ABSTRACT
Circ_0020123 was highly expressed in NSCLC tissues and cell lines, and knockdown of circ_0020123 abolished cell growth, migration and invasion in vitro and hindered tumor growth in nude mice. Mechanically, circ_0020123 directly targeted miR-940, and KIAA1522 was a target of miR-940. Thereafter, a series of rescue experiments showed that circ_0020123 served its biological functions by miR-940/KIAA1522 axis. In all, circ_0020123 acted as an oncogene to promote the tumorigenesis of NSCLC via miR-940/KIAA1522 axis, suggesting a potential therapeutic target for NSCLC treatment.
Introduction
As the first among all malignancies in cancer-related deaths in the world and in China, lung cancer is a serious threat to human health [Citation1,Citation2]. It is reported that about four fifths of lung cancer case is non-small cell lung carcinomas (NSCLC) [Citation3], and the 5-year survival rate for advanced NSCLC remains poor largely with approximately 15% due to the emergence of drug resistance, as well as high rate of metastasis and recurrence despite advances in diagnosis and treatment [Citation4–6]. Hence, further investigation on the molecular mechanism of NSCLC pathogenesis is necessary.
Circular RNAs (circRNAs) are a class of non-coding RNAs molecules possessing covalent closed-loop structures formed by the join of 3’ and 5ʹends of circRNAs, which make them resistant to RNase R decay, and stably represent in eukaryotic cells [Citation7–9]. Currently, altered circRNAs expression profiles have been shown in many types of cancers, and circRNAs, as transcriptional/post-transcriptional regulators, play critical roles in cancer progression through regulating cellular crucial biological behaviors related to carcinogenesis, drug resistance and metabolism [Citation10–12]. For example, circHIPK3 was up-regulated in colorectal cancer, which served as an independent prognostic factor of poor prognosis, and could facilitate cancer progression [Citation13]. Yang et al. found circ-ITCH was decreased in bladder cancer, low circ-ITCH expression predicted poor prognosis and enforced-expression of circ-ITCH reduced cell viability and metastasis in cancer [Citation14]. The down-regulation of circRNA_0000285 restrained cervical cancer growth and metastasis by regulating FUS [Citation15]. Thus, circRNAs may be the ideal candidates for cancer diagnosis and treatment. Circ_0020123 is a novel identified circRNA, it was reported to function as a competitive endogenous RNA (ceRNA) for microRNA (miRNA/miR)-488-3p or miR-144 to promote the growth and metastasis of NSCLC [Citation16,Citation17], suggesting the potential regulatory effects of circ_0020123 in the tumorigenesis of NSCLC. However, large-scale identification of circ_0020123 in NSCLC was not yet reported.
Herein, this study focused on investigating novel molecular mechanism of circ_0020123 with regard to tumorigenesis and the progression of NSCLC to provide a better understanding of NSCLC pathogenesis, thus helping the development of novel therapeutic targets for NSCLC.
Materials and methods
Clinical specimens
The tumor tissues and matched non-cancerous tissues (N = 77) were obtained from patients with NSCLC by resection at Tumor Hospital Affiliated to Nantong University, and then instantly stored at −80°C. None of the patients had received chemoradiotherapy before surgery. This research was approved by the Ethics Committee of Tumor Hospital Affiliated to Nantong University, and all patients were completely informed and signed the written informed consents before this study. The clinicopathological features of 77 cases of NSCLC patients are given in .
Table 1. Correlation between clinicopathological characteristics and circ_0020123 expression level in NSCLC patients
Cell culture
Human non-cancerous lung cell lines (BEAS-2B) and NSCLC cell lines (A549 and PC9) were obtained from Beijing Institute for Cancer Research Collection (Beijing, China). All cells were cultivated in RPMI-1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Gibico, CA, USA) and 1% penicillin-streptomycin in an incubator with 5% CO2 at 37°C.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Whole-RNA was isolated using Trizol reagent (Thermo Fisher Scientific) from targeted tissues or cells. The first strand complementary DNAs (cDNAs) were generated using the RNA to cDNA EcoDry Premix Kit (Clontech, Otsu, Japan) with Random or Oligo (dT)18 primers. Then, the expression levels of circ_0020123, linear PDZD8 mRNA and KIAA1522 mRNA were analyzed by qRT-PCR with SYBR Green PCR Kit (Qiagen). For miR-940 detection, the cDNA was synthesized using the miScript II RT Kit (Qiagen), and then quantified by qRT-PCR with the miScript SYBR Green PCR Kit (Qiagen). The relative expression was tested using the 2−ΔΔCt method and normalized to RNU6B (U6) or glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Primer sequences used for PCR amplification were listed: circ_0020123: F 5’-ATCCTCGTATGCACTCTGGC-3’, R 5’-CGATAAGTCGATCTCCCCGC-3’; PDZD8: F 5’-ACTAGTTTGGGCTGGTTTTGT-3’; R 5’-TACCCCAACAACAGTCGAGC-3’; KIAA1522: F 5’-CAAGAGGGCCAAGGGCAAAG-3’, R, 5’-GGTCGCCCACTGGGAAAGAA-3’; GADPH: F 5’-CCCACATGGCCTCCAAGGAGTA-3’, R 5’-GTGTACATGGCAACTGTGAGGAGG-3’; U6: F 5’-AAAGCAAATCATCGGACGACC-3’, R 5’-GTACAACACATTGTTTCCTCGGA-3’; miR-940: F 5’-CAGTGCAGGGTCCGAGGTA-3’, R 5’-GCATAAGGCAGGGCCCC-3’.
