ABSTRACT
Laryngeal squamous cell carcinoma (LSCC) is an aggressive malignancy with highly mortality rate. Long non-coding RNA (lncRNA) AGAP2-AS1 is an identified oncogene in several types of cancers. However, the role of AGAP2-AS1 in LSCC remains unclear. The expression levels of AGAP2-AS1 in LSCC tissues and cell lines were measured using qRT-PCR. AGAP2-AS1 was knocked down in LSCC cells through transfection with siRNA-AGAP2-AS1. Cell proliferation and invasion were detected using MTT and transwell assays. Dual-luciferase reporter gene assay was performed to confirm the interaction with AGAP2-AS1 and downstream genes. Our results showed that AGAP2-AS1 expression was remarkably increased in human LSCC tissues and cell lines. Knockdown of AGAP2-AS1 significantly inhibited the proliferation and invasion of LSCC cells. In addition, AGAP2-AS1 sponged miR-193a-3p and regulated its expression in LSCC cells. Inhibition of miR-193a-3p reversed the effects of AGAP2-AS1 knockdown on LSCC cells. Furthermore, Lysyl oxidase-like 4 (LOXL4) was a target gene of miR-193a-3p and the role of miR-193a-3p was mediated by LOXL4. In conclusion, these findings suggest that knockdown of AGAP2-AS1 inhibited the proliferation and invasion of LSCC cells through regulating the miR-193a-3p/LOXL4 axis.
1. Introduction
Laryngeal squamous cell carcinoma (LSCC) is a highly aggressive malignancy that accounts for 5% of all systemic malignant tumors [Citation1]. Although encouraging progress in the LSCC treatment has been achieved in the past decades, the overall survival rate of supraglottic and subglottic LSCC remains unfavorable [Citation2,Citation3]. Lymphatic metastasis is an important reason for the poor prognosis of LSCC [Citation4]. Hence, preventing lymphatic metastasis has become a hot approach in the treatment of LSCC.
Long non-coding RNAs (lncRNAs) are arbitrarily defined by their size exceeding 200 nts and apparent lack of protein-coding capacity [Citation5]. LncRNAs exert their functions through interactions with other components such as proteins, RNAs and DNAs [Citation6]. Accumulating evidences indicate that lncRNAs are implicated in various human cancers and involved in the tumorigenesis and metastasis [Citation7,Citation8]. Previous studies have suggested the involvement of lncRNAs in LSCC, however, the overall pathophysiological contribution of lncRNAs to LSCC is largely unknown [Citation9].
LncRNA AGAP2-AS1 is an antisense lncRNA transcribed from a gene located at 12q14.1 with the length of 1567 nt. It has been demonstrated that AGAP2-AS1 is associated with the tumorigenesis of several types of cancers, such as gastric cancer [Citation10], non-small-cell lung cancer (NSCLC) [Citation11,Citation12], breast cancer [Citation13,Citation14], glioblastoma multiforme [Citation15], ovarian carcinoma [Citation16], pancreatic cancer [Citation17], hepatocellular carcinoma (HCC) [Citation18]. AGAP2-AS1 expression is up-regulated in HCC tissues, especially in metastatic and recurrent cases. AGAP2-AS1 promotes cell proliferation, migration, invasion, epithelial-mesenchymal transition (EMT) progression and inhibits apoptosis of HCC cells [Citation18]. AGAP2-AS1 is highly expressed in the gastric cancer tissues and associated with poor prognosis and short overall survival. Knockdown of AGAP2-AS1 significantly inhibits cell proliferation, migration, and invasion in gastric cancer cells and suppresses tumor growth in vivo [Citation10]. AGAP2-AS1 expression level was found to be significantly upregulated in NSCLC tissues and negatively correlated with poor prognostic outcomes. Loss-of-function assays reveal that AGAP2-AS1 knockdown inhibits cell proliferation, migration and invasion, and induces cell apoptosis [Citation12]. These findings suggest that AGAP2-AS1 may be an oncogene and promotes metastasis in these cancers.
In the present study, we investigated the role of AGAP2-AS1 in LSCC. The results showed that AGAP2-AS1 expression was upregulated in LSCC tissues. Knockdown of AGAP2-AS1 inhibited cell proliferation and invasion in LSCC cells, implying that AGAP2-AS1 promotes the metastasis of LSCC.
2. Materials and methods
2.1 Clinical tissues and cell culture
The present study was approved by the Ethics Committee of First Affiliated Hospital, Xi’an Jiaotong University College of Medicine (No. 2,016,021; Xi’an, China). A total of 23 patients with LSCC underwent partial or total laryngectomy was recruited in the present study. The LSCC tissues and corresponding adjacent non-tumor tissues were collected from the patients at First Affiliated Hospital, Xi’an Jiaotong University College of Medicine between March 2016 and February 2018. All involved patients have signed informed consent. Tissue samples were resected and then immediately frozen in liquid nitrogen, followed by storage at −80°C until RNA extraction.
