ABSTRACT
Intervertebral disc degeneration (IDD) constitutes the pathological foundation of most musculoskeletal disorders of the spine. Previous studies have noted that cell proliferation is a common feature of IDD. Bioinformatics indicated that aberrantly expressed long non-coding RNAs (lncRNAs) were involved in the development of IDD. In this study, we aimed to investigate the function of lncRNA HOTAIR in the proliferation of human nucleus pulposus (NP) cells of IDD in vitro and further clarified its mechanism. The expression of HOTAIR and miR-130b was quantified by qRT-PCR in nucleus pulposus (NP) tissues. Furthermore, NP cells proliferation were assayed by CCK8 and Immunostaining. Dual-luciferase reporter and RIP assay were used to examine the expression of HOTAIR, PTEN, and their co-target gene miR-130b. Western blotting was used to test AKT expression. Our in vitro experiments on human normal NP cells observed that HOTAIR was significantly dysregulated in IDD. Further, HOTAIR can suppress proliferation by directly targeting miR-130b. In addition, Both HOTAIR and PTEN were confirmed to target miR-130b, and miR-130b upregulation reversed the phenomenon of ectopic expression of HOTAIR. More importantly, HOTAIR upregulation significantly reduced CyclinD1 protein expression by PTEN/AKT signaling pathway. Our findings suggest that HOTAIR may bind to miR-130b and subsequently increased CyclinD1 expression via PTEN/Akt pathway. Thereby, HOTAIR could become a potential target for the treatment of IDD.
Abbreviations : IDD; intervertebral disc degeneration ncRNAs; non-coding RNAs lncRNAs; long non-coding RNAs miRNAs; microRNAs NP; nucleus pulposus qRT-PCR; quantitative reverse transcription–PCR LBP; Low back pain ORF; open reading frame HOTAIR; Hox transcript antisense intergenic RNA FAF1; Fas-associated protein factor-1 Erk; extracellular signal-regulated kinase TUG1; Taurine Up-regulated Gene 1 HIF1A hypoxia-inducible factor 1-alpha PI3K; phosphoinositide-3 kinase AIS; adolescent idiopathic scoliosis ECM; extracellular matrix LN;lupus nephritis CT;computed tomography MRI; magnetic resonance imaging PBS; phosphate-buffered salin PBS; phosphate-buffered salin PVDF; polyvinylidene fluoride TBST; Tris-buffered saline Tween ECL; enhanced chemiluminescence RIP; RNA immunoprecipitation
1. Introduction
Low back pain (LBP) is one of the most common symptoms of spinal abnormalities, with an annual point prevalence averaging 30% [Citation1]. Currently, it has become a major factor affecting quality of life and leads to an enormous socioeconomic burden worldwide. Although the etiology of LBP remains to be defined, intervertebral disc degeneration (IDD) is thought to be the most common cause. Morphologically, degenerated disc is marked by degradation of the extracellular matrix (ECM), by increased vascularization and innervation of anulus fibrosus, and by the appearance of cell clusters [Citation2]. Current therapy for IDD aims to relieve pain and control symptoms rather than interfere with its pathophysiology. Although several biological strategies capable of slowing mild and moderate disc degeneration have been developed, their efficacy and safety remain to be evaluated in clinical settings [Citation3]. Therefore, a better understanding of IDD’s pathogenesis is essential to develop effective treatments.
Healthy NP cells are sparsely distributed and relatively small, but their activity in synthesizing extracellular matrix (ECM) is crucial to disc function. Cell clusters of nucleus pulposus are a common feature of intervertebral disc degeneration [Citation2]. Exceptional proliferation has been considered as the main cause of cell cluster formation and delineated the complex mechanisms underlying IDD progression [Citation4]. Long non-coding RNAs (lncRNAs) are defined as RNA > 200 nucleotides in length without an open reading frame (ORF). Accumulating evidence suggests that lncRNAs are differentially expressed in human degenerative NP tissue and may play a critical role in modulating the progression of disc degeneration [Citation5]. Recently, Zhou and colleagues observed that the expression of FAF1 was significantly higher in IDD and positively correlated with the grade of disc degeneration. They further found overexpression of FAF1 promoted NP cell proliferation by activating of the extracellular signal-regulated kinase (Erk) signaling pathway [Citation6]. Similarly, another study revealed that silencing lncRNA Taurine Up-regulated Gene 1 (TUG1) may appreciably enhanced the proliferation ability of TNF-a induced human NP cells through inhibiting Wnt/b-catenin signaling pathway [Citation7]. Collectively, these studies have shown that lncRNAs is implicated in IDD progression by regulating NP cell proliferation.
Hox transcript antisense intergenic RNA (HOTAIR) is a trans-acting lncRNA that was first identified by Rinn et al. in 2007 [Citation8]. This lncRNA arises from the transcription of antisense strand of HoxC gene, which specifically locates between HoxC11 and HoxC12 on chromosome 12q13.13. Increased HOTAIR expression has been frequently reported in hepatocellular cancer, gastric cancer, non-small cell lung cancer and acute myeloid leukemia [Citation9–11]. In contrast, several researches have indicated that HOTAIR is significantly decreased in IDD, revealing a link of HOTAIR to IDD [Citation12,Citation13]. In recent years, it has been reported that HOTAIR can control the function of miRNAs by acting as a competition with endogenous RNA (ceRNA) in IDD [Citation14]. However, the detail functions of HOTAIR modulate the progression of IDD still remain little known.
The Akt pathway plays a pivotal regulatory role in promoting NP cell proliferation [Citation15]. As the inhibitor of the Akt pathway, PTEN is also a direct target of miR-130b [Citation16]. It has been demonstrated that the ectopic expression of miR-130b promotes cell proliferation by activating the AKT pathway via silencing of PTEN in multiple-type cells, including breast cancer cells [Citation17] and osteosarcoma cells [Citation18]. Cyclin D1, which is located in 11q13, is a important mediator in cell cycle transition from G1-to-S-phase [Citation19]. Interestingly, activation of the Akt signaling pathway can promote cell proliferation by up-regulating the expression of Cyclin D1 in liver cancer cells [Citation20]. Additionally, Zhao et al. found that Cyclin D1 expression is notably augmented with the progression of IDD [Citation21]. However, it remains unclear whether miR-130b also regulates NP cell proliferation by activation of PTEN/Akt/Cyclin D1 pathway in IDD.