RNase R and actinomycin D digestion
For RNase R assay, 2 µg of RNA extracts were incubated without or with 3 U/µg RNase R for 20 min at room temperature, and then purified using the RNeasy MinElute Cleanup Kit (Qiagen, Tokyo, Japan). For the actinomycin D treatment, 2 µg of total RNA was treated with 2 μg/mL actinomycin D for 0, 4, 8, 12 and 24 h, respectively. Finally, qRT-PCR assay was employed to test the abundances of circ_0020123 and PDZD8 mRNA.
Subcellular fractionation
The NE-PER™ Nuclear and Cytoplasmic Extraction Reagents Kit (Thermo Fisher Scientific) was applied to isolate and purify the RNA from cytosolic and nuclear fractions. Afterward, the relative expression of circ_0020123 in A549 and PC9 cells was analyzed using qRT-PCR assay with GAPDH or U6 as control transcript.
Cell transfection
The circ_0020123-specific siRNA (si-circ_0020123, 5’-GACCAGCTTACGTTG AATTAAdtdt-3’) and negative control siRNA (si-NC, 5’-TTCTCCGAACGTGTCAC GT-3’), pcDNA3.1 vector encoding KIAA1522 (KIAA1522) and plasmids containing scrambled sequences (pcDNA), miR-940 mimic (miR-940, 5’-AAGGCAGGGCCCCC GCUCCCC-3’), miR-940 inhibitor (anti-miR-940, 5’-GGGGAGCGGGGCCCUGCC UU-3’) or their negative controls (miR-NC 5’-UUCUCCGAACGUGUCACGUTT-3’ or anti-miR-NC 5’-CAGUACAUUGGUUCUGCAA-3’) were synthesized by Invitrogen (San Diego, CA, USA). Lentiviral expressing short hairpin RNA (shRNA) targeting circ_0020123 (sh-circ_0020123) or scrambled control (sh-NC) were procured from ribobio company (Guangzhou, China). The transfection was carried out using Lipofectamine 2000 (Invitrogen).
Cell proliferation analysis
Transfected A549 and PC9 cells (5 × 103) seeded in 96-well plates were incubated with cell counting kit-8 (CCK-8) solution (10 μL, Dojindo Labora tories, Kumamoto, Japan) for 2 h. Finally, the absorbance at 450 nm was tested using a microplate reader at indicated times.
For colony formation assay, transfected cells were seeded at a density of 1000/well into a 6-well plate with RPMI 1640 medium and allowed to culture for 14 days. Then cell colonies were fixed with methanol and stained with 0.1% crystal violet (Sigma-Aldrich, Irvine, Ayrshire, UK). At last, the colonies were photographed and counted using a microscope.
Cell cycle assay
After transfection, A549 and PC9 cells were fixed in 75% ice-cold ethanol for 24 h, followed by staining with FxCycle PI/RNase Staining Solution (Thermo Fisher Scientific) in the dark referring to the producer’s guidance. Finally, the percentage of cells in the S, G0/G1, and G2/M phases were quantified using a flow cytometer.
Cell apoptosis assay
The transfected A549 and PC9 cells (1 × 105) were washed with phosphate-buffered saline (PBS) (Sigma-Aldrich), and then double stained with FITC-Annexin V (10 μL) and propidium iodide (PI) (10 μL) (BD Biosciences, San Jose, CA, USA) for 30 min. Lastly, apoptotic cells were examined using a flow cytometer.
Cell migration and invasion assay
Transwell chambers (8 µm pore size) (Sangon Biotech, Shanghai, China), whose membranes were pre-coated without or with Matrigel (BD Biosciences), were employed to determine cell migration and invasion. Equal numbers of transfected A549 and PC9 cells (1 × 104 for migration, and 1 × 105 for invasion) in serum-free medium were plated into the upper chamber, and lower chamber was filled with medium containing 10% FBS. 24 h later, cells on the lower surface of the membranes were fixed and stained with Giemsa. Finally, migrated and invaded cells were imaged (×100) and counted.
Dual-luciferase reporter assay
The specific sequences of Wild-type (WT) or mutated (MUT) circ_0020123 and KIAA1522 3’-UTR at the putative miR-940 binding sites were amplified and inserted into the psiCHECK-2 vector (Promega, Madison, WI, USA) to generate the corresponding luciferase reporter constructs, termed as circ_0020123-WT/MUT, KIAA1522 3’-UTR-WT/MUT. Thereafter, A549 and PC9 cells were co-transfected with reporter constructs and miR-940 mimic or miR-NC. After 24 h of transfection, relative luciferase activity was evaluated by the dual-luciferase reporter analysis system (Promega).
RNA immunoprecipitation (RIP) assay
A549 and PC9 cells were homogenized by RIPA lysis buffer, then cell lysate was incubated with magnetic bead-conjugated with Ago2 antibody and negative control IgG antibody (Millipore, Billerica, MA, USA). After proteinase K buffer treatment, the immunoprecipitated RNA was isolated, purified and measured using qRT-PCR.