LSCC cell lines including Tu-212, AMC-HN-8 and Tu-177 cells, as well the normal bronchial epithelial cell line (16HBE) were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured in the RPMI-1640 medium (Gibco, Grand Island, NY, USA) supplemented with 0.1% penicillin/streptomycin and 10% fetal bovine serum (FBS; Hyclone, Logan, UT, USA).
2.2 Cell transfection
Small interfering RNA (siRNA) targeting AGAP2-AS1 (si-AGAP2-AS1) and control siRNA (si-NC) were chemical synthesized by RiboBio Co., Ltd. (Guangzhou, China). MiR-193a-3p mimics, miR-193a-3p inhibitor, and negative controls were synthesized by GenePharma (Shanghai, China). Lysyl oxidase-like 4 (LOXL4)-overexpressing plasmid (pcDNA 3.1-LOXL4) was constructed in our laboratory. For cell transfection, AMC-HN-8 and Tu-177 cells (5 × 105 cells/well) were seeded in 6-well plates, followed by overnight incubation at 37°C. Then cells were transfected with si-AGAP2-AS1, si-AGAP2-AS1+ miR-193a-3p inhibitor, miR-193a-3p mimics, miR-193a-3p mimics+pcDNA 3.1-LOXL4 and si-AGAP2-AS1+ pcDNA 3.1-LOXL4 using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Waltham, MA, USA) following to the manufacturer’s instructions. The concentrations of miR-193a-3p mimics, miR-193a-3p inhibitor, and negative controls for transfection were 50 nM. The concentrations of si-AGAP2-AS1 and pcDNA 3.1-LOXL4 for transfection were 100 nM.
2.3 Cell proliferation assay
After 24 h post-transfection, AMC-HN-8 and Tu-177 cells (1 × 104 cells/well) were seeded into 96-well plates and cultured for 0, 24, 48 and 72 h. At the end period of the indicated time-point, 0.5 mg/ml MTT solution (Sigma-Aldrich, St. Louis, MO, USA) was added and incubated for 4 h. After that, the medium was removed and 150 µl dimethyl sulfoxide (DMSO) was added, followed by shaking for 15 min at room temperature. A microplate reader (Thermo) was used to measure the absorbance at the wavelength of 490 nm.
2.4 Cell invasion assay
To detect cell invasion, the chamber was precoated with Matrigel (BD Biosciences). AMC-HN-8 and Tu-177 cells (2 × 104/well) suspended in 200 µl serum-free RPMI-1640 medium were seeded into the upper chamber of a Transwell apparatus (BD Biosciences, Franklin Lakes, NJ, USA). The lower chamber was filled with normal medium containing 15% FBS. After incubation for 24 h, the cells invaded the membrane were fixed with 95% ethanol for 30 min and stained with 0.1% crystal violet. Three independent fields were photographed, and the cell numbers were counted.
2.5 RNA extraction and qRT-PCR assay
Total RNA was isolated using the TRIzol reagent (Invitrogen) and the concentration was measured with a DS-11 Plus Spectrophotometer (DeNovix, Wilmington, Delaware, USA). 5 μg RNA of each sample was reverse-transcribed to cDNA using the PrimeScript® cDNA synthesis kit (Takara, Dalian, China) according to the manufacturer’s instructions. qPCR was performed in a final volume of 10 μl, which consisted of 5 μl SsoFast™ EvaGreen Supermix (Bio-Rad Laboratories, Inc., Hercules, CA, USA), 1 μl cDNA (1:50 dilution) and 2 μl each of the forward and reverse primers (1 mM). The qRT-PCR was performed on ABI7700 mRNA detector (Applied Biosystems, Foster, CA, USA). The relative expression levels were calculated by 2−ΔΔCt method with the references of GAPDH or U6. The primers were shown as follows: AGAP2-AS1, forward primers 5ʹ-TACCTTGACCTTGCTGCTCTC-3ʹ and reverse primers 5ʹ-TGTCCCTTAATGACCCCATCC-3ʹ; GAPDH, forward primers 5ʹ- GGGAGCCAAAAGGGTCAT-3ʹ and reverse primers 5ʹ- GAGTCCTTCCACGATACCAA-3ʹ; miR-193a-3p, forward primers 5ʹ- ACTGGCCTACAAAGTCCCAGT-3ʹ and reverse primers 5ʹ- GTGCAGGGTCCGAGGT-3ʹ; U6, forward primers 5ʹ-CTCGCTTCGGCAGCACA-3ʹ and reverse primers 5ʹ-AACGCTTCACGAATT TGCGT-3ʹ. The PCR conditions included an initial denaturation step of 94°C for 2 min, followed by 35 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 2 min, and a final elongation step of 72°C for 10 min.
2.6 Western blot
The whole LSCC cells lysates were prepared using RIPA lysis buffer (Sangon Biotech, Shanghai, China). The protein concentration was measured using a bicinchoninic acid (BCA) protein assay kit (Sangon Biotech). Proteins (50 μg) were separated on 10% SDS-PAGE, and then electrophoretically transferred onto polyvinylidene difluoride (PVDF) membranes (Whatman, Maidstone, UK). Next, the membranes were blocked in 5% skim milk for 1 h and then incubated with the primary antibodies against LOXL4 or GAPDH (Invitrogen) overnight at 4°C. Then, the membranes were incubated with secondary antibody (Invitrogen) for 1 h at room temperature. Finally, the immunoreactivity was visualized by enhanced chemiluminescence (ECL) Western blotting Substrate (Thermo).
2.7 Target prediction
The putative target miRNAs of AGAP2-AS1 were predicted using the Starbase (http://starbase.sysu.edu.cn/). The putative target genes of miR-193a-3p were predicted using the TargetScan (http://www.targetscan.org) and miRanda (http://www.microrna.org/microrna/).
2.8 Luciferase reporter assay
Based on the results from bioinformatics analysis, miR-193a-3p was found to be putative target miRNA of AGAP2-AS1. The wild type (Wt) or mutant (Mut) sequences of AGAP2-AS1 was inserted into pGL3 vector to generate pGL3-AGAP2-AS1-Wt and pGL3-AGAP2-AS1-Mut. The pGL3-AGAP2-AS1-Wt or pGL3-AGAP2-AS1-Mut together with miR-193a-3p mimics or control mimics were co-transfected into cells.
Bioinformatics analysis identified LOXL4 as a potential target gene of miR-193a-3p. The Wt or Mut sequences of 3ʹ-untranslated region (UTR) of LOXL4 harboring the binding sites was cloned into pGL3 vector to construct pGL3-LOXL4-Wt and pGL3-LOXL4-Mut. The pGL3-LOXL4-Wt/pGL3-LOXL4-Mut and miR-193a-3p mimics/control mimics were co-transfected into cells.
The transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Luciferase activities were determined using the Dual-Luciferase Reporter Assay system (Promega) at 48 h post-transfection.
2.9 Statistical analysis
All of the statistical analyses were performed using the SPSS 20.0 software (IBM, New York, NY, USA). Data were reported as means ± standard deviation (SD) from at least three independent experiments. The Shapiro-Wilk test was used for analyzing data normality. Data were analyzed using Students t-test for two groups or one-way analysis of variance (ANOVA) for multiple groups. Differences were considered statically significant when p values were less than 0.05.
3. Results
3.1 AGAP2-AS1 is highly expressed in LSCC tissues and cell lines
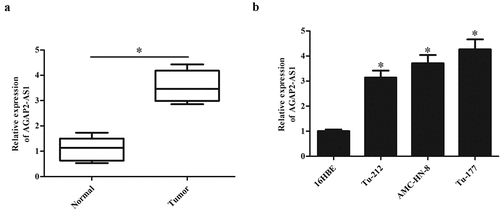
The expression levels of AGAP2-AS1 were evaluated by qRT-PCR in 23 paired LSCC tissues and adjacent normal tissues. As shown in ), AGAP2-AS1 expression was significantly higher in LSCC tissues than that in corresponding adjacent non-tumor tissues (p < 0.05). In addition, compared with the normal bronchial epithelial cell line (16HBE), AGAP2-AS1 expressions were markedly higher in LSCC cell lines, especially in AMC-HN-8 and Tu-177 cells ()).
Figure 1. AGAP2-AS1 expression in clinical tissues and cell lines. (a) The expression levels of AGAP2-AS1 were evaluated by qRT-PCR in 23 paired LSCC tissues and adjacent non-tumor tissues. (b) AGAP2-AS1 expression in the normal bronchial epithelial cell line 16HBE and LSCC cell lines including Tu-212, AMC-HN-8 and Tu-177 cells. * p < 0.05.
3.2 Downregulation of AGAP2-AS1 inhibited the proliferation and invasion of LSCC cells
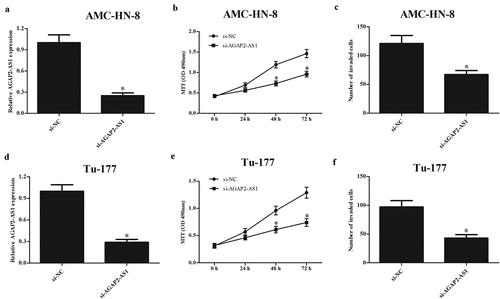
To investigate the role of AGAP2-AS1 in LSCC, AMC-HN-8 and Tu-177 cells were transfected with si-AGAP2-AS1. The qRT-PCR showed that AGAP2-AS1 expression was greatly decreased in si-AGAP2-AS1-transfected AMC-HN-8 and Tu-177 cells, respectively (-d)). In addition, the results of MTT assay indicated that the proliferation of AMC-HN-8 and Tu-177 cells was dramatically decreased by si-AGAP2-AS1 (-e)). Furthermore, knockdown of AGAP2-AS1 also significantly suppressed the invasion of AMC-HN-8 and Tu-177 cells, respectively ( and 2f).
