ABSTRACT
LINC01234 has been suggested to correlate with the survival of ovarian cancer (OS), but its role in the properties of OC stem cells (OCSCs) has been rarely described. We aim to investigate the effect of LINC01234 on the differentiation and self-renewal of OCSCs through adsorption of microRNA (miR)-27b-5p to target sirtuins 5 (SIRT5). Expression of LINC01234 and SIRT5 in OC and normal samples included in TCGA and GTEx was searched through the GEPIA2 database. Bioinformatics analysis was conducted to predict the relation of LINC01234, miR-27b-5p and SIRT5. Expression of LINC01234, miR-27b-5p and SIRT5 in OC tissues and cells was detected. OCSCs were cultured and identified. CD133+ OCSCs were introduced with related oligonucleotides or vectors of LINC01234 or miR-27b-5p and SIRT5 to figure out their roles in OCSCs progression and tumorigenesis in vivo. The interaction of miR-27b-5p with LINC01234 or SIRT5 was analyzed. Bioinformatics analysis suggested that LINC01234 was very likely to influence SIRT5 and regulate the development of OC through miR-27b-5p. Up-regulated LINC01234 exhibited in OC tissues and cells. Down-regulated LINC01234 or elevated miR-27b-5p suppressed OCSCs progression and tumorigenesis in vivo. LINC01234 could restore SIRT5 expression by binding to miR-27b-5p. Down-regulated miR-27b-5p reversed the effect of silenced LINC01234 on OCSCs development and tumorigenesis in vivo. Up-regulation of SIRT5 reduced the effects of elevated miR-27b-5p on OCSCs progression and tumorigenesis in vivo. LINC01234 regulates miR-27b-5p to induce the migration, invasion and self-renewal of OCSCs through targeting SIRT5.
Introduction
Ovarian cancer (OC) is one of the malignant tumors with high incidence, and in 2020, the incidence rate worldwide was 6.6% based on Global Cancer Observatory, and 55,342 cases were diagnosed in China. In addition, the symptoms are not distinct in early OC, and OC is short of simple and practical diagnostic approaches [Citation1]. The poor prognosis of patients with OC results from the inoperative control of advanced cancer growth, metastasis and recurrence [Citation2]. The OC therapy at present includes debulking surgery and platinum- and taxane-based chemotherapy, but most patients have a recurrence within short years [Citation3]. Therefore, improving the prognosis of OC remains a vital focus of research efforts at present.
Long non-coding RNA (lncRNA) genes transcribed but not translated, longer than 200 nucleotides, are classified as a separate class of non-protein coding genes that could encode small and functional peptides [Citation4,Citation5]. LINC01234 is an identified lncRNA that is associated with the oncological processes, such as non-small-cell lung cancer cell metastasis [Citation6] and hepatocellular carcinoma cell proliferation [Citation7]. A study has revealed that depression of LINC01234 inhibits esophageal cancer (EC) development [Citation8]. In addition, a study has emerged that LINC01234 facilitates oral squamous cell carcinoma (OSCC) development and might be a possible therapeutic target for OSCC [Citation9] and LINC01234 could effectively classify OC patients with wild-type BRCA1/2, and LINC01234 is correlated to the progression and prognosis of OC patients [Citation10]. While the mechanism of LINC01234 in OC has not been elucidated.
Through a bioinformatic website, we found that there existed binding sites between LINC01234 and microRNA-27b-5p (miR-27b-5p). MiRNAs, evolutionarily conserved small noncoding RNA, take a crucial role in modulating gene expression [Citation11]. A study has suggested that miR-27b is a novel inhibitor of OC cell-mediated vascular mimicry, offering a new possible drug candidate for anti-OC therapy [Citation12]. In the meantime, a study has manifested that elevated miR-27b-5p can cancel Mono-2-ethyhexyl phthalate-induced cell proliferation and proliferating cell nuclear antigen expression in OSCC cells [Citation13]. It has been reported that miR-27b-5p expression is low in OC, and miR-27b-5p may inhibit the development of ovarian cancer by targeting downstream [Citation14].
Importantly, we predicted a targeting relationship between miR-27b-5p and sirtuins 5 (SIRT5). SIRT, also named as Sir2-like proteins, is part of the class III histone deacetylases and is extensively conserved in all kinds of species. Belonging to the SIRT family, SIRT5 is featured by obvious lysine-demalonylase, -deglutarylase, and -desuccinylase activities [Citation15]. A study has revealed that SIRT5 enhances cisplatin resistance in OC [Citation16] and it is a possible target for accurate therapy for OC patients [Citation17].
Cancer stem cells (CSCs) are frequently resistant to common cancer therapies [Citation18]. Targeting CSCs represents a promising strategy for overcoming therapy resistance and reducing mortality of OC, but further efforts must be made to improve our understanding of the mechanisms involved in therapy resistance [Citation19]. Therefore, we aim to investigate the effect of LINC01234 on the differentiation and self-renewal of OCSCs through the adsorption of miR-27b-5p to target SIRT5.
Materials and methods
Bioinformatics analysis
Through the GEPIA2 database (http://gepia2.cancer-pku.cn/#index), the expression of LINC01234 and SIRT5 in OC and normal samples included in TCGA and GTEx was searched, and the highly-expressed genes were obtained. Through the RNA22 database (http://gepia2.cancer-pku.cn/#index), the target binding site of LINC01234 and miR-27b-5p was predicted. On the TargetScan database (http://www.targetscan.org/vert_71/) and mirDIP database (http://ophid.utoronto.ca/mirDIP/index.jsp#r), the target genes of miR-27b-5p were predicted. Through the GeneMANIA database (http://genemania.org/), the association analysis of candidate target genes was performed and SIRT5 associated genes were analyzed. Using the KOBAS3.0 database (http://kobas.cbi.pku.edu.cn/kobas3/help/), KEGG pathway enrichment analysis on SIRT5 related genes was conducted. Subsequently, the target binding site of miR-27b-5p and SIRT5 was obtained through the TargetScan database [Citation20,Citation21].
Ethical approval
All human materials were acquired in line with consent regulation in the Declaration of Helsinki and approved by the Ethics Review Committee of the Third Affiliated Hospital of Zhengzhou University (ethical number: 201,720,523). Participants provided written informed consent to participate in this study. In addition, this study met the ethical requirements of animal experiments and was approved by the Institutional Animal Care and Use Committee of the Third Affiliated Hospital of Zhengzhou University (ethical number: 201,740,617).
Cells and clinical specimens
Four OC cell lines (SKOV3 CAOV3, HO8910, A2780) and normal ovarian cell line (IOSE80) were all purchased from Shanghai Institute of Cell Biology, Chinese Academy of Science (Shanghai, China).
OC tissues and adjacent normal tissues from 104 patients who accepted OC surgery in the Third Affiliated Hospital of Zhengzhou University were collected. The tissues were frozen at −80°C, dehydrated, embedded and preserved in paraffin [Citation22,Citation23].
Culture of A2780 OCSCs
OC cells A2780 were detached with 0.25% trypsin, joined with Dulbecco’s Modified Eagle Medium (DMEM)/F12 (Gibco, CA, USA) containing 10% fetal bovine serum (TBD, Tianjin, China) and centrifuged. The cells were joined with serum-free DMEM/F12 containing human epidermal growth factor (PeproTech, Rocky Hill, NJ, USA), basic fibroblast growth factor (PeproTech), bovine serum albumin (BSA) (Sigma, Missouri, USA) and human insulin (Sigma), triturated and filtered. Then the cells were counted, seeded and incubated. When A2780 cells were suspended in a serum-free medium with a large number, the suspended cell mass was mechanically triturated with a straw to prepare a single-cell suspension, which was then seeded and cultured, and the solution was changed every other day. Finally, the cells were passaged after 3–4 d culture, and those at passage 3 were used for subsequent experiments [Citation24].
Identification of OCSCs
Flow cytometry detection: A2780 cells and A2780 stem cells were centrifuged and harvested. The cell suspension (1 × 107 cells/mL, 100 μL) was joined with 1 μg fluorescein-labeled mouse anti-human CD133 antibody (Miltenyi Biotec, Bergisch Gladbach, Germany), and added with the same type of immunoglobulin G (IgG) antibody (Adlitteram Diagnostic laboratories, Phoenix, USA) in the control group. The cells were suspended in 200 μL ice phosphate buffer saline (PBS) and detected on the machine.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR) was applied to detect the expression of stem cell-related genes CD44, Nanog and Oct4. The CSCs were enriched through tumor sphere formation experiments [Citation22].
Cell transfection
The cells were seeded into the 6-well plate 24 h before treatment. With 50% cell confluence, OCSCs were transfected via lipofectamine 2000 reagent (Invitrogen, CA, USA) for 6 h. OCSCs continued to be cultured for 48 h. Stably transfected OCSCs were selected by puromycin (2.5 μg/mL, Sigma Aldrich, MO, USA) and then harvested for subsequent experiments [Citation22]. All constructs were provided by ThermoFisher Scientific (Waltham, MA, USA).
RT-qPCR
The total RNA of the cells and tissues was extracted via Trizol reagent (ThermoFisher Scientific). After determination of the concentration, the RNA was reversely transcribed into cDNA using Prime Script RT kits (Takara Bio Inc., Shiga, Japan) or miRcute Plus miRNA first strand cDNA synthesis kit (TIANGEN, Beijing, China). With cDNA as the template, the detection was conducted via real-time fluorescence quantitative PCR kit (ThermoFisher Scientific). Data were analