year survival rate is estimated to be less than 15% [Citation4]. Therefore, the development of novel therapeutic targets is of particular importance for the treatment of NSCLC.
Circular RNAs (circRNAs), a novel class of non-coding RNAs, are characterized by a covalent bond linking the 3ʹ and 5ʹ ends generated by backsplicing [Citation5]. Although they do not function as vehicles for protein translation, functional circRNAs have been shown to act as cytoplasmic microRNA sponges, RNA-binding protein sequestering agents, or nuclear transcriptional regulators, thereby participating in the regulatory networks governing gene expression [Citation6]. It has been documented that circRNAs are involved in various biological events, such as intercellular communication, cell development and differentiation, cell signaling, and immune system functions. An increasing number of studies have identified the origin, biogenesis and function of circRNAs as well as their roles in various diseases, especially in cancers [Citation7]. CircRNAs regulate development of cancer through a wide range of mechanisms, including acting as miRNA sponges, modulating the Wnt signaling pathway and regulating epithelial-mesenchymal transition [Citation8–10]. Previous studies have provided evidence supporting the roles of various circRNAs in carcinogenesis as well as in the development of clinical diagnostic and therapeutic strategies.
CircSLC8A1 is a circRNA derived from the SLC8A1 gene. It has been reported that the expression of circSLC8A1 is significantly down-regulated in bladder cancer tissues and cell lines, and is positively correlated with the clinical stage and histological grade of bladder cancer [Citation11]. However, the role of circSLC8A1 in NSCLC progression remains largely unknown. In the present study, we evaluated the function of circSLC8A1 in NSCLC and elucidated its functional mechanism.
2. Materials and methods
2.1. Clinical tissues and cell culture
The Clinical Research Ethics of HanZhong Central Hospital (HanZhong, China) approved the use of human samples in this study. Informed consent for the use of human tissues and research purposes was obtained from each patient recruited in this study. A total of 37 pairs of NSCLC tissues and corresponding adjacent non-tumor tissues were obtained from patients diagnosed with NSCLC and undergoing surgery at HanZhong Central Hospital. None of the patients received any local or systemic treatment prior to the surgery. All tissue samples were snap-frozen in liquid nitrogen and stored at −80°C until use.
Human NSCLC cell lines (A549, H1299, H358, PC9 and SPCA1 cells) and bronchial epithelial cells (16HBE) were purchased from the Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The cells were cultured in RPMI-1640 (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco Laboratories, Grand Island, NY, USA) and antibiotics (Gibco) at 37°C in a humidified atmosphere with 5% CO2.
2.2. Cell transfection
To elevate the expression of circSLC8A1 in A549 and H1299 cells, circSLC8A1 was cloned into the pcDNA3.1 vector (Invitrogen) to generate the pcDNA3.1-circSLC8A1 construct. MiR-106b-5p mimics, miR-106b-5p inhibitor and negative controls were obtained from RiboBio (Guangzhou, China). FOXJ3 siRNA (si-FOXJ3) and negative siRNA (si-NC) were obtained from GenePharma (Shanghai, China). Transfection was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Cells were collected 48 h after transfection for the transfection efficiency.
2.3. Cell proliferation assay
Cell proliferation assay was undertaken using the Cell Counting Kit-8 (CCK-8, Dojindo, Japan). Briefly, A549 and H1299 cells (2.0 × 103 cells per well) were seeded in 96-well plates and cultured for 0, 24, 48 and 72 h. At the indicated time points, CCK-8 solution was added to each well and incubated for 3 h at 37°C. The absorbance was measured at 450 nm using a microplate reader (Bio-Tek, Winooski, VT, USA). The growth rates were recorded from three independent experiments.
2.4. Cell invasion assay
For the invasion assay, the membrane was coated with 20% Matrigel (Sigma-Aldrich). A549 and H1299 cells (5 × 105 cells) in serum-free media were cultured in the upper chamber with an 8 μm pore size membrane (Millipore). The lower chambers were filled with 600 μl RPMI 1640 medium containing 20% FBS. After 24 h of incubation, the cells that passed through the membranes were fixed with 4% paraformaldehyde and then stained with 0.1% crystal violet (Sigma) for 20 min. The cells were photographed, and the cell numbers in three random fields were counted under a microscope.