Figure 2. Downregulation of AGAP2-AS1 inhibited the proliferation and invasion of LSCC cells. (a and d) AGAP2-AS1 was knocked down in AMC-HN-8 and Tu-177 cells through transfection with si-AGAP2-AS1. (b and e) MTT assay was performed to detect the proliferation of AMC-HN-8 and Tu-177 cells. (c and f) Cell invasion of AMC-HN-8 and Tu-177 cells was evaluated by transwell assay. * p < 0.05.
3.3 AGAP2-AS1 targeted miR-193a-3p in LSCC cells
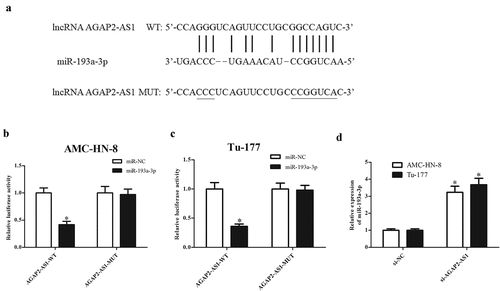
The potential molecular mechanism underlying the role of AGAP2-AS1 was analyzed by exploring its direct target miRNAs. According to the predicted results from bioinformatics analysis, miR-193a-3p was found to be putative target miRNA of AGAP2-AS1 ()). Results from luciferase reporter assay showed that miR-193a-3p mimics reduced the luciferase activity of pGL3-AGAP2-AS1-Wt in AMC-HN-8 and Tu-177 cells, respectively (-c)). Moreover, miR-193a-3p expression was obviously increased in si-AGAP2-AS1-transfected AMC-HN-8 and Tu-177 cells, indicating that AGAP2-AS1 might act as a sponge of miR-193a-3p ()).
Figure 3. AGAP2-AS1 acted as a sponge of miR-193a-3p in LSCC cells. (a) Predicted results from bioinformatics analysis showed that miR-193a-3p might be a putative target miRNA of AGAP2-AS1. (b) Luciferase reporter assay was performed to confirm the interaction with AGAP2-AS1 and miR-193a-3p in AMC-HN-8 cells. * p < 0.05 vs. AMC-HN-8 cells co-transfected with pGL3-AGAP2-AS1-Mut and miR-193a-3p mimics. (c) Luciferase reporter assay was performed to confirm the interaction with AGAP2-AS1 and miR-193a-3p in Tu-177 cells. * p < 0.05 vs. Tu-177 cells co-transfected with pGL3-AGAP2-AS1-Mut and miR-193a-3p mimics. (d) Knockdown of AGAP2-AS1 elevated miR-193a-3p expression in AMC-HN-8 and Tu-177 cells. *p < 0.05 vs. si-NC.
3.4 The expression of miR-193a-3p in LSCC tissues and cell lines
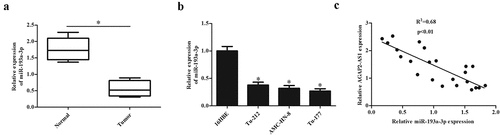
Next, we evaluated the miR-193a-3p expression in LSCC tissues and adjacent non-tumor tissues. Expression of miR-193a-3p was significantly downregulated in LSCC tissues ()). In addition, miR-193a-3p expressions were markedly down-regulated in LSCC cell lines, including Tu-212, AMC-HN-8, and Tu-177 cells ()). Furthermore, Spearman rank correlation assessment indicated an inverse correlation between AGAP2-AS1 and miR-193a-3p in LSCC tissues ()). The results showed that miR-193a-3p expression levels in LSCC tissues and cell lines were negatively associated with AGAP2-AS1.
Figure 4. MiR-193a-3p expression in clinical tissues and cell lines. (a) The expression levels of miR-193a-3p in 23 paired LSCC tissues and adjacent normal tissues. (b) MiR-193a-3p expression in 16HBE cells and LSCC cell lines (Tu-212, AMC-HN-8 and Tu-177 cells). * p < 0.05. (c) The correlation between AGAP2-AS1 and miR-193a-3p in LSCC tissues was using qRT-PCR.