Pull-down assay
The biotin-labeled RNA probes targeting miR-940 (Bio-miR-940) or miR-NC (Bio-NC) were synthesized by GenePharma Company (Shanghai, China). Approximately 1 × 107 cells were lysed in lysis buffer, and incubated with the biotin-labeled probes for 3 h at 4°C. Next, the complexes were mixed with streptavidin magnetic beads (Thermo Fisher Scientific). Following incubation for 2 h, the RNA complexes bound to the beads were eluted and isolated, and then determined using qRT-PCR assay.
Western blot
The proteins were isolated from tissues and cells using RIPA lysis solution (Beyotime, Shanghai, China). Extracted proteins were loaded onto a sodium dodecyl sulfate polyacrylamide gel electrophoresis minigel for separation and then shifted to polyvinylidene difluoride membranes (Millipore). Afterward, membranes were interacted with primary antibody against KIAA1522 (1:1000, ab122203) and β-actin (1:5000, ab8226), obtained from Abcam (Cambridge, MA, USA), followed by incubation with HRP-conjugated secondary antibody (1:3000, Sangon, Shanghai, China). The blots were quantified using an enhanced chemiluminescence kit (Sangon).
Xenograft experiments in vivo
The study in nude mice was manipulated in line with the procedure approved by the Ethics Committee of Tumor Hospital Affiliated to Nantong University. BALB/c nude mice (4–6 weeks old) obtained from Shanghai Laboratory Animal Center were randomly divided into two groups (N = 7). All mice were grown in controlled 40–70% humidity and ambient temperature (24 ± 2°C) with a 12-h light/dark cycle. Standard pelleted chow and drinking water were available ad libitum. All mice were acclimatized for at least 1 week prior to study, and the health and behavior of mice were observed every day. We also exclude the male mice with body weight below 15 g or over 25 g at 6 weeks of age. Mice with weakness, mental fatigue, ill looking, teeth problem or hair diseases are also excluded from study. A549 cells (2 × 106 cells/mouse) stably transfected with sh-NC or sh-circ_0020123 were subcutaneously injected into the right flank of randomly assigned mice in two groups to establish murine xenograft models. Tumor volume was calculated every week after 7 days postinoculation. At day 35, the mice were sacrificed by cervical dislocation after deep anesthesia with 2% isoflurane, and tumors were excised, weighed and harvested for further determination.
Statistical analysis
Data from thrice-repeated experiments were showed as mean ± standard deviation (SD). All statistical analyses were conducted using the GraphPad Prism 7.0 software. The comparison was performed with Student’s t-test or one-way analysis of variance (ANOVA) as appropriate. The linear relationship was determined via Pearson’s correlation coefficient. Differences suggested statistically significant at P < 0.05.
Results
The expression profile of circ_0020123 in NSCLC
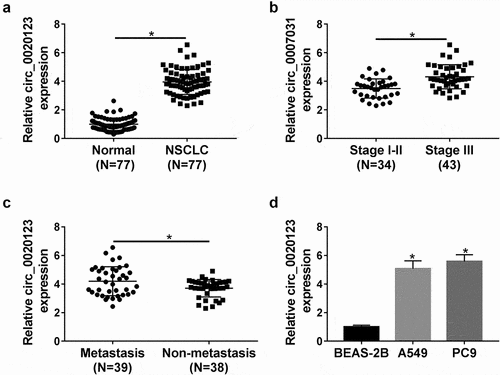
First, the expression levels of circ_0020123 in NSCLC were assessed. The qRT-PCR analysis showed circ_0020123 expression was significantly elevated in NSCLC tissues relative to para-carcinoma tissues (), which was also confirmed by IHC staining (Fig. S1). Besides that, it was also uncovered that higher circ_0020123 expression was more commonly observed in patients with advanced TNM stages () and with metastasis (). Moreover, the detailed correlation between circ_0020123 expression and clinical characteristics in patients with NSCLC is shown in . The high expression of circ_0020123 was significantly correlated with tumor size, pathological type, clinical stage, differentiation, and lymph node metastasis. After that, circ_0020123 expression in two NSCLC cell lines (A549 and PC9) was also found to be up-regulated by contrast with BEAS-2B cells, a non-cancerous lung cell line (). These results revealed circ_0020123 was differentially expressed in NSCLC, and might be associated with the progression of NSCLC.
Figure 1. The expression profile of circ_0020123 in NSCLC. (a) qRT-PCR analysis of circ_0020123 expression level in 77 paired NSCLC tissues and adjacent normal tissues. (b, c) qRT-PCR analysis of circ_0020123 expression level in NSCLC tissues based on TNM staging and lymph node metastasis. (d) qRT-PCR analysis of circ_0020123 expression level in two NSCLC cell lines (A549 and PC9) and non-cancerous lung cell line BEAS-2B. *P < 0.05.