2.5. qRT-PCR analysis
Total RNA was isolated from frozen NSCLC tissues or cultured cells using Trizol reagent (Invitrogen). Then the RNA was used to synthesize cDNA using a reverse transcription kit (Takara, Dalian, China). Subsequently, SYRB Real-time PCR Master Mix (Takara) was used for real-time PCR. The results were normalized to the expression of GAPDH or U6. The specific primers were as follows: circSLC8A1 forward: 5ʹ-ATCGAAGGGACTGCCAGAGG-3ʹ and reverse: 5ʹ- GGTGAAAGACTTAATCGCCGC-3ʹ; miR-106b-5p forward: 5ʹ- TGCGGCAAC ACCAGTCGATGG-3ʹ and reverse: 5ʹ-CCAGTGCAGGGTCCGAGGT-3; FOXJ3 forward: 5ʹ- CACGAGCTCCGGCAACCATTATTTGTAACTGTTT-3ʹ and reverse: 5ʹ-TTTCTCGAGTGCCATTCATGCATATCAGTTCTGG-3; GAPDH forward: 5ʹ-GTGGACCTGACCTGCCGTCT-3ʹ and reverse: 5ʹ-GGAGGAGTG GGTGTCGCTGT-3ʹ; U6 forward: 5ʹ-CGCTTCGGCAGCACATATAC-3ʹ and reverse: 5ʹ-TTCACGAATTTGCGTGTCAT-3ʹ. The relative expressions of genes were calculated using comparative Ct method. The comparison Ct (2−ΔΔCt) method was used to analyze the data.
2.6. Western blot
Proteins were extracted from NSCLC cells using RIPA buffer (KeyGEN, Shanghai, China) and the protein concentration was calculated using a Pierce BCA Protein Assay Kit (Sangon Biotech, Shanghai, China). The protein samples were separated by 10% SDS-PAGE and then transferred onto PVDF membranes (Millipore, Billerica, MA, USA). Thereafter, the membranes were blocked with 5% nonfat milk buffer and incubated with primary antibodies against FOXJ3 and GAPDH (1: 500; Abcam, Cambridge, MA, USA) at 4°C overnight. Next, the membranes were incubated with secondary antibody (1: 3000; Abcam) for 2 h. Finally, the proteins were visualized using an Image Quant LAS 4000 mini system (GE Healthcare, Little Chalfont, United Kingdom).
2.7. RNase R treatment assay
For RNase R treatment, 2 µg of total RNA was incubated with RNase (3 units per μg, Sigma) for 30 min. Then, the relative levels of circSLC8A1 and SLC8A1 were assayed by qRT-PCR, normalizing to those measured in mock group.
2.8. Luciferase reporter assay
NSCLC cells were plated in 12-well plates and co-transfected with miR-106b-5p mimic/control mimic and reporter plasmids (pmirGLO-circSLC8A1-WT, pmirGLO-circSLC8A1-MUT, pmirGLO-FOXJ3-WT, pmirGLO-FOXJ3-MUT) as indicated in the figures. Cells were collected 48 h after transfection, and luciferase activity was assayed using the dual-luciferase reporter assay kit (Promega, Fitchburg, WI, USA).
2.9. Statistical analysis
Results are expressed as the mean ± standard deviation (SD). Student’s t-test and one-way analysis of variance (ANOVA) were used to analyze data using GraphPad Prism 6.0 software (GraphPad, La Jolla, CA, USA). P values of less than 0.05 were considered statistically significant.
3. Results
3.1. CircSLC8A1 expression is down-regulated in NSCLC tissues and cell lines
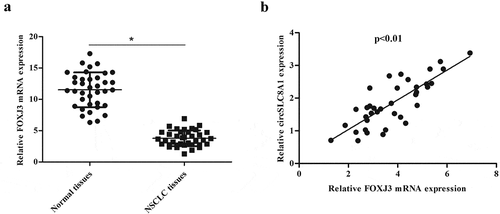
To explore the potential relevance of circSLC8A1 in NSCLC, we examined the expression of circSLC8A1 in NSCLC tissues. The results of qRT-PCR showed that circSLC8A1 expression was significantly down-regulated in NSCLC tissues compared to that in normal tissues ()). Moreover, the expression of circSLC8A1 was noted in NSCLC cell lines; the expression of circSLC8A1 was lower in A549, H1299, H358, PC9, and SPCA1 cells as compared to 16HBE cells ()). A549 and H1299 cell lines were chosen for the gain-of-function study because they have the lower expression of circSLC8A1. Furthermore, the incubation of RNase R led to a reduction in the level of SLC8A1 linear mRNA, not the levels of circSLC8A1 ()), suggesting that circSLC8A1 was more stable than its linear RNA. These results indicate that circSLC8A1 may be involved in the tumorigenesis of NSCLC.