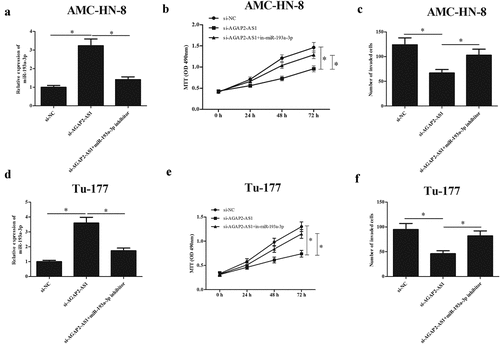
3.5 MiR-193a-3p inhibitor reversed the effects of AGAP2-AS1 knockdown on LSCC cell proliferation and invasion
To determine whether the inhibitory effects of si-AGAP2-AS1 were mediated by miR-193a-3p, we co-transfected miR-193a-3p inhibitor and si-AGAP2-AS1 in AMC-HN-8 and Tu-177 cells. As indicated in and 5d, transfection with miR-193a-3p inhibitor reversed the promotive effect of si-AGAP2-AS1 on miR-193a-3p expression in AMC-HN-8 and Tu-177 cells, respectively. Additionally, we found that the si-AGAP2-AS1-caused decrease in cell proliferation was mitigated by inhibition of miR-193a-3p in AMC-HN-8 and Tu-177 cells, respectively ( and 5e). Moreover, the reduced cell invasion of si-AGAP2-AS1-transfected AMC-HN-8 and Tu-177 cells were elevated by miR-193a-3p inhibition ( and 5f).
Figure 5. MiR-193a-3p inhibitor reversed the effects of si-AGAP2-AS1 on proliferation and invasion of LSCC cells. (a and d) MiR-193a-3p expression was suppressed in AMC-HN-8 and Tu-177 cells through transfection with miR-193a-3p inhibitor and si-AGAP2-AS1. (b and e) MTT assay was performed to detect the proliferation of AMC-HN-8 and Tu-177 cells. (c and f) Cell invasion of AMC-HN-8 and Tu-177 cells was evaluated by transwell assay. * p < 0.05.
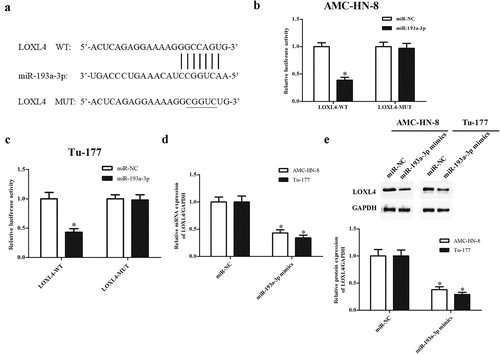
3.6 MiR-193a-3p targeted LOXL4 in LSCC cells
Bioinformatics analysis was then carried out to predict the downstream gene of miR-193a-3p, and the results indicated that LOXL4 might be potential target gene of miR-193a-3p ()). Luciferase reporter assay demonstrated that luciferase activity of pGL3-LOXL4-Wt was markedly reduced by co-transfection with miR-193a-3p mimics in AMC-HN-8 and Tu-177 cells, while miR-193a-3p mimics did not affect the luciferase activity of pGL3-LOXL4-Mut (-c)). Furthermore, the expression of LOXL4 at both mRNA and protein levels was significantly decreased by miR-193a-3p mimics in AMC-HN-8 and Tu-177 cells, respectively (). These findings suggested that miR-193a-3p directly targeted LOXL4 and suppressed its expression.
Figure 6. LOXL4 was a target gene of miR-193a-3p targeted in LSCC cells. (a) Bioinformatics analysis predicted that LOXL4 might be potential target gene of miR-193a-3p. (b) Luciferase reporter assay was performed to confirm the interaction with miR-193a-3p and LOXL4 in AMC-HN-8 cells. * p < 0.05 vs. AMC-HN-8 cells co-transfected with pGL3-LOXL4-Mut and miR-193a-3p mimics. (c) Luciferase reporter assay was performed to confirm the interaction with miR-193a-3p and LOXL4 in Tu-177 cells. * p < 0.05 vs. Tu-177 cells co-transfected with pGL3-LOXL4-Mut and miR-193a-3p mimics. (d) MiR-193a-3p suppressed the mRNA expression levels of LOXL4 in AMC-HN-8 and Tu-177 cells. (e) MiR-193a-3p suppressed the protein expression levels of LOXL4 in AMC-HN-8 and Tu-177 cells. *p < 0.05 vs. miR-NC.