In the current study, we found HOTAIR was significantly down-regulated both in NP tissues and cells, whereas ectopic expression of it diminished cell proliferation in IDD. In addition, the interaction among HOTAIR, miR-130b, and PTEN was also studied in order to reveal the underlying mechanisms. We identified that HOTAIR may act as a ceRNA of miR-130b to modulate the expression of CyclinD1, which may regulate the activity of PTEN/Akt signaling pathway. Our research also demonstrates that HOTAIR may be a therapeutic target of IDD.
2. Results
2.1. Decreased HOTAIR expression in human degenerative NP tissues
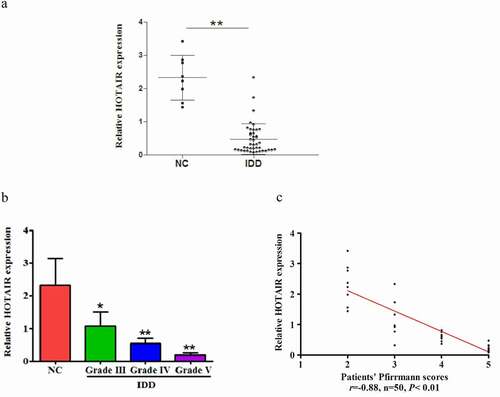
To explore the role of HOTAIR in the pathogenesis of IDD, we first detected the relative expression of HOTAIR in 42 degenerative NP tissues samples and eight healthy controls from adolescent idiopathic scoliosis (AIS) NP tissues, normalized to β-actin. Compared with healthy controls, the expression levels of HOTAIR were significantly decreased in degenerative NP tissues ()). Moreover, its expression was gradually downregulated with exacerbation of disc degeneration degree ()). Additionally, as shown in ), HOTAIR expression inversely correlated with Pfirrmann grade (r= −0.88, P< 0.01).
Figure 1. Decreased HOTAIR expression in NP tissues: Downregulation of HOTAIR expression in human degenerative NP tissues. (a) Expression of HOTAIR in NP tissue samples from 8 AIS patients (No. 1 to 8) and 42 IDD patients (No. 8 to 50) using qRT-PCR assay. (b) Comparison of HOTAIR expression between NC and IDD patients with various Pfirrmann grades. (c) Correlation between HOTAIR expression and Pfirrmann scores. Data are denoted in terms of the mean ± SD. *P< 0.05, **P < 0.01 vs. NC group.
2.2. Overexpression of HOTAIR inhibits human NP cell proliferation
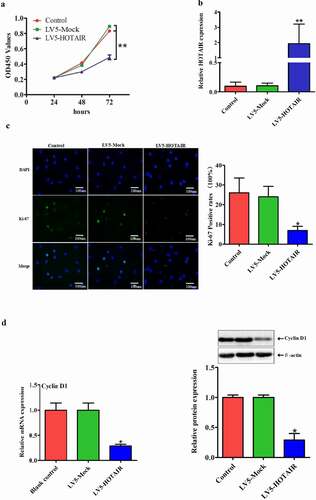
Excessive proliferation of NP cell is thought to be a key contributor to disc degeneration. To explore the effect of HOTAIR on human NP cell proliferation, cells were transfected with LV5-HOTAIR or LV5-Mock, which showed a higher transfection efficiency ()). Moreover, HOTAIR overexpression markedly inhibited cell proliferation, as shown by the CCK-8 proliferation assay ()). This effect was also confirmed by immunohistochemical staining of Ki-67 ()). Given that CyclinD1 has been used widely as one of the proliferation markers. We next examined the changes of CyclinD1 expression. Consistent with the above results, overexpression of HOTAIR decreased the protein and mRNA levels of CyclinD1 ()). Taken together, these data suggest HOTAIR as a negative regulator of human NP cell proliferation.
Figure 2. Overexpression of HOTAIR inhibits cell proliferation: Upregulation of HOTAIR inhibited the proliferation of human degenerative NP cells in vitro. (a) Relative expression levels of HOTAIR after NP cells transfected with LV5-HOTAIR and LV5-MOCK. (b) CCK-8 assay was performed to detect the proliferation effect of LV5-HOTAIR and LV5-MOCK transfected degenerative NP cells. (c) The mRNA and protein levels of CyclinD1 were measured by qRT-PCR and Western blot assay. (d) Immunostaining assay to evaluate the effect of HOTAIR on cell proliferation (Magnification, ×400, scale bar = 100 um). Data are presented as mean ± SD from three independent experiments. *P < 0.05, **P< 0.01 vs. LV5-MOCK group.
2.3. HOTAIR is physically associated with miR-130b in human NP cells
Next, we explored the potential mechanisms underlying cell proliferation suppression after HOTAIR upregulated. Intriguingly, HOTAIR has recently begun to emerge as miRNA sponge, thereby increasing mRNA expression and imposing an additional level of post-transcriptional modulation in some cancers [Citation10]. Thus, we considered whether HOTAIR has the same effect in IDD. By using starBase v2.0 data (http://starbase.sysu.edu.cn/mirLncRNA.php), we predicted the potential binding sites of miR-130b in HOTAIR transcripts ()). We subsequently investigated the potential interaction between HOTAIR and miR-130b in human degenerated NP cells. Firstly, by using qRT-PCR assay, we found that the expression of miR-130b was highly expressed in human NP tissues compared with normal tissues ()). Additionally, we also found that miR-130b level in NP tissues was positively correlated with Pfirrmann grade ()).