Figure 1. Expression of circSLC8A1 in clinical tissues and cell lines. (a). Relative expression of circSLC8A1 in NSCLC tissues compared with normal tissues. *p < 0.05 vs. normal tissues. (b). Relative expression of circSLC8A1 in NSCLC cell lines A549, H1299, H358, PC9 and SPCA1 cells, as compared to the bronchial epithelial cells 16HBE. *p < 0.05 vs. 16HBE cells. (c). The expression level of circSLC8A1 and SLC8A1 were detected in A549 and H1299 cells by qRT-PCR after treatment with actinomycin D and RNase R, respectively. *p < 0.05
3.2. CircSLC8A1 inhibits the proliferation and invasion of NSCLC cells
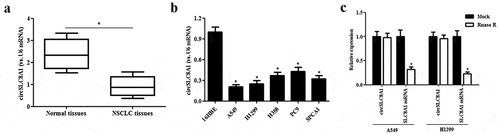
To investigate the precise biological function of circSLC8A1 in NSCLC, gain-of-function experiments were performed by overexpressing circSLC8A1. The circSLC8A1 overexpressing vector pcDNA3.1-circSLC8A1 was transfected into A549 and H1299 cells to elevate the expression of circSLC8A1 ()). Then we examined the effects of circSLC8A1 on cell proliferation and invasion. The CCK-8 assay showed that circSLC8A1 overexpression significantly inhibited the proliferation of A549 and H1299 cells ( and c)). Moreover, transwell invasion assay revealed that circSLC8A1 overexpression also greatly decreased the invasive ability of A549 and H1299 cells, respectively ()).
Figure 2. Effect of circSLC8A1 on the proliferation and invasion of NSCLC cells. (a). Relative expression of circSLC8A1 in A549 and H1299 cells after transfection with pcDNA3.1-circSLC8A1 or vector. (b and c). CCK-8 assay was performed to examine the effects of circSLC8A1 on the proliferation in A549 and H1299 cells. (d). Transwell assay was carried out to measure the effect of circSLC8A1 on cell invasion in A549 and H1299 cells. *p < 0.05 vs. vector group
3.3. circSLC8A1 targets miR-106b-5p in NSCLC cells
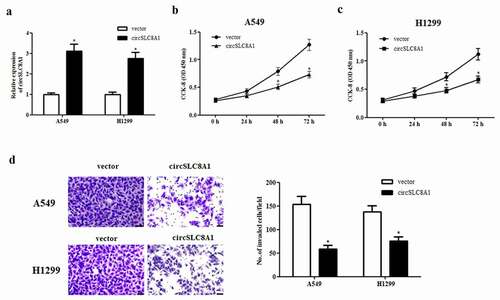
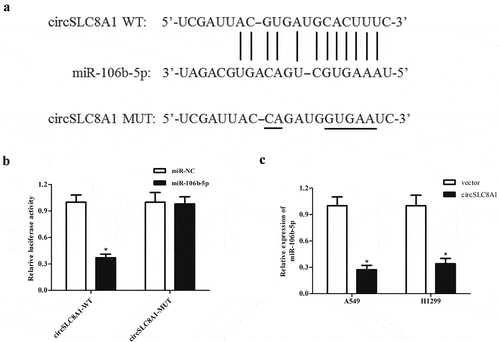
It is well known that many circRNAs act as sponges for miRNAs, therefore, we used the online software starBase (http://starbase.sysu.edu.cn/) to predict the target miRNA of circSLC8A1. Based on the predicted results, miR-106b-5p might be a downstream target of circSLC8A1 ()). Subsequently, we used a luciferase reporter assay to confirm whether miR-106b-5p is the target miRNA of circSLC8A1. The data showed that circSLC8A1 was able to bind to miR-106b-5p, thereby inhibiting the luciferase activity ()). In addition, circSLC8A1 overexpression significantly suppressed miR-106b-5p expression in A549 and H1299 cells ()).