Identification of circ_0020123 in NSCLC cells
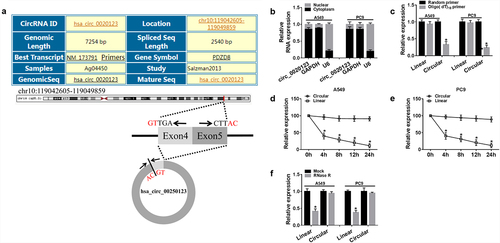
Next, the characterization of circ_0020123 was analyzed. Circ_0020123 arose from its host gene PDZD8 and consisted of the head-to-tail splicing of exon 4 and 5. Circ_0020123 is located at chr10:119,042,605–119,049,859 and the genomic length is 7254 bp (). Then, the location of circ_0020123 in NSCLC cells was explored, results showed circ_0020123 was prominently distributed in the cytoplasmic fraction in A549 and PC9 cells (). To validate that circ_0020123 was indeed circular transcript, Random and Oligo(dT)18 primers in reverse transcription experiments were used, and we found that circ_0020123 expression was lower than linear transcript (). Additionally, actinomycin D and RNase R exonuclease were further adopted. Results revealed that circ_0020123 could resistant to actinomycin D and RNase R exonuclease in A549 and PC9 cells (). All these data confirmed that circ_0020123 was indeed circular.
Figure 2. Identification of circ_0020123 in NSCLC cells. (a) The information of circ_0020123. (b) Comparison of the abundance of circ_0020123 in the nuclear and cytoplasmic using qRT-PCR. (c) qRT-PCR analysis of the expression of circ_0020123 and linear PDZD8 mRNA in reverse transcription using Random and Oligo(dT)18 primers. (d, e) qRT-PCR analysis of the abundances of circ_0020123 and PDZD8 mRNA in A549 and PC9 cells after treatment with Actinomycin D for 0, 4, 8, 12 and 24 h, respectively. (f) qRT-PCR analysis of the circ_0020123 and PDZD8 mRNA in A549 and PC9 cells treated with or without RNase R. *P < 0.05.
Effects of circ_0020123 on NSCLC tumorigenesis in vitro
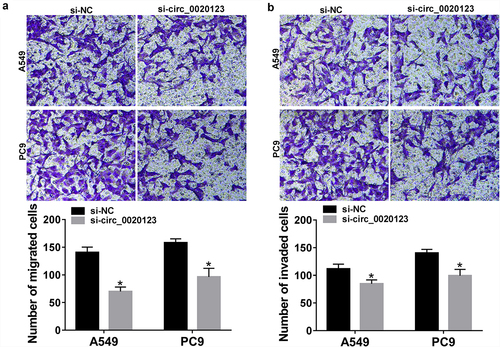
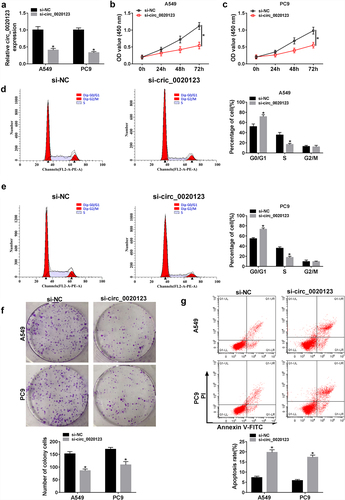
The role of circ_0020123 in NSCLC progression was studies. A549 and PC9 cells were transfected with constructed si-circ_0020123 or si-NC, as expected, circ_0020123 expression was significantly reduced in cells after si-circ_0020123 introduction (). Then, the effects of circ_0020123 on NSCLC cell growth were investigated using CCK-8 assay, flow cytometry, and colony formation assay. Results exhibited that knockdown of circ_0020123 suppressed cell viability (), induced the increase of the proportions of cells in G0/G1 phase and decrease in S phase (), attenuated the numbers of visible colonies (), and promoted the percentages of apoptotic cells () in A549 and PC9 cells, suggesting that circ_0020123 silencing repressed cell growth in NSCLC. Additionally, transwell migration and invasion assays demonstrated that circ_0020123 knockdown impaired the migration and invasion abilities of A549 and PC9 cells (). Cumulatively, circ_0020123 knockdown inhibited cell growth, migration and invasion in NSCLC.
Figure 3. Effects of circ_0020123 on NSCLC tumorigenesis in vitro. A549 and PC9 cells were transfected with si-circ_0020123 or si-NC. After transfection, (a) qRT-PCR analysis of circ_0020123 expression in cells; (b, c) CCK-8 assay of cell viability at different time points of 0, 24, 48, and 72 h; (d, e) flow cytometry assay of cell cycle; (f) colony formation assay of cell proliferation; (g) cell apoptosis analysis using flow cytometry assay. *P < 0.05.