Figure 3. HOTAIR is physically associated with miR-130b: HOTAIR directly targeted miR-130b in NP cells. (a) Putative miR-130b binding sites within the HOTAIR 3ʹ-UTR are shown. The position of the binding sites (green) was numbered relative to the first nucleotide of the 3ʹ-UTR. Mutations (read) were introduced into HOTAIR 3ʹ-UTR that matched the seed region of miR-130b as shown in HOTAIR Mut. (b) MiR-130b was highly expressed in NP tissues. (c and d) miR-130b expressed levels were inversely correlated with Pfirrmann grades of IDD. (e) Dual-luciferase assay was performed in HEK293T co-transfected with luciferase constructs containing the HOTAIR wild-type or Mut 3ʹ-UTR and miR-130b mimics or scrambled oligonucleotides as the negative control. (f) RIP assay was used to verify the combination between HOTAIR and miR-130b. (g) The efficiency of miR-130b expression levels after NP cells transfected with miR-130b NC, miR-130b mimics, mimic control, miR-130b inhibitors and inhibitor control. (h) The relative gene expressions of miR-130b were analyzed by qRT-PCR after transfected with LV5-HOTAIR. (i) Relative gene expressions of HOTAIR were detected by qRT-PCR after transfected with miR-130b mimic, mimic control, miR-130b inhibitor and inhibitor control. (j) Pearson’s correlation analysis of the relationship between HOTAIR expression and miR-130b level in NP tissue samples from 8 AIS patients and 42 IDD patients. Each value is the mean ± SD of three experiments. Each value is the mean ± SD of three experiments.. *P < 0.05, **P < 0.01 compared to the control or LV5-HOTAIR group.
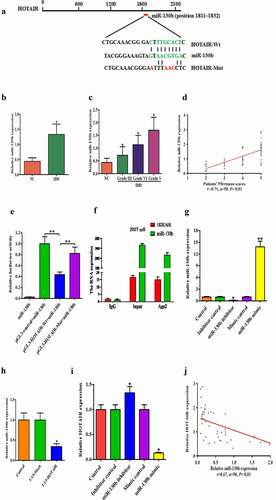
Then, we constructed luciferase reporter plasmids containing the wild-type HOTAIR (pGL3-HOTAIR-WT) and mutant HOTAIR with mutations of predicted miR-130b binding site (pGL3-HOTAIR-MUT). Dual-luciferase reporter assays showed that miR-130b over-expression led to a marked decrease in luciferase activity in pGL3-HOTAIR-WT, without changing the luciferase activity of pGL3-HOTAIR-MUT in 293 T cells ()). Meanwhile, RNA immunoprecipitation (RIP) experiment demonstrated that both HOTAIR and miR-130b could be pulled down by Ago2 antibody ()). These results suggested a direct interaction between HOTAIR and miR-130b. To explore the impact of miR-130b on cell proliferation, human NP cells were transfected with miR-130b inhibitor/mimic and their negative controls, which showed higher transfection efficiency ()). Meanwhile, real-time PCR showed that endogenous HOTAIR was reduced in miR-130b mimic-transfected human NP cells, but miR-130b inhibitor increased HOTAIR levels ()). Moreover, we further found that HOTAIR overexpression significantly decreased the expression level of endogenous miR-130b ()) and was inversely correlated with the expression of miR-130b in 50 human lumbar NP tissue samples ()). These findings suggested that miR-130b was HOTAIR targeting endogenous miRNAs in human degenerated NP cells.
2.4. Repression miR-130b restores LV5-HOTAIR induced inhibitory effects on human NP cell proliferation
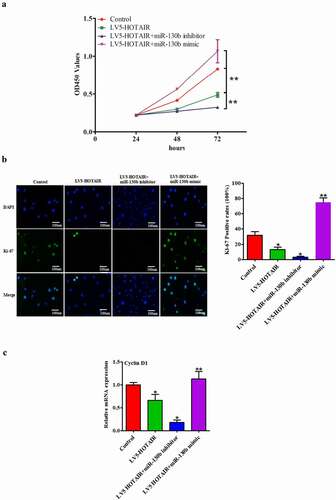
In order to investigate whether HOTAIR exerts biological functions through miR-130b, we performed a rescue experiment. A CCK-8 proliferation assay was performed to detect the effect of co-transfected with HOTAIR and miR-130b mimics or miR-130b inhibitors on NP cells. We found that HOTAIR overexpression combined with miR-130b inhibitors group was strongly inhibited cell proliferation, and miR-130b mimics reversed the proliferation of HOTAIR overexpression NP cells ()). Immunohistochemical staining of Ki-67 was used to further assess the proliferation ability. The results showed that HOTAIR overexpression combined with miR-130b knockdown resulted in significant reduction of proliferation of NP cells, whereas miR-130b mimics rescued the proliferation ability in HOTAIR up-regulation NP cells ()). In addition, HOTAIR overexpression combined with miR-130b knockdown group was strongly reduced CyclinD1 mRNA level, and miR-130b mimics reversed the effect of HOTAIR ()). Therefore, these results suggest that HOTAIR can inhibit cell proliferation and CyclinD1 level though miR-130b in human NP cells.
Figure 4. Repression miR-130b restores LV5-HOTAIR induced inhibitory effects: MiR-130b abolished the LV5-HOTAIR induced inhibitory effects on NP cells. (a) CCK-8 assay was performed to detect the proliferation after co-transfected with LV5-HOTAIR and miR-130b mimics or miR-130b inhibitors. (b) Immunostaining assay in NP cells was used to determine the proliferation after co-transfected with LV5-HOTAIR and miR-130b mimics or miR-130b inhibitors (Magnification, ×400, scale bar = 100 um). (c) The mRNA levels of CyclinD1 were measured by qRT-PCR after co-transfected with LV5-HOTAIR and miR-130b mimics or miR-130b inhibitors. All results are present as the mean ± SD from three independent experiments, each performed in triplicate. *P < 0.05, **P < 0.01 compared to the control or LV5-HOTAIR group.
2.5. HOTAIR controls cell proliferation and contributes to the activation of the miR-130b/PTEN/AKT pathways
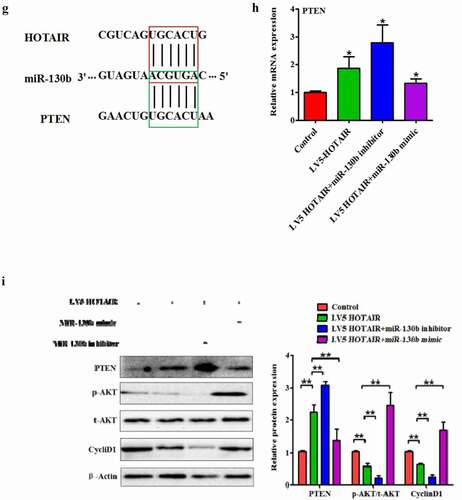
Among the many targets of miR-130b, we concentrated on PTEN since modulates cell proliferation, survival, apoptosis and metabolism via its target molecules, hosphoinositide-3 kinase (PI3K) and Akt [Citation22]. According to the TargetScan algorithm, sequence alignment of the miR-130b complementary site in the 3ʹ-UTR of PTEN mRNA allowed us to identify the conserved binding site, position 2961–2967 ()). To further determine whether miR-130b targeted PTEN directly by 3ʹ-UTR interaction, we cloned the fragment of the PTEN 3ʹ-UTR mRNA containing the binding site into the luciferase reporter vectors and transfected them into cells in the presence of either miR-130b mimic or control. As anticipated, the luciferase assay showed that miR-130b efficiently inhibited luciferase activity of pGL3-PTEN 3ʹ-UTR reporter, whereas the mutant reporter was not affected by miR-130b ()).
Figure 5. HOTAIR and AKT pathways: miR-130b directly targeted the 3′-UTR of PTEN and inhibits its expression in human degenerated NP cells. (a) Predicted pairing between miR-130b and the 3ʹ-UTR of human PTEN gene, and schematic representation of WT and MUT PTEN 3ʹ-UTR-pGL-REPORT vector constructs. Dual-Luciferase reporter system was then used to detect luciferase activity. (b) Luciferase activity in NP cells co-transfected with miR-130b mimics and luciferase reporters containing PTEN WT or MUT 3′-UTR. (c and d) Pearson’s correlation analysis of the relationship between PTEN and miR-130b, PTEN and HOTAIR in NP tissue samples from 8 AIS patients and 42 IDD patients. (e and f) qRT-PCR and Western blot assays were performed to detect the mRNA and protein levels of PTEN. Each value is the mean ± SD of three experiments. (g) The predicted of HOTAIR and PTEN co-binding sites in the 3′-UTR region of miR-130b was shown. (h)Relative gene expressions of PTEN were analyzed by qRT-PCR. (i and j) PTEN, CyclinD1 and phosphorylation of Akt were analyzed by Western blotting. (k) Immunostaining assay in NP cells was performed to examine NP cell proliferation (Magnification, ×200, scale bar = 100um). *P < 0.05, **P < 0.01 compared to the control or LV5-HOTAIR group. Each value is the mean ± SD of three experiments.
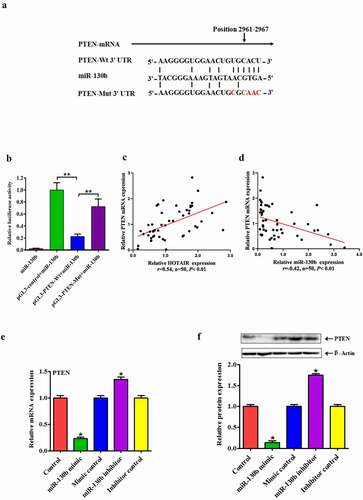
We subsequently investigated the correlation among HOTAIR, PTEN, and miR-130b in human NP cells. The mRNA expression of PTEN was inversely correlated with miR-130b and positively correlated with HOTAIR, respectively (). In addition, we also detected the expression levels of PTEN in human degenerated NP cells with miR-130b by Western blot and qRT-PCR assay. MiR-130b upregulation efficiently reduced both mRNA and protein expression levels of PTEN compared to miR-130b NC while inhibited miR-130b expression presented the opposite results (). These findings suggest that miR-130b inhibits PTEN expressions in human degenerated NP cells by directly targeting the 3′UTR of PTEN. It is known that lncRNA can act as ceRNA to influence post-transcriptional regulation by interfering with the miRNA pathways. By using lnCeDB database, we found that HOTAIR and PTEN were the common targeting of miR-130b ()). To investigate whether HOTAIR could act as a ceRNA for PTEN by regulating miR-130b in human NP cells, qRT-PCR and Western blot assays were performed to detect the mRNA and protein levels of PTEN after cells transfected with LV5-HOTAIR combined with miR-130b mimic or miR-130b inhibitor. We found that transfection with LV5-HOTAIR increased the level of PTEN protein and mRNA, whereas the level of miR-130b mRNA decreased ()).
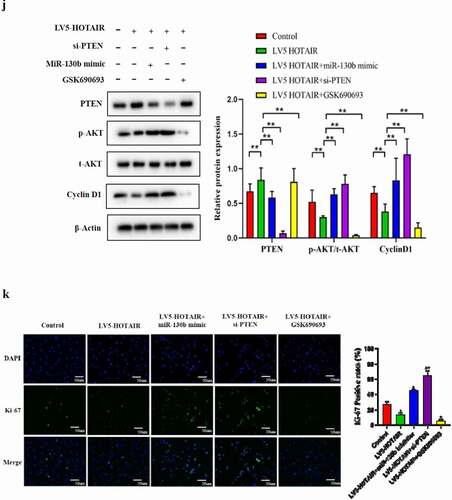
To further uncover the underlying principles of HOTAIR/miR-130b/PTEN regulating cell proliferation, comprehensive Western blot and immunostaining of cell proliferation assay was carried out. Overexpression HOTAIR significantly reduced Akt phosphorylation and CyclinD1 protein expression in human degenerated NP cells, which was reversed by miR-130b mimic ()). In addition, knockdown of PTEN by si-PTEN partially reversed the proliferation induced by HOTAIR overexpression ()). GSK690693 is a small molecule ATP-competitive inhibitor of the pro-survival kinase Akt [Citation23]. Futhermore, inhibition of Akt activity by GSK690693 abolished the effect of HOTAR on NP cell proliferation ()). These data indicate that HOTAIR can function as an endogenous “sponge” by binding miR-130b, thus abolishing the miRNA-induced repressing activity on the PTEN 3ʹ-UTR and contributes to the inactivation of the Akt pathways via miR-130b.
3. Discussion
In recent years, numbers of reports revealed that lncRNAs are differentially expressed in IDD, but the understanding of their regulatory role is just beginning to emerge [Citation24–26]. In 2014, Wan et al. first reported the expression profiling of lncRNAs in human IDD and revealed that HOTAIR (NR_003716) is the top decreased lncRNAs among the deregulated lncRNAs [Citation12]. In a later study, the expression of HOTAIR expression is significantly decreased in degenerative NP tissue samples compared with normal ones [Citation9]. Here, our results were consistent with the previous researches suggesting that HOTAIR could play a crucial role in IDD and may potentially be novel therapeutic targets in the treatment of IDD.
Emerging evidence suggests that the lncRNA-miRNA-mRNA ceRNA regulatory network has emerged in the study of IDD development [Citation27–29]. The hypothesis of ceRNA function assumes that some specific lncRNAs can function as miRNA sponges to control miRNAs available for binding with their targets, functionally liberating mRNA transcripts targeted by certain miRNAs. Notably, HOTAIR can regulate the degenerative diseases including Parkinson, osteoarthritis, and IDD by a complex network of binding with other RNAs [Citation14,Citation30,Citation31]. For example, Shao and colleagues reported that HOTAIR can decrease NP cell apoptosis via sponging miR-34a-5p, and downregulated miR-34a-5p disinhibits apoptosis by inactivating Notch1 expression [Citation13]. Similarly, HOTAIR/miR-34a/BCL-2 axis can also inhibit the level of NP cell apoptosis [Citation14]. MiR-130b is one of the well-documented miRNAs involved in multiple cell processes, including proliferation, apoptosis, and invasion [Citation16,Citation32]. Herein, our present study confirmed that miR-130b was significantly increased in NP tissues and inversely correlated with HOTAIR. Interestingly, we further found HOTAIR is also directly target of miR-130b. Moreover, cell proliferation was inhibited by HOTAIR, and this inhibition was further enhanced by miR-130b down-regulation, indicating that cell proliferation was regulated by the HOTAIR/miR-130b/PTEN axis.
Dysregulation of AKT signaling pathways has been shown to be implicated in IDD [Citation33]. It is worth noting that PTEN is the specific inhibitor targeting Akt, which can regulate many biological processes, including cell proliferation, apoptosis, cytokine release [Citation34–36]. As expected, our results found that PTEN was significantly decreased by miR-130b and resulted in the activation of Akt. However, restored expression of HOTAIR or Akt suppression (GSK690693) reversed the mitogenic effect of miR-130b. CyclinD proteins (D1, D2, and D3) are essentially interchangeable due to substantive functional redundancy in normal proliferating human tissues [Citation37]. Up-regulated CyclinD1 expression can increase cell proliferation. A study by Liu et al. found that ectopic expression of miR-21 upregulation in intervertebral disc degeneration could inhibit PTEN, which would activate the Akt pathway and then upregulated CyclinD1 expression, thus promoting cell proliferation [Citation38]. To further explore whether HOTAIR/miR-130b/PTEN signaling network regulates the cell proliferation of IDD by modulating AKT pathway. We demonstrated that CyclinD1 expression was inhibited by HOTAIR, and this inhibition was further enhanced by miR-130b inhibitor, indicating that CyclinD1 expression level was regulated by the HOTAIR/miR-130b/PTEN axis. All these data suggest that HOTAIR can influence human NP cell proliferation by binding miR-130b and subsequently upregulating PTEN, leading to inhibiting Akt pathway.
There are some limitations in our research. The present work only focused on the ability of HOTAIR/miR-130b/PTEN signaling network to influence NP cell proliferation in vitro. Whether it can reverse the onset and progression of IDD in vivo needs to clarify and confirm. Recently, using nanoparticles or stem cell-derived exosomes injected into tumor served as effective delivery vehicles for lncRNA [Citation39]. For instance, the use of magnetic nanoparticles of HOTAIR siRNA have been shown to effectively inhibit the proliferation and invasion of human glioma stem cells in vivo [Citation40]. Thus, targeting HOTAIR for therapeutic purposes is highly attractive and available further studies in IDD.
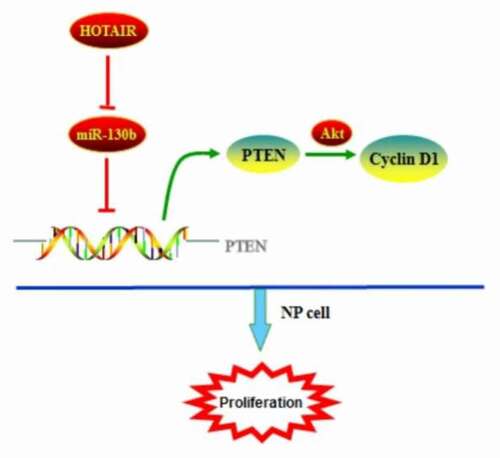
In conclusion, HOTAIR expression levels are decreased in human degenerated NP tissues than healthy controls, and an excellent biomarker of IDD. Upregulation of HOTAIR suppressed cell proliferation by directly targeting miR-130b and negatively regulating its expression in human degenerated NP cells. The positive regulation between HOTAIR and PTEN may be partially attributed to the ceRNA regulatory network via miR-130b, which may regulate the activity of Akt signaling pathway (). Thus, the identification of the ceRNA signaling network of HOTAIR/miR-130b/PTEN will undoubtedly enhance our knowledge of how lncRNAs function, allowing us to better understand the pathogenesis and development of IDD and ultimately facilitate the development of lncRNA-directed diagnostics and therapeutics against this degenerative disease.