Figure 3. CircSLC8A1 targets miR-106b-5p in NSCLC cells. (a). Schematic representation of the binding sites between circSLC8A1 and miR-106b-5p. (b). Luciferase reporter assay was used to confirm whether miR-106b-5p was a target miRNA of circSLC8A1. *p < 0.05 vs. cells transfected with miR-106b-5p mimics and pmirGLO-circSLC8A1-MUT. C. Effect of circSLC8A1 overexpression on miR-106b-5p expression in A549 and H1299 cells. *p < 0.05 vs. vector group
3.4. MiR-106b-5p expression is up-regulated in NSCLC tissues and cell lines
Subsequently, we detected the expression of miR-106b-5p in NSCLC tissues and cell lines. As shown in ), the expression of miR-106b-5p was markedly increased in NSCLC tissues compared with that in normal tissues. In addition, higher expression of miR-106b-5p was also found in the NSCLC cell lines (A549, H1299, H358, PC9 and SPCA1 cells) than in the bronchial epithelial cells 16HBE ()). Furthermore, Spearman rank correlation assessment indicated an inverse correlation between circSLC8A1 and miR-106b-5p in NSCLC tissues ()).
Figure 4. Expression levels of miR-106b-5p in clinical tissues and cell lines. (a). Relative expression of miR-106b-5p in NSCLC tissues compared with normal tissues. *p < 0.05 vs. normal tissues. (b). Relative expression of miR-106b-5p in NSCLC cell lines including A549, H1299, H358, PC9 and SPCA1 cells, as compared to the bronchial epithelial cells 16HBE. *p < 0.05 vs. 16HBE cells. (c). Expression correlation between circSLC8A1 and miR-106b-5p in NSCLC tissues by qRT-PCR
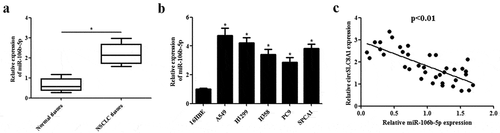
3.5. MiR-106b-5p mimics reverses the inhibitory effects of circSLC8A1 overexpression on cell proliferation and invasion
Considering that circSLC8A1 might execute its roles by sponging miR-106b-5p, we next investigated the effects of miR-106b-5p overexpression on the circSLC8A1-mediated decrease in cell proliferation and invasion. As shown in ), miR-106b-5p mimics greatly increased miR-106b-5p expression in A549 and H1299 cells. In addition, we observed that the overexpression of miR-106b-5p reversed the inhibitory effects of circSLC8A1 overexpression on cell proliferation and invasion (–d)).
Figure 5. Effect of miR-106b-5p mimics on the cell proliferation and invasion in circSLC8A1-overexpressing NSCLC cells. (a). Relative expression of miR-106b-5p in A549 and H1299 cells after transfection with miR-106b-5p mimics or control mimics. NSCLC cells were co-transfected with circSLC8A1 and miR-106b-5p mimics. (b and c). CCK-8 assay was performed to examine cell proliferation in A549 and H1299 cells. (d). Transwell assay was carried out to measure cell invasion in A549 and H1299 cells. *p < 0.05 vs. vector+miR-NC group, #p < 0.05 vs. circSLC8A1+ miR-NC group
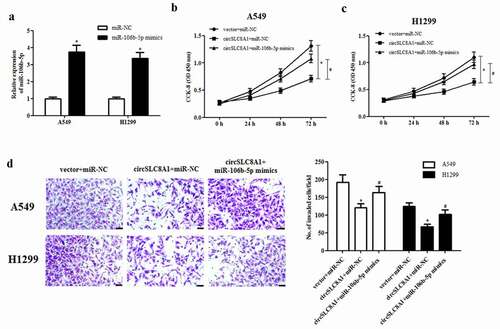
3.6. MiR-106b-5p targets FOXJ3 in NSCLC cells
MiRNAs have been documented to exert their roles by regulating the expression of target mRNAs. According to the results predicted by the online software TargetScan (http://www.targetscan.org), FOXJ3 was a potential target gene of miR-106b-5p ()). Further luciferase reporter assay demonstrated that miR-106b-5p directly bound to the 3ʹUTR of FOXJ3 and inhibited luciferase activity ()). Additionally, the expression of FOXJ3 at both mRNA and protein levels in A549 and H1299 cells was significantly decreased after transfection with miR-106b-5p mimics ( and d)).