Circ_0020123 directly bind to miR-940
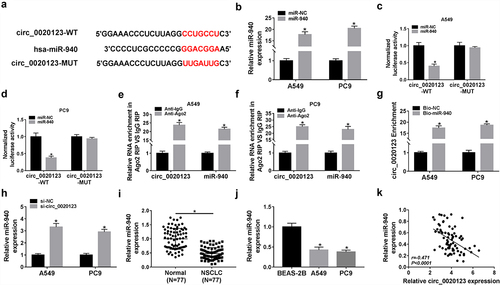
Considering that circRNAs have been reported to function as miRNA sponges to regulate the abundance of available miRNAs, and circ_0020123 prominently located in the cytoplasm, we speculated that circ_0020123 might regulate NSCLC tumorigenesis by sponging miRNAs. Through searching circinteractome online database, miR-940 was found to have the complementary sites in circ_0020123 (). Then the potential interaction between miR-940 and circ_0020123 was analyzed. First, it was found miR-940 mimic transfection significantly increased miR-940 expression in A549 and PC9 cells (). Next, results of dual-luciferase reporter assay showed that miR-940 mimic markedly reduced the luciferase activity of A549 and PC9 cells transfected with circ_0020123-WT (). RIP assay suggested that miR-940 and circ_0020123 were highly enriched in the complex precipitated by Ago2 antibody relative to nonspecific IgG control antibody in A549 and PC9 cells (). Moreover, pull-down assay exhibited that circ_0020123 was enrichment in miR-940-biotin-labeled probe (Bio-miR-940) compared with the negative control in A549 and PC9 cells (). In addition, we also observed that miR-940 expression was elevated in circ_0020123-decreased A549 and PC9 cells (). After that, the expression profile of miR-940 in NSCLC was analyzed, qRT-PCR analysis displayed that miR-940 was lowly expressed in NSCLC tissues and cell lines (), and a negative correlation between miR-940 and circ_0020123 expression in NSCLC tissues was detected (). All these data confirmed that circ_0020123 directly targeted miR-940 in NSCLC cells and negatively regulated its expression.
Figure 5. Circ_0020123 directly bind to miR-940. (a) Schematic illustration exhibiting the binding sites between circ_0020123 and miR-940. (b) qRT-PCR analysis of miR-940 expression in A549 and PC9 cells transfected with miR-NC or miR-940. (c, d) Luciferase activity analysis in A549 and PC9 cells co-transfected with the luciferase reporter plasmid and the indicated miRNAs using the dual-luciferase reporter assay. (e, f) The enrichment of circ_0020123 and miR-940 in the immunoprecipitated RNA of A549 and PC9 cells using anti-Ago2 or anti-IgG. (g) qRT-PCR analysis of circ_0020123 enrichment in A549 and PC9 cells with Bio-NC or Bio-miR-940. (h) qRT-PCR analysis of miR-940 in circ_0020123-silenced A549 and PC9 cells. (i, j) qRT-PCR analysis of miR-940 expression level in 77 paired NSCLC tissues and adjacent normal tissues, as well as in NSCLC cell lines (A549 and PC9) and non-cancerous lung cell line BEAS-2B. (k) The correlation between miR-940 level and circ_0020123 expression in NSCLC tissues using the Spearman test. *P < 0.05.
MiR-940 mediates the effects of circ_0020123 on NSCLC tumorigenesis
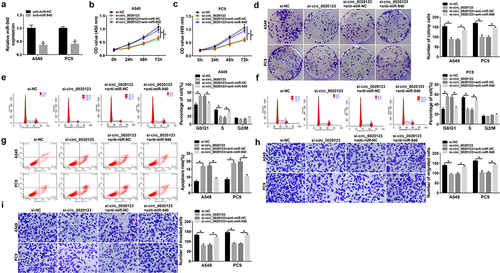
We then attempted to investigate whether miR-940 mediated the effect of circ_0020123 on the progression of NSCLC. The results indicated that the introduction of miR-940 inhibitor significantly triggered reduction of miR-940 level in A549 and PC9 cells (), then the anti-miR-NC or anti-miR-940 was transfected into circ_0020123-silenced A549 and PC9 cells to conduct rescue assay. We found that miR-940 down-regulation reversed si-circ_0020123-evoked cell proliferation inhibition (), cell cycle arrest (), cell apoptosis enhancement () as well as cell migration and invasion abilities suppression () in A549 and PC9 cells. Taken together, inhibition of miR-940 reversed the anticancer effects of circ_0020123 knockdown on NSCLC.
Figure 6. MiR-940 mediates the effects of circ_0020123 on NSCLC tumorigenesis. (a) qRT-PCR analysis of miR-940 expression in A549 and PC9 cells transfected with anti-miR-NC or anti-miR-940. The anti-miR-NC or anti-miR-940 was transfected into circ_0020123-silenced A549 and PC9 cells. After transfection, (b-d) cell proliferation analysis using CCK-8 assay and colony formation assay; (e-g) flow cytometry assay of cell cycle and cell apoptosis; (h, i) cell migration and invasion ability analysis using transwell assay. *P < 0.05.