3.7 LOXL4 reversed the effects of miR-193a-3p mimics or AGAP2-AS1 knockdown on LSCC cell proliferation and invasion
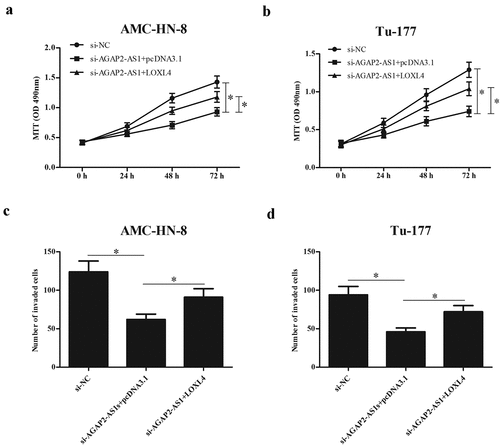
Since LOXL4 was a target gene of miR-193a-3p, we then determined whether or not the role of miR-193a-3p was mediated by LOXL4. We examined the expression of LOXL4 in LSCC tissues and found that the expression levels of LOXL4 were significantly up-regulated in LSCC tissues ()). Then, to determine whether the regulatory effects of miR-193a-3p were mediated by LOXL4, miR-193a-3p mimics and LOXL4-overexpressing plasmid (pcDNA 3.1-LOXL4) were co-transfected into AMC-HN-8 and Tu-177 cells. As indicated in ), overexpression of LOXL4 greatly up-regulated the protein expression of LOXL4 in miR-193a-3p mimics-transfected AMC-HN-8 and Tu-177 cells, respectively. Furthermore, we showed that overexpression of miR-193a-3p significantly inhibited the proliferation and invasion of AMC-HN-8 and Tu-177 cells. However, the inhibitory effects of miR-193a-3p on cell proliferation and invasion were reversed by LOXL4 overexpression (). Similarly, the AGAP2-AS1-knockdown-mediated effects were also partially reversed by LOXL4 overexpression ().
Figure 7. LOXL4 reversed the effects of miR-193a-3p mimics on proliferation and invasion of LSCC cells. (a) The expression levels of LOXL4 were evaluated by qRT-PCR in 23 paired LSCC tissues and adjacent non-tumor tissues. miR-193a-3p mimics and LOXL4-overexpressing plasmid (pcDNA 3.1-LOXL4) were co-transfected into AMC-HN-8 and Tu-177 cells. (b) The protein expression level of LOXL4 was measured using Western blot. (c and d) MTT assay was performed to detect the proliferation of AMC-HN-8 and Tu-177 cells. (e and f) Cell invasion of AMC-HN-8 and Tu-177 cells was evaluated by transwell assay. * p < 0.05.
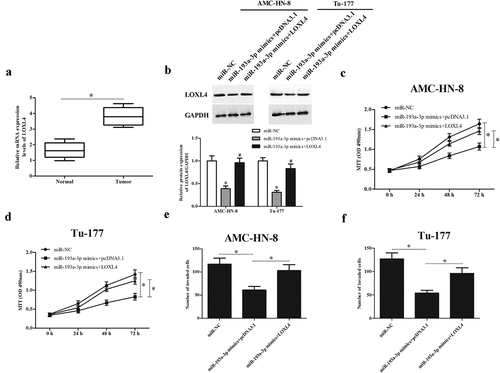
Figure 8. LOXL4 reversed the effects of AGAP2-AS1 knockdown on proliferation and invasion of LSCC cells. Si-AGAP2-AS1 and LOXL4-overexpressing plasmid (pcDNA 3.1-LOXL4) were co-transfected into AMC-HN-8 and Tu-177 cells. (a and b) MTT assay was performed to detect the proliferation of AMC-HN-8 and Tu-177 cells. (c and d) Cell invasion of AMC-HN-8 and Tu-177 cells was evaluated by transwell assay. * p < 0.05.
4. Discussion
Metastasis, characterized as transfer of tumor cells from the primary site to one distant site, is considered as the most important reason for death [Citation19]. In recent years, a number of researchers have attempted to reveal the biological and clinical relevance of lncRNAs in the tumorigenesis and metastasis of LSCC. Significant downregulation of lncRNA AC008440.10 has been detected in LSCC tumor and metastatic lymph node in advanced stage of patient. The downregulation of lncRNA AC008440.10 is correlated with the increasing risk of metastasis, poor prognosis and patient survival [Citation20]. The expression of lncRNA SNHG1 in LSCC tissues is significantly increased compared with that in para-carcinoma tissues. The up-regulation of SNHG1 contributes to proliferation and metastasis in LSCC [Citation21]. LncRNA RP11–169D4.1 expression is markedly decreased in LSCC tissues and cell lines. Overexpression of RP11–169D4.1 inhibits the proliferation, migration and invasion of LSCC cell lines, indicating that RP11–169D4.1 acts as a metastasis suppressor in LSCC [Citation22].
In the present study, we aimed to evaluate the role of an oncogenic lncRNA, AGAP2-AS1, in the metastasis of LSCC. We found that AGAP2-AS1 expression was highly expressed in LSCC tissues and cell lines. Then we investigate the function of AGAP2-AS1 in vitro using loss-of-function assay. Metastasis is a biologically multifaceted process involving a range of cellular processes. Promoted cell proliferation, migration, invasion, angiogenesis and EMT are considered as vital processes for the metastatic expansion. Our results proved that knockdown of AGAP2-AS1 inhibited cell proliferation and invasion in LSCC cells, implying that AGAP2-AS1 might promote the metastasis of LSCC.