Figure 6. MiR-106b-5p targets FOXJ3 in NSCLC cells. (a). Schematic representation of the binding sites between miR-106b-5p and FOXJ3. (b). Luciferase reporter assay was used to confirm whether FOXJ3 was a target gene of miR-106b-5p. *p < 0.05 vs. cells transfected with miR-106b-5p mimics and pmirGLO-FOXJ3-MUT. (c). Effect of miR-106b-5p mimics on FOXJ3 mRNA expression in A549 and H1299 cells. (d). Effect of miR-106b-5p mimics on FOXJ3 protein expression in A549 and H1299 cells. *p < 0.05 vs. miR-NC group
3.7. Expression correlation between circSLC8A1 and FOXJ3 in NSCLC tissues
Next, we examined the expression of FOXJ3 in NSCLC tissues and the results indicated that FOXJ3 expression was significantly decreased in NSCLC tissues compared with that in adjacent non-tumor tissues ()). We also demonstrated a positive correlation between FOXJ3 and circSLC8A1 in NSCL tissues ()).
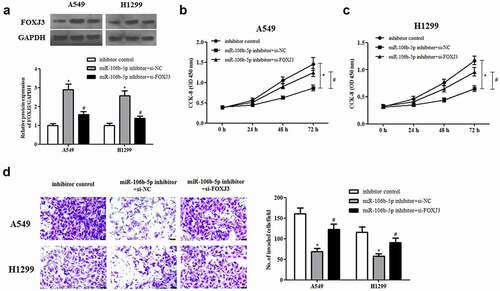
3.8. Si-FOXJ3 reverses the inhibitory effects of miR-106b-5p inhibitor on cell proliferation and invasion
We also explored the functions of miR-106b-5p and FOXJ3 in NSCLC cells. As indicated in ), transfection with si-FOXJ3 greatly rescued miR-106b-5p inhibitor-increased FOXJ3 expression in A549 and H1299 cells. Moreover, we found that inhibition of miR-106b-5p significantly decreased cell proliferation and invasion in A549 and H1299 cells. However, the inhibitory effects were mitigated by knockdown of FOXJ3 (–d)).
Figure 8. Effect of FOXJ3 knockdown on cell proliferation and invasion in miR-106b-5p-downregulated NSCLC cells. A549 and H1299 cells were transfected with si-FOXJ3/si-NC and/or miR-106b-5p inhibitor/inhibitor control. (a). The protein expression of FOXJ3 was detected using western blot. (b and c). CCK-8 assay was performed to examine cell proliferation. (d). Transwell assay was carried out to measure cell invasion. *p < 0.05 vs. inhibitor control group, #p < 0.05 vs. miR-106b-5p inhibitor +si-NC group
4. Discussion
Studies have shown that many circRNAs are differentially expressed in lung cancer tissues, suggesting the involvement of circRNAs in the occurrence and development of lung cancer [Citation12–15]. The study by Jiang et al. [Citation16] reported that human circRNAs microarray revealed a total of 957 abnormal circRNA expression in NSCLC tissue, compared to adjacent normal tissues. The circular microarray in plasma samples validated 1377 differentially expressed circRNAs between gefitinib effective and ineffective groups, indicating that these may be used as biomarkers for predicting the efficacy of gefitinib therapy for NSCLC [Citation17]. Microarray analysis has identified 15,504 circRNAs are differentially expressed in AZD9291-resistant NSCLC cell lines, implying that circRNAs are involved in the drug resistance of NSCLC [Citation18]. Additionally, some specific circRNAs such as circP4HB, circ_0003645, circ_0020123, and circVANGL1 have been discovered and proven to be closely related to the occurrence, development, tumor node metastasis staging, pathological grade, and lymph node metastasis in NSCLC [Citation19–22]. These findings suggest that circRNAs may be employed as potential markers for the diagnosis, treatment, and prognosis of NSCLC. As more and more circRNAs are being associated with NSCLC, exploring the roles of circRNAs will continue to receive more attention.