KIAA1522 is a target of miR-940
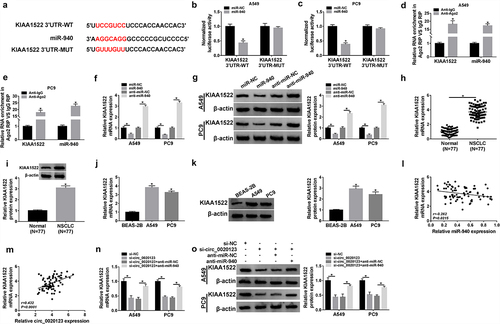
The downstream genes underlying miR-940 were further predicted through using miRDB online database, and KIAA1522 was identified to be a potential target of miR-940 (). Later, the dual-luciferase reporter assay indicated that miR-940 overexpression significantly reduced the luciferase activity of KIAA1522 3’ UTR-WT but not the KIAA1522 3’ UTR-MUT in A549 and PC9 cells (). Meanwhile, RIP assay exhibited the enrichment of Ago2 antibody on KIAA1522 and miR-940 levels in A549 and PC9 cells compared with IgG control antibody (). Besides that, it was also observed that miR-940 re-expression suppressed KIAA1522 expression, while its down-regulation increased KIAA1522 expression in A549 and PC9 cells (). All these results revealed that miR-940 targetedly repressed KIAA1522 expression in NSCLC cells. Subsequently, KIAA1522 was found to be distinctly up-regulated in NSCLC tissues and cell lines (), and its expression was negatively correlated with miR-940 (), but positively correlated with circ_0020123 (). Importantly, suggested that the levels of KIAA1522 in A549 and PC9 cells were reduced by circ_0020123 knockdown both at mRNA and protein level, which were recovered by the introduction of miR-940 inhibitor. Thus, we confirmed that circ_0020123 could regulate KIAA1522 expression via miR-940 in NSCLC.
Figure 7. KIAA1522 is a target of miR-940. (a) Schematic illustration presenting the binding sites between KIAA1522 and miR-940. (b, c) Luciferase activity analysis in A549 and PC9 cells co-transfected with the luciferase reporter plasmid and miR-NC or miR-940 using the dual-luciferase reporter assay. (d, e) The enrichment of KIAA1522 and miR-940 in anti-Ago2 or anti-IgG in A549 and PC9 cells. (f, g) qRT-PCR and Western blot analysis of KIAA1522 expression in miR-940-overexpressed or miR-940-down-regulated A549 and PC9 cells. (h-k) qRT-PCR and Western blot analysis of KIAA1522 expression level in 77 paired NSCLC tissues and adjacent normal tissues, as well as in NSCLC cell lines (A549 and PC9) and non-cancerous lung cell line BEAS-2B. (l, m) The correlation between KIAA1522 expression and circ_0020123 or miR-940 level in NSCLC tissues using the Spearman test. (n, o) qRT-PCR and Western blot analysis of KIAA1522 expression in A549 and PC9 cells transfected with si-NC, si-circ_0020123, si-circ_0020123 + anti-miR-NC, or si-circ_0020123 + anti-miR-940. *P < 0.05.
MiR-940 suppresses NSCLC tumorigenesis through targeting KIAA1522
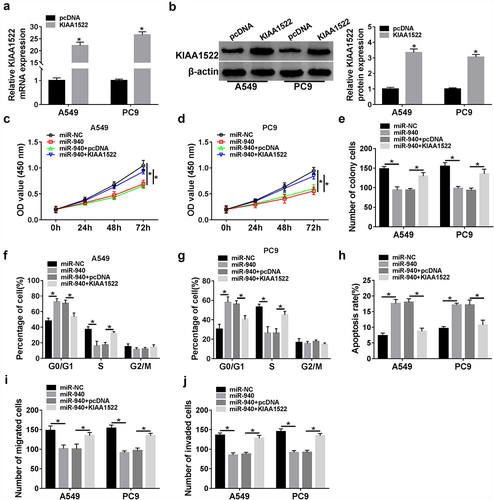
Based on the aforementioned results, miR-940 directly targeted KIAA1522, we then studied whether KIAA1522 might function as an important factor in miR-940-mediated NSCLC progression. First, A549 and PC9 cells were transfected with KIAA1522 or pcDNA, results showed that KIAA1522 expression was significantly up-regulated in cells after KIAA1522 introduction in comparison with homologous controls (). Next, rescue assay was conducted by transfecting miR-NC, miR-940, miR-940 + pcDNA, or miR-940 + KIAA1522 into NSCLC cells. In the CCK-8 and colony formation assays, miR-940 re-expression decreased cell proliferation, which were strikingly reversed by KIAA1522 up-regulation in A549 and PC9 cells (). The data from flow cytometry indicated that miR-940 overexpression induced cell cycle arrest and promoted cell apoptosis, while these effects were attenuated by KIAA1522 up-regulation in A549 and PC9 cells (). Moreover, miR-940 restoration-elicited inhibition of migration and invasion was significantly abolished after KIAA1522 up-regulation in 549 and PC9 cells (). Altogether, miR-940 suppressed NSCLC cell growth, migration, and invasion through targeting KIAA1522.
Figure 8. MiR-940 suppresses NSCLC tumorigenesis through targeting KIAA1522. (a, b) qRT-PCR and Western blot analysis of KIAA1522 expression in A549 and PC9 cells transfected with pcDNA or KIAA1522. A549 and PC9 cells were transfected with miR-NC, miR-940, miR-940 + pcDNA, or miR-940 + KIAA1522. After transfection, (c-e) cell proliferation analysis using CCK-8 assay and colony formation assay; (f-h) flow cytometry assay of cell cycle and cell apoptosis; (i, j) transwell assay of cell migration and invasion ability. *P < 0.05.
Circ_0020123 silencing hinders tumor growth in vivo
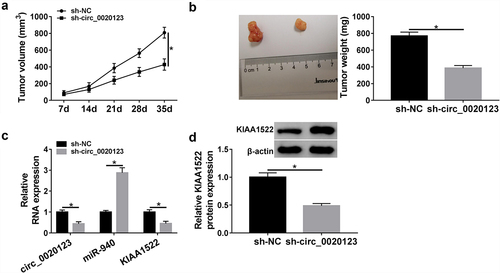
The underlying effects and mechanisms of circ_0020123 on tumor growth in vivo were further investigated. As presented in A and B, down-regulation of circ_0020123 reduced tumor volume and weight compared with the control group. Further molecular analyses suggested that the expression levels of circ_0020123 and KIAA1522 were lower, while miR-940 expression level was higher in tumors excised from sh-circ_0020123 murine xenograft than those of sh-NC murine xenograft (). However, the opposite functions were observed after circ_0020123 overexpression in xenograft model. After confirming the transfection efficiency of lentivirus mediated circ_0020123 (Fig. S2A), we found that overexpression of circ_0020123 promoted tumor growth in nude mice (Fig. S2B, C). Besides that, circ_0020123 and KIAA1522 were higher, while miR-940 was lower in xenograft tumors of circ_0020123 group than those in Vector group (Fig. S2D, E). Collectively, we verified that knockdown of circ_0020123 could impede tumor growth in vivo via regulating miR-940 and KIAA1522.
Figure 9. Circ_0020123 silencing hinders tumor growth in vivo. (a) The tumor volume was evaluated every week after 7 days postinoculation. (b) Images of tumors and tumor weight after 35 days were calculated. (c, d) qRT-PCR and Western blot analysis of circ_0020123, miR-940, and KIAA1522 expression in excised tumors. *P < 0.05.
Discussion
NSCLC is one of the most life-threatening malignancy in the world, which is mainly ascribed to high rate of distant metastasis, recurrence and drug resistance [Citation4–6]. Therefore, further investigations of novel diagnostic, therapeutic and prognostic indicators for NSCLC are urgent. This study first demonstrated that circ_0020123/miR-940/KIAA1522 network may contribute to the tumorigenesis of NSCLC.
CircRNAs are a group of endogenous RNAs widely expressed in various cell types, they are characterized by a ring structure and high specie-, tissue- and cell-specific expressed [Citation18,Citation19], which make them have high stability and specificity, suggesting that circRNAs may serve as a biomarker candidates in many diseases, including cancers [Citation20]. In NSCLC, some circRNAs have been identified to be involved in the progression and development of this cancer. For example, Chen et al. revealed that circRNA 100146 was significantly increased in NSCLC, and promoted cell malignant biological behaviors [Citation21]. Jin et al. showed an up-regulation of circARHGAP10 expression in NSCLC, and circARHGAP10 knockdown inhibited cell glycometabolism, proliferation and metastasis in cancer [Citation22]. Zhang’s team demonstrated that hsa_circRNA_101237 was increased and contributed to cell proliferative, migratory and invasive capacities in NSCLC. The above data indicates that circRNAs may be promising biomarkers for NSCLC therapy. In this study, circ_0020123 showed a high expression level in NSCLC, especially in patients with advanced TNM stages and with lymph node metastasis. Subsequent analysis suggested that circ_0020123 was also up-regulated in NSCLC cells, functionally, silencing of circ_0020123 repressed cell proliferative, invasive, migratory capacities, induced cell cycle arrest, and promoted apoptosis in vitro. Importantly, clinical relevance of circ_0020123 was validated by the observed promotion in NSCLC tumorigenic in vivo using xenograft tumor assay. Thus, circ_0020123 exerts oncogenic functions in NSCLC, and circ_0020123 siRNA may be a useful therapeutic molecular for NSCLC molecular targeted therapy.
The subcellular localization of circ_0020123 was identified to be mainly expressed in cytoplasm, previous studies have reported that circRNAs located in cytoplasm can modulate gene expression through functioning as ceRNA, whereby circRNAs can act as “sponges” of miRNAs, thus abrogating their function to target mRNAs [Citation23–25]. Hence, the circ_0020123-miRNA-mRNA network, which is involved in the progression of NSCLC was investigated. We first confirmed that miR-940 was a target of circ_0020123 in NSCLC. Gu et al. suggested miR-940 was decreased in NSCLC, and lowly expressed miR-940 indicated a shorter overall survival rate, and restoration of miR-940 reduced cell viability through targeting FAM83 F in NSCLC [Citation26]. Jiang et al. showed miR-940 overexpression in NSCLC cells suppressed TGF-β-induced epithelial–mesenchymal transition (EMT) through directly interacting with Snail [Citation27]. In our study, we also observed a down-regulated expression of miR-940 in NSCLC, and re-expression of miR-940 suppressed cell growth, invasion and migration in vitro; besides that, it was proved that silencing of miR-940 reversed the regulatory effects of circ_0020123 knockdown on NSCLC tumorigenesis. Thus, we firstly identified the circ_0020123/miR-940 axis in NSCLC progression.