4. Materials and methods
4.1. Patients and samples
Human lumbar NP tissues were collected from patients with IDD undergoing discectomy in the Department of Spine Surgery, the First Affiliated Hospital from June 2016 to November 2019. This study enrolled a total of 50 patients (24 males, 26 females) ranging from 9 to 65 years old. Selection criteria were as follows: (1) patients typically had clinical symptoms of lumbar disc herniation; (2) preoperative X-ray, computed tomography (CT) and magnetic resonance imaging (MRI) determined patients with lumbar disc herniation or IDD. Exclusion criteria were patients with infections, idiopathic scoliosis, and previous lumbar disc surgery. By grading from T2-weighted images using Pfirrmann classiffication, patients with IDD were classified into eight cases in grade I, nine cases in grade III, 12 cases in grade IV, and 21 cases in grade V. Eight normal NP tissues were obtained from patients with AIS (grade I). All NP tissue specimens dissected carefully during surgery were washed thrice with phosphate-buffered salin (PBS), followed by collection using a stereotaxic microscope and freeze in the liquid nitrogen. This study was approved by the Ethics Committee of the First Affiliated Hospital, University of South China.
4.2. Isolated and cultured of human NP cells
NP cells were isolated as previously described. After isolation, NP cells were resuspended in DMEM/F12 containing 10% FBS (GIBCO, NY, USA), 100 mg/ml streptomycin, and 100 U/ml penicillin and then incubated at 37uC in a humidified atmosphere with 95% (v/v) air and 5% (v/v) CO2. The confluent cells were detached by trypsinization, seeded into 25-mm2 T-Flasks Vented Caps in complete culture medium (DMEM/F12 supplemented with 10% FBS, 100 mg/ml streptomycin, and 100 U/ml penicillin) in a 37°C, 5% CO2 (v/v) environment. The complete culture medium was changed twice a week. The second passage was used for subsequent experiments.
4.3. Antibodies and reagents
Rabbit antibodies against cyclinD1, PTEN, p-Akt, t-Akt, and β-actin as well as horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG were provided by Santa Cruz Biotechnology (Santa Cruz, CA, USA). MiR-130b mimic/inhibitor and their negative controls were synthesized by Ribobio (Shanghai, China). For constructing HOTAIR, full length of HOTAIR was synthesized and cloned into the mammalian expression vector LV5 (Invitrogen, USA).
4.4. RNA isolation and quantitative real-time PCR (qRT-PCR)
Total RNAs were extracted from NP tissues or cells using TRIzol reagent (Invitrogen, CA, USA) and RNeasy mini kit (Qiagen,CA, USA) according to the manufactures’ instructions. These RNAs were then eluted in 50ul of nuclease-free water, and stored at −80°C for Real-time reverse transcription PCR (qRT-PCR) assay. A NanoDrop ND-1000 spectrophotometer (Thermo scientific, Rockford, IL, USA) was used to detect the purity and concentration of RNAs. By using Power SYBR Green (Takara, Dalian China), the mRNA expression levels of genes were measured. qRT-PCR was performed on ABI 7500 Real-time PCR system (Thermo Scientific, Rockford, IL, USA), quantitative measurements using the 2−∆∆Ct method were determined. All primer sequences are presented in .
Table 1. Primer sequences used in this study
4.5. Transfection of NP cells
The plasmid vectors were transfected according to the manufacturer’s instructions. The day before transfection, human NP cells were seeded at 1 × 105–1 × 106 cells/well into 6-well dishes, and then 2 ml of complete tissue culture medium was added to each well. Cell density had to be around 85% for subsequent transfection. Human NP cells were treated with miR-130b mimic (20 nM), mimic control (20 nM), miR-130b inhibitor (40 nM) or inhibitor control (40 nM) for 24 h. Then, the LV5-HOTAIR (50 nM) was transfected into NP cells, and miR-130b mimic/inhibitor was transfected into NP cells using Lipofectamine 2000 for 24 h. At 48–96 h after transfection, qRT-PCR or Western blot analyses were used to evaluate the expression of gene and protein.
4.6. Western blot assay
Human NP cells were lysed using RIPA Lysis Buffer (Thermo Scientific, Rockford, IL, USA). Cells protein lysates were resolved by 12% sodium dodecyl sulfate-polyacrylamide (SDS-PAGE) gel and transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA, USA). The membrane was blocked by 5% (v/v) low-fat skim milk in Tris-buffered saline Tween (TBST) at room temperature for 2 h, and then incubated with the primary antibodies at 4°C overnight. The next day, the membrane was washed and subsequently incubated with HRP-conjugated secondary antibody at room temperature for 1 h. According to the manufacturer’s instructions, the protein expression was measured using the enhanced chemiluminescence (ECL) substrate kit (Amersham Biosciences, Inc.) and the enhanced chemiluminescence detection system (Amersham Biosciences, Piscataway, New Jersey, USA). The results were normalized using β-actin.
4.7. Cell proliferation assay
Cell proliferation was performed using CCK-8 (Sigma, St. Louis, Mo) assay according to the standard instruction. Briefly, human NP cells were placed into 96-well plates and treated with various reagents. Cell vitality was assessed every 24 h using CCK-8 solution. By using a microplate spectrophotometer, the optical density was read at 450 nm. Each experiment was performed in triplicate
4.8. Bioinformatics methods
The potential microRNAs binding sites of HOTAIR predicted by computer-aided algorithms were obtained from starBase v2.0 data (http://starbase.sysu.edu.cn/mirLncRNA.php), Targetscan (http://www.targetscan.org/vert_71/) and microRNA.org-target program (www. microRNA.org).
4.9. RIP assay
RIP assay was performed using 293 T cells as described. Magna RIP RNA‐Binding Protein Immunoprecipitation Kit (Millipore, Billerica, Massachusetts) was used to experimentize RIP assay according to the manufacturer’s protocol. AGO2 antibodies and control IgG (Millipore) were performed for RIP assays as the manufacturer’s instructions. The total RNAs were the input controls. The qRT‐PCR analysis was used to detect the cDNA, which was synthesized by the coprecipitated RNAs.