In the present study, we identified the tumor-suppressive role of circSLC8A1 in NSCLC. CircSLC8A1 was found to be downregulated in NSCLC tissues and cell lines. Overexpression of circSLC8A1 inhibits the proliferation and invasion of NSCLC cells. This result was consistent with a recent study on bladder cancer, in which results reveal that circSLC8A1 was down-regulated in bladder cancer tissues and cell lines, and circSLC8A1 inhibited bladder cancer cell migration, invasion and proliferation [Citation11]. These findings together with our results suggest that circSLC8A1 exerts anti-tumor activity in NSCLC. According to the previous studies, circSLC8A1 executes its roles via several mechanisms. Importantly, circSLC8A1 acts as an miRNA sponge and regulates the expression of its target gene, thus contributing to the inhibition of tumorigenesis. Therefore, we used an online software to predict the potential target miRNAs of circSLC8A1. The results indicated that an oncogenic miRNA, miR-106b-5p, might be a target of circSLC8A1. Further investigations showed that circSLC8A1 was able to bind to miR-106b-5p, thereby inhibiting the miR-106b-5p expression in NSCLC cells. These results indicate that circSLC8A1 might exert its tumor-suppressive role by sponging miR-106b-5p. Thus, we revealed a novel regulatory axis constituted by circSLC8A1/miR-106b-5p in NSCLC.
MiR-106b-5p has been identified as an oncogene in several cancers. miR-106b-5p is frequently overexpressed in hepatocellular carcinoma tissues. Overexpression of miR-106b-5p markedly promotes hepatocellular carcinoma cell proliferation and invasion in vitro [Citation23]. Another study revealed that the expression of miR-106b-5p is up-regulated in clear cell renal cell carcinoma (ccRCC). miR-106b-5p increases the spheres formation ability, as well as promotes tumor growth and the number of metastatic colonies in the lungs [Citation24]. Additionally, it has been shown that its expression is significantly upregulated in glioma cell lines and plays a crucial role in the development and progression of glioma [Citation25]. In accordance with previous studies, our findings also showed that miR-106b-5p expression was markedly increased in NSCLC tissues and NSCLC cell lines. Moreover, miR-106b-5p reversed the inhibitory effects of circSLC8A1 on cell proliferation and invasion, suggesting that circSLC8A1 may inhibit NSCLC progression via sponging of miR-106b-5p.
As miRNAs have been demonstrated to perform their biological functions by targeting their target genes, identifying the target genes of miR-106b-5p is crucial for exploring the functional mechanism of miR-106b-5p in NSCLC. The results predicted by TargetScan indicated that FOXJ3 might be a target gene of miR-106b-5p. MiR-106b-5p directly bound to the 3ʹUTR of FOXJ3 and suppressed its expression. Notably, FOXJ3 has been implicated in the progression of NSCLC [Citation26]. In this study, our results showed that knockdown of FOXJ3 reversed the inhibitory effects of miR-106b-5p inhibitor on cell proliferation and invasion. The results indicated that miR-106b-5p exerted its tumor promotive effects via targeting FOXJ3 in NSCLC.
In conclusion, we found that circSLC8A1 is frequently lowly expressed in NSCLC tissues. Overexpression of circSLC8A1 suppressed NSCLC cell proliferation and invasion in vitro. Further experiments revealed that the tumor suppressive activity of circSLC8A1 was mediated by the miR-106b-5p/FOXJ3 axis. These data imply that the circSLC8A1/miR-106b-5p/FOXJ3 axis might represent a promising therapeutic target for the management of NSCLC.
Ethical approval
This study was approved by the Ethics Committee at HanZhong Central Hospital (HanZhong, China).
Informed consent from participants
Written informed consent was obtained from the patients for their anonymized information to be published in this article.
Data availability statement for basic data sharing policy
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132.
- Hong QY, Wu GM, Qian GS, et al. Prevention and management of lung cancer in China. Cancer. 2015;121(Suppl 17):3080–3088.
- Liu X, Pu W, and He H et al,Novel ROR1 inhibitor ARI-1 suppresses the development of non-small cell lung cancer Cancer Lett. 2019;458:76–85.
- Collins L, Haines C, Perkel R, et al. Lung cancer: diagnosis and management. Am Fam Physician. 2007;75:56–63.
- Chen I, Chen CY, Chuang TJ. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev RNA. 2015;6:563–579.