It has been acknowledged that AGAP2-AS1 exerts its roles via sponging miRNAs. AGAP2-AS1 promotes the proliferation and inhibits apoptosis of glioma cells by sponging miR-15a/b-5p [Citation23]. AGAP2-AS1 promotes cell proliferation, migration, invasion, EMT progression and inhibited apoptosis of HCC cells through acting as a competing endogenous RNA (ceRNA) by sponging miR-16–5p [Citation18]. We used bioinformatics analysis to predict the target miRNA of AGAP2-AS1. The results showed that AGAP2-AS1 directly bound to miR-193a-3p and suppressed the miR-193a-3p expression.
Previously, miR-193a-3p has been demonstrated to be a tumor suppressor in various cancers. MiR-193a-3p expression levels are significantly decreased in human colorectal cancer (CRC) cell lines compared with that in normal colonic epithelium cell line. MiR-193a-3p inhibits cell growth, migration and angiogenesis of CRC cells [Citation24]. MiR-193a-3p expression is obviously decreased in the NSCLC tumor tissues. Enforced expression of miR-193a-3p inhibits tumor formation and suppresses cell proliferation and cell migration [Citation25]. MiR-193a-3p was found to serve as useful biomarker for the diagnosis of osteosarcoma and may be potential candidate for the treatment of metastatic osteosarcoma [Citation26]. In the current study, we demonstrated that AGAP2-AS1 expressions were markedly up-regulated in LSCC tissues and cell lines, which were negatively associated with AGAP2-AS1. In addition, miR-193a-3p inhibitor reversed the effects of AGAP2-AS1 knockdown on LSCC cell proliferation and invasion. These findings suggest that AGAP2-AS1 promotes LSCC metastasis via sponging miR-193a-3p.
LOXL4 is a member of lysyl oxidase (LOX) family that has diverse critical biological functions, especially its roles in tumor development and metastasis [Citation27,Citation28]. Up-regulation of LOXL4 expression is a frequent event in gastric cancer progression, contributes to tumor cell proliferation and metastasis [Citation29]. Li et al. reported that LOXL4 promotes HCC cell invasion and metastasis, and angiogenesis in HCC [Citation30]. LOXL4 is selective up-regulated in head and neck squamous cell carcinomas (HNSCC) and significantly correlated with lymph node metastases and higher tumor stages [Citation31]. Additionally, LOXL4 is upregulated in tumor tissue samples of LSCC [Citation32]. Our results showed that LOXL4 was a target gene of miR-193a-3p and regulated by miR-193a-3p in LSCC cells. LOXL4 reversed the inhibitory effects of miR-193a-3p mimics or AGAP2-AS1 knockdown on LSCC cell proliferation and invasion. The results indicated that the role of miR-193a-3p was mediated by LOXL4.
In conclusion, the results demonstrated that AGAP2-AS1 is highly expressed in LSCC tissues and cell lines. AGAP2-AS1 promoted the proliferation and invasion of LSCC cells through regulating miR-193a-3p/LOXL4 axis. These findings suggested that AGAP2-AS1 might be a therapeutic target for LSCC treatment.
Acknowledgments
This study was funded by Natural Science Foundation of China (No. 81803317, 81971766), Natural Science Basic Research Plan in Shaanxi Province (No.2019JQ-957), and Fundamental Research Funds for the Central Universities, China (xjj2018094).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Nadal A, Cardesa A. Molecular biology of laryngeal squamous cell carcinoma. Virchows Arch. 2003;442(1):1–7.
- Marioni G, Marchese-Ragona R, Kleinsasser NH, et al. Partial laryngeal surgery in recurrent carcinoma. Acta Oto-Laryngologica. 2015;135(2):119–124.
- Sewnaik A, van Den Brink JL, Wieringa MH, et al. Surgery for recurrent laryngeal carcinoma after radiotherapy: partial laryngectomy or total laryngectomy for a better quality of life? Otolaryngol Head Neck Surg. 2005;132(1):95–98.
- Schlüter A, Weller P, Kanaan O, et al. CD31 and VEGF are prognostic biomarkers in early-stage, but not in late-stage, laryngeal squamous cell carcinoma. BMC Cancer. 2018;18(1):272.
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159.
- Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346.
- Nina H, Damjan G. Long non-coding RNA in cancer. Int J Mol Sci. 2013;14(3):4655–4669.
- Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9(6):703–719.
- Yang QQ, Deng YF. Long non-coding RNAs as novel biomarkers and therapeutic targets in head and neck cancers. Int J Clin Exp Pathol. 2014;7(4):1286–1292.
- Qi F, Liu X, Wu H, et al. Long noncoding AGAP2-AS1 is activated by SP1 and promotes cell proliferation and invasion in gastric cancer. J Hematol Oncol. 2017;10:48.
- Fan KJ, Liu Y, Yang B, et al. Prognostic and diagnostic significance of long non-coding RNA AGAP2-AS1 levels in patients with non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2017;21(10):2392–2396.
- Li W, Sun M, Zang C, et al. Upregulated long non-coding RNA AGAP2-AS1 represses LATS2 and KLF2 expression through interacting with EZH2 and LSD1 in non-small-cell lung cancer cells. Cell Death Dis. 2016;7(5):e2225.