KIAA1522 is a newly cloned, large protein-coding gene of uncharacterized functions. However, former studies have shown KIAA1522 was elevated in multiple cancers, such as esophageal cancer, hepatocellular carcinoma and breast cancer, and functioned as an oncogene to promote malignant progression of cancers [Citation28–30]. Furthermore, KIAA1522 was also proved to be increased in NSCLC, and was positively associated with poor overall survival of patients, and accelerated cell growth [Citation31]. All these data suggested the oncogenic roles of KIAA1522 in malignancies. In the present study, we verified that miR-940 directly targeted KIAA1522, and circ_0020123 acted as a ceRNA to bind to miR-940, and then up-regulated KIAA1522 expression in vitro and in vivo. What’s more, we validated that miR-940 performed anticancer effects via regulating KIAA1522. Thus, a circ_0020123/miR-940/ KIAA1522 regulatory network in NSCLC progression was confirmed.
In conclusion, this study uncovered that circ_0020123 contributed to NSCLC tumorigenesis and growth via regulating miR-940/KIAA1522 axis, indicating the clinical applicability of circ_0020123 siRNA as the anticancer drug in NSCLC.
Supplemental Material
Download Zip (3.7 MB)Acknowledgments
None
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Supplementary material
Supplemental data for this article can be accessed here
Additional information
Funding
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30.
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132.
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30.
- Rocco D, Della Gravara L, Battiloro C, et al. The role of combination chemo-immunotherapy in advanced non-small cell lung cancer. Expert Rev Anticancer Ther. 2019;19(7):561–568.
- MacDonagh L, Gray SG, Breen E, et al. Lung cancer stem cells: the root of resistance. Cancer Lett. 2016;372(2):147–156.
- Ettinger DS, Wood DE, Aisner DL, et al. Non-Small cell lung cancer, version 5.2017, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2017;15(4):504–535.
- Rybak-Wolf A, Stottmeister C, Glažar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58(5):870–885.
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338.
- Zhou R, Wu Y, Wang W, et al. Circular RNAs (circRNAs) in cancer. Cancer Lett. 2018;425:134–142.
- Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691.
- Yu T, Wang Y, Fan Y, et al. CircRNAs in cancer metabolism: a review. J Hematol Oncol. 2019;12(1):90.
- Hua X, Sun Y, Chen J, et al. Circular RNAs in drug resistant tumors. Biomed Pharmacother. 2019;118:109233.
- Zeng K, Chen X, Xu M, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9(4):417.
- Yang C, Yuan W, Yang X, et al. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol Cancer. 2018;17(1):19.
- Chen RX, Liu HL, Yang LL, et al. Circular RNA circRNA_0000285 promotes cervical cancer development by regulating FUS. Eur Rev Med Pharmacol Sci. 2019;23(20):8771–8778.
- Zhang S, Mo Q, Wang X. Oncological role of HMGA2 (Review). Int J Oncol. 2019;55(4):775–788.
- Qu D, Yan B, Xin R, et al. A novel circular RNA hsa_circ_0020123 exerts oncogenic properties through suppression of miR-144 in non-small cell lung cancer. Am J Cancer Res. 2018;8(8):1387–1402.
- Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148.
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338.
- Zhao ZJ, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 2017;14(5):514–521.
- Chen L, Nan A, Zhang N, et al. Circular RNA 100146 functions as an oncogene through direct binding to miR-361-3p and miR-615-5p in non-small cell lung cancer. Mol Cancer. 2019;18(1):13.
- Jin M, Shi C, Yang C, et al. Upregulated circRNA ARHGAP10 predicts an unfavorable prognosis in NSCLC through regulation of the miR-150-5p/GLUT-1 Axis. Mol Ther Nucleic Acids. 2019;18:219–231.
- Du WW, Zhang C, Yang W, et al. Identifying and characterizing circRNA-Protein interaction. Theranostics. 2017;7(17):4183–4191.
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388.
- Lv X, Wang M, Qiang J, et al. Circular RNA circ-PITX1 promotes the progression of glioblastoma by acting as a competing endogenous RNA to regulate miR-379-5p/MAP3K2 axis. Eur J Pharmacol. 2019;863:172643.
- Gu GM, Zhan YY, Abuduwaili K, et al. MiR-940 inhibits the progression of NSCLC by targeting FAM83F. Eur Rev Med Pharmacol Sci. 2018;22(18):5964–5971.
- Jiang K, Zhao T, Shen M, et al. MiR-940 inhibits TGF-β-induced epithelial-mesenchymal transition and cell invasion by targeting Snail in non-small cell lung cancer. J Cancer. 2019;10(12):2735–2744.
- Xie ZH, Yu J, Shang L, et al. KIAA1522 overexpression promotes tumorigenicity and metastasis of esophageal cancer cells through potentiating the ERK activity. Onco Targets Ther. 2017;10:3743–3754.
- Li Y, Wang Y, Fan H, et al. miR-125b-5p inhibits breast cancer cell proliferation, migration and invasion by targeting KIAA1522. Biochem Biophys Res Commun. 2018;504(1):277–282.
- Jiang S, Zhang Y, Li Q, et al. KIAA1522 promotes the progression of hepatocellular carcinoma via the activation of the Wnt/β-Catenin signaling pathway. Onco Targets Ther. 2020;13:5657–5668.
- Liu YZ, Yang H, Cao J, et al. KIAA1522 is a novel prognostic biomarker in patients with non-small cell lung cancer. Sci Rep. 2016;6(1):24786.