- Ebbesen KK, Kjems J, Hansen TB. Circular RNAs: identification, biogenesis and function. Biochim Biophys Acta. 2016;1859:163–168.
- Wang Y, Liu J, Ma J, et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer. 2019;18:116.
- Kristensen LS, Hansen TB, Venø MT, et al. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565.
- He J, Xie Q, Xu H, et al. Circular RNAs and cancer. Cancer Lett. 2017;396:138–144.
- Bi W, Huang J, Nie C, et al. CircRNA circRNA_102171 promotes papillary thyroid cancer progression through modulating CTNNBIP1-dependent activation of β-catenin pathway. J Exp Clin Cancer Res Cr. 2018;37:275.
- Lu Q, Liu T, Feng H, et al. Circular RNA circSLC8A1 acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer progression via regulating PTEN. Mol Cancer. 2019;18:111.
- Di X, Jin X, Li R, et al. CircRNAs and lung cancer: biomarkers and master regulators. Life Sci. 2019;220:177–185.
- Chen D, Wei M, Ke Z, et al. CircRNA hsa_circ_100395 regulates miR-1228/TCF21 pathway to inhibit lung cancer progression. Cell Cycle. 2018;17:2080–2090.
- Liu W, Ma W, Yuan Y, et al. Circular RNA hsa_circRNA_103809 promotes lung cancer progression via facilitating ZNF121-dependent MYC expression by sequestering miR-4302. Biochem Biophys Res Commun. 2018;500:846–851.
- Zhang H, Wang X, Hu B, et al. Circular RNA ZFR accelerates non-small cell lung cancer progression by acting as a miR-101-3p sponge to enhance CUL4B expression. Artifi Cells. 2019;47. 3410–3416.
- Jiang -M-M, Mai Z-T, Wan S-Z, et al. Microarray profiles reveal that circular RNA hsa_circ_0007385 functions as an oncogene in non-small cell lung cancer tumorigenesis. J Cancer Res Clin Oncol. 2018;144:667–674.
- Liu YT, Han XH, Xing PY, et al. Circular RNA profiling identified as a biomarker for predicting the efficacy of Gefitinib therapy for non-small cell lung cancer. J Thorac Dis. 2019;11:1779–1787.
- Chen T, Luo J, Gu Y, et al. Comprehensive analysis of circular RNA profiling in AZD9291-resistant non-small cell lung cancer cell lines. Thorac Cancer. 2019;10:930–941.
- An J, Shi H, Zhang N, et al. Elevation of circular RNA circ_0003645 forecasts unfavorable prognosis and facilitates cell progression via miR-1179/TMEM14A pathway in non-small cell lung cancer. Biochem Biophys Res Commun. 2019;511:921–925.
- Wan J, Hao L, Zheng X, et al. Circular RNA circ_0020123 promotes non-small cell lung cancer progression by acting as a ceRNA for miR-488-3p to regulate ADAM9 expression. Biochem Biophys Res Commun. 2019;515:303–309.
- Wang T, Wang X, Du Q, et al. The circRNA circP4HB promotes NSCLC aggressiveness and metastasis by sponging miR-133a-5p. Biochem Biophys Res Commun. 2019;513:904–911.
- Wang L, Ma H, Kong W, et al. Up-regulated circular RNA VANGL1 contributes to progression of non-small cell lung cancer through inhibition of miR-195 and activation of Bcl-2. Biosci Rep. 2019;39:BSR20182433.
- Gu H, Gu S, Zhang X, et al. miR6b promotes aggressive progression of hepatocellular carcinoma via targeting RUNX3. Cancer Med. 2019;8:6756–6767.
- Lu J, Wei JH, Feng ZH, et al. miR-106b-5p promotes renal cell carcinoma aggressiveness and stem-cell-like phenotype by activating Wnt/β-catenin signalling. Oncotarget. 2017;8:21461.
- Liu F, Gong J, Huang W, et al. MicroRNA-106b-5p boosts glioma tumorigensis by targeting multiple tumor suppressor genes. Oncogene. 2014;33:4813–4822.
- Jin J, Zhou S, Li C, et al. MiR-517a-3p accelerates lung cancer cell proliferation and invasion through inhibiting FOXJ3 expression. Life Sci. 2014;108:48–53.