4.10. Dual luciferase reporter assay
The cDNA fragments from HOTAIR containing the predicted miR-130b binding site were amplified by PCR. The putative sequences of the binding site were then cloned into a pGL-3 Dual-luciferase miRNA Target Expression Vector (Promega, Madison, WI, USA) to form the reporter vector pGL3-HOTAIR-wild-type (HOTAIR-WT). To mutate the putative-binding site of miR-130b in HOTAIR gene, the sequence of putative-binding site was replaced as indicated and was named as pGL3-HOTAIR-mutated-type (HOTAIR-MUT). Subsequently, 293 T cells (5 × 104 cells/well) were co-transfected with above plasmids (1 µg) in combination with the wild type (HOTAIR-WT) or mutant (HOTAIR-Mut) using Lipofectamine 2000 for 48 h according to the manufacturer’s protocol. Dual-Luciferase reporter system (Promega, Madison, WI) used to detect the luciferase activity.
Similarly, the full 3ʹ-UTR of PTEN was amplified by RT-PCR from human NP cells. These amplified products were then cloned into a pGL-3 luciferase reporter vector using restriction sites XbaI and BamHI (Promega, Madison, WI, USA). Additionally, Quik Change II XL Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA) was used to mutate the PTEN seed sequence located in the 3ʹ UTR of PTEN mRNA to produce mutant PTEN 3ʹUTR luciferase reporter construct (MUT PTEN 3ʹ UTR-pGL-3 vector). The transfection procedure and measurement of Luciferase activities were handled similarly as described above. All assays were independently performed in triplicate.
4.11. Immunostaining of the proliferative marker Ki-67
Coverslips were placed into 6-well plate where NP cells were plated for 48–72 h. Afterward, medium was removed and the cells were washed twice with PBS and fixed with 3.5% formaldehyde for 30 min at 37uC. The cells were rinsed with PBS for 3 times, permeabilized with 0.1% (v/v) Triton X-100 in PBS for 20 min and blocked with 3% (w/v) BSA and 0.05% (v/v) Tween 20 in PBS for 30 min at room temperature. After blocking, the cells were incubated overnight at 4uC with rabbit monoclonal anti-Ki-67 antibody (Bioworlde, USA; 1:500). The cells were then treated with fluorescent anti-rabbit secondary antibody (Bioworlde, USA; 1:500) for 2 h at room temperature. Nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI). Fluorescence images were acquired with a Leica TCS SP2 confocal microscopy (Leica, Mannheim, Germany) using the Leica Confocal Software.
4.12. Statistical analysis
Results were expressed as means ± SD of multiple experiments. Statistical analysis was performed with Student’s t-test for comparison between two groups or an analysis of variance (ANOVA) followed by the Turkey’s t-test for comparison of multiple groups. P value less than 0.05 was considered as statistically significance.
Authors’ contributions
Wen-Kang Chen designed the study, performed experiments, analyzed the data, and wrote the manuscript; Cheng Wang and Ming-Xiang Zou designed the study and performed experiments and interpretation of data; Yi-Guo Yan designed the study Zhang Hang-Jing and Xin-Li Zhan revised the article; Wen-Jun Wang and Xue-Lin Li designed the study, interpreted the data, and wrote the manuscript. All authors read and approved the final manuscript.
Availability of data and materials
The dataset supporting the conclusions of this manuscript is included within the manuscript.
Ethics approval
All human IVD tissue sample studies were approved by the Ethics Committee of the First Affiliated Hospital, University of South China.
Ethical review committee statement
All protocols involving human subjects were approved by Ethics Committee, the First Affiliated Hospital of University of South China, with all subjects providing informed consents.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Urban JPG, Fairbank JCT. Current perspectives on the role of biomechanical loading and genetics in development of disc degeneration and low back pain; a narrative review. J Biomech. 2020;102:109573.
- Johnson WE, Eisenstein SM, Roberts S. Cell cluster formation in degenerate lumbar intervertebral discs is associated with increased disc cell proliferation. Connect Tissue Res. 2001;42(3):197–207.
- Ji ML, Jiang H, Zhang XJ, et al. Preclinical development of a microRNA-based therapy for intervertebral disc degeneration. Nat Commun. 2018;9(1):5051.
- Yu X, Li Z, Shen J, et al. MicroRNA-10b promotes nucleus pulposus cell proliferation through RhoC-Akt pathway by targeting HOXD10 in intervetebral disc degeneration. PloS One. 2013;8(12):e83080.
- Guo HY, Guo MK, Wan ZY, et al. Emerging evidence on noncoding-RNA regulatory machinery in intervertebral disc degeneration: a narrative review. Arthritis Res Ther. 2020;22(1):270.
- Mi D, Cai C, Zhou B, et al. Long non‑coding RNA FAF1 promotes intervertebral disc degeneration by targeting the Erk signaling pathway. Mol Med Rep. 2018;17(2):3158–3163.
- Chen J, Jia YS, Liu GZ, et al. Role of LncRNA TUG1 in intervertebral disc degeneration and nucleus pulposus cells via regulating Wnt/β-catenin signaling pathway. Biochem Biophys Res Commun. 2017;491(3):668–674.
- Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323.
- Yang L, Peng X, Li Y, et al. Long non-coding RNA HOTAIR promotes exosome secretion by regulating RAB35 and SNAP23 in hepatocellular carcinoma. Mol Cancer. 2019;18(1):78.
- Qu X, Alsager S, Zhuo Y, et al. HOX transcript antisense RNA (HOTAIR) in cancer. Cancer Lett. 2019;454:90–97.
- Li T, Mo X, Fu L, et al. Molecular mechanisms of long noncoding RNAs on gastric cancer. Oncotarget. 2016;7(8):8601–8612.
- Wan ZY, Song F, Sun Z, et al. Aberrantly expressed long noncoding RNAs in human intervertebral disc degeneration: a microarray related study. Arthritis Res Ther. 2014;16(5):465.