- Zheng Z, Chen M, Xing P, et al. Increased Expression of Exosomal AGAP2-AS1 (AGAP2 Antisense RNA 1) In Breast Cancer Cells Inhibits Trastuzumab-Induced Cell Cytotoxicity. Med Sci Monit. 2019;25:2211–2220.
- Dong H, Wang W, Mo S, et al. SP1-induced lncRNA AGAP2-AS1 expression promotes chemoresistance of breast cancer by epigenetic regulation of MyD88. J Exp Clin Cancer Res. 2018;37(1):202.
- Tian Y, Zheng Y, Dong X. AGAP2-AS1 serves as an oncogenic lncRNA and prognostic biomarker in glioblastoma multiforme. J Cell Biochem. 2019;120(6):9056–9062.
- Chen J, Peng X, Dai Y. The Long Non-Coding RNA (lncRNA) AGAP2-AS1 is Upregulated in Ovarian Carcinoma and Negatively Regulates lncRNA MEG3. Med Sci Monit. 2019;25:4699–4704.
- Hui B, Ji H, Xu Y, et al. RREB1-induced upregulation of the lncRNA AGAP2-AS1 regulates the proliferation and migration of pancreatic cancer partly through suppressing ANKRD1 and ANGPTL4. Cell Death Dis. 2019;10(3):207.
- Liu Z, Wang Y, Wang L, et al. Long non-coding RNA AGAP2-AS1, functioning as a competitive endogenous RNA, upregulates ANXA11 expression by sponging miR-16–5p and promotes proliferation and metastasis in hepatocellular carcinoma. J Exp Clin Cancer Res. 2019;38(1):194.
- Talebi A, Masoodi M, Mirzaei A, et al. Biological and clinical relevance of metastasis-associated long noncoding RNAs in esophageal squamous cell carcinoma: a systematic review. J Cell Physiol. 2019;235(2):848–868.
- Chen J, Shen Z, Deng H, et al. Long non-coding RNA biomarker for human laryngeal squamous cell carcinoma prognosis. Gene. 2018;671:96–102.
- Lin SX, Jiang H, Xiang GZ, et al. Up-regulation of long non-coding RNA SNHG1 contributes to proliferation and metastasis in laryngeal squamous cell carcinoma. Eur Rev Med Pharmacol Sci. 2018;22(5):1333–1341.
- Zhao J, Lv K, Li Z-H, et al. Functional significance of the long non-coding RNA RP11–169D4.1 as a metastasis suppressor in laryngeal squamous cell carcinoma by regulating CDH1. Oncol Rep. 2017;38(1):211–220.
- Zheng Y, Lu S, Xu Y, et al. Long non-coding RNA AGAP2-AS1 promotes the proliferation of glioma cells by sponging miR-15a/b-5p to upregulate the expression of HDGF and activating Wnt/beta-catenin signaling pathway. Int J Biol Macromol. 2019;128:521–530.
- Lin M, Zhang Z, Gao M, et al. MicroRNA-193a-3p suppresses the colorectal cancer cell proliferation and progression through downregulating the PLAU expression. Cancer Manag Res. 2019;11:5353–5363.
- Fan Q, Hu X, Zhang H, et al. MiR-193a-3p is an Important Tumour Suppressor in Lung Cancer and Directly Targets KRAS. Cell Physiol Biochem. 2017;44(4):1311–1324.
- Pu Y, Zhao F, Cai W, et al. MiR-193a-3p and miR-193a-5p suppress the metastasis of human osteosarcoma cells by down-regulating Rab27B and SRR, respectively. Clin Exp Metastasis. 2016;33(4):359–372.
- Wang TH, Hsia SM, Shieh TM. Lysyl Oxidase and the Tumor Microenvironment. Int J Mol Sci. 2016;18(1):62.
- Barker HE, Cox TR, Erler JT. The rationale for targeting the LOX family in cancer. Nat Rev Cancer. 2012;12(8):540–552.
- Li RK, Zhao WY, Fang F, et al. Lysyl oxidase-like 4 (LOXL4) promotes proliferation and metastasis of gastric cancer via FAK/Src pathway. J Cancer Res Clin Oncol. 2015;141(2):269–281.
- Li R, Wang Y, Zhang X, et al. Exosome-mediated secretion of LOXL4 promotes hepatocellular carcinoma cell invasion and metastasis. Mol Cancer. 2019;18(1):18.
- Weise JB, Rudolph P, Heiser A, et al. LOXL4 is a selectively expressed candidate diagnostic antigen in head and neck cancer. Eur J Cancer. 2008;44(9):1323–1331.
- Yilmaz M, Suer I, Karatas OF, et al. Differential expression of LOXL4 in normal and tumour tissue samples of laryngeal squamous cell carcinoma. Clin Otolaryngol. 2016;41(3):206–210.