- Shao T, Hu Y, Tang W, et al. The long noncoding RNA HOTAIR serves as a microRNA-34a-5p sponge to reduce nucleus pulposus cell apoptosis via a NOTCH1-mediated mechanism. Gene. 2019;715:144029.
- Yu Y, Zhang X, Li Z, et al. LncRNA HOTAIR suppresses TNF-α induced apoptosis of nucleus pulposus cells by regulating miR-34a/Bcl-2 axis. Biomed Pharmacother. 2018;107:729–737.
- Li Z, Li X, Chen C, et al. Melatonin inhibits nucleus pulposus (NP) cell proliferation and extracellular matrix (ECM) remodeling via the melatonin membrane receptors mediated PI3K-Akt pathway. J Pineal Res. 2017;63(3):e12435.
- Lu Q, Liu T, Feng H, et al. Circular RNA circSLC8A1 acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer progression via regulating PTEN. Mol Cancer. 2019;18(1):111.
- Miao Y, Zheng W, Li N, et al. MicroRNA-130b targets PTEN to mediate drug resistance and proliferation of breast cancer cells via the PI3K/Akt signaling pathway. Sci Rep. 2017;7:41942.
- Han W, Liu J. LncRNA-p21 inhibited the proliferation of osteosarcoma cells via the miR-130b/PTEN/AKT signaling pathway. Biomed Pharmacother. 2018;97:911–918.
- Thomasova D, Anders HJ. Cell cycle control in the kidney. Nephrol Dial Transplant. 2015;30(10):1622–1630.
- Yang X, Feng Y, Liu Y, et al. Fuzheng Jiedu Xiaoji formulation inhibits hepatocellular carcinoma progression in patients by targeting the AKT/CyclinD1/p21/p27 pathway. Phytomedicine. 2021;87:153575.
- Zhao YT, Qin Y, Yang JS, et al. Wharton’s Jelly-derived mesenchymal stem cells suppress apoptosis of nucleus pulposus cells in intervertebral disc degeneration via Wnt pathway. Eur Rev Med Pharmacol Sci. 2020;24(19):9807–9814.
- Ghafouri-Fard S, Abak A, Shoorei H, et al. Regulatory role of microRNAs on PTEN signaling. Biomed Pharmacother. 2021;133:110986.
- Zhong Q, Chen Y, Chen Z. LncRNA MINCR regulates irradiation resistance in nasopharyngeal carcinoma cells via the microRNA-223/ZEB1 axis. Cell Cycle. 2020;19(1):53–66.
- Zhu J, Zhang X, Gao W, et al. lncRNA/circRNA‑miRNA‑mRNA ceRNA network in lumbar intervertebral disc degeneration. Mol Med Rep. 2019;20(4):3160–3174.
- Zhu J, Yu W, Wang Y, et al. lncRNAs: function and mechanism in cartilage development, degeneration, and regeneration. Stem Cell Res Ther. 2019;10(1):344.
- Chen WK, Yu XH, Yang W, et al. lncRNAs: novel players in intervertebral disc degeneration and osteoarthritis. Cell Prolif. 2017;50(1):e12313.
- Sun Z, Tang X, Wang H, et al. LncRNA H19 aggravates intervertebral disc degeneration by promoting the autophagy and apoptosis of nucleus pulposus cells through the miR-139/CXCR4/NF-κB axis. Stem Cells Dev. 2021;30(14):736–748.
- Tang N, Dong Y, Xiao T, et al. LncRNA TUG1 promotes the intervertebral disc degeneration and nucleus pulposus cell apoptosis though modulating miR-26a/HMGB1 axis and regulating NF-κB activation. Am J Transl Res. 2020;12(9):5449–5464.
- Chen X, Li Z, Xu D, et al. LINC01121 induced intervertebral disc degeneration via modulating miR-150-5p/MMP16 axis. J Gene Med. 2020;22(10):e3231.
- Zhao J, Li H, Chang N. LncRNA HOTAIR promotes MPP+-induced neuronal injury in Parkinson’s disease by regulating the miR-874-5p/ATG10 axis. EXCLI J. 2020;19:1141–1153.
- Tu J, Huang W, Zhang W, et al. The emerging role of lncRNAs in chondrocytes from osteoarthritis patients. Biomed Pharmacother. 2020;131:110642.
- Liao Y, Wang C, Yang Z, et al. Dysregulated Sp1/miR-130b-3p/HOXA5 axis contributes to tumor angiogenesis and progression of hepatocellular carcinoma. Theranostics. 2020;10(12):5209–5224.
- Ouyang ZH, Wang WJ, Yan YG, et al. The PI3K/Akt pathway: a critical player in intervertebral disc degeneration. Oncotarget. 2017;8(34):57870–57881.
- Xi Y, Ma J, Chen Y. PTEN promotes intervertebral disc degeneration by regulating nucleus pulposus cell behaviors. Cell Biol Int. 2020;44(2):583–592.
- Sun K, Luo J, Guo J, et al. The PI3K/AKT/mTOR signaling pathway in osteoarthritis: a narrative review. Osteoarthritis Cartilage. 2020;28(4):400–409.
- Lin Y, Guo W, Chen KW, et al. VO-OHpic attenuates intervertebral disc degeneration via PTEN/Akt pathway. Eur Rev Med Pharmacol Sci. 2020;24(6):2811–2819.
- Tiedemann RE, Mao X, Shi CX, et al. Identification of kinetin riboside as a repressor of CCND1 and CCND2 with preclinical antimyeloma activity. J Clin Invest. 2008;118(5):1750–1764.
- Liu H, Huang X, Liu X, et al. miR-21 promotes human nucleus pulposus cell proliferation through PTEN/AKT signaling. Int J Mol Sci. 2014;15(3):4007–4018.
- Barile L, Vassalli G. Exosomes: therapy delivery tools and biomarkers of diseases. Pharmacol Ther. 2017;174:63–78.
- Fang K, Liu P, Dong S, et al. Magnetofection based on superparamagnetic iron oxide nanoparticle-mediated low lncRNA HOTAIR expression decreases the proliferation and invasion of glioma stem cells. Int J Oncol. 2016;49(2):509–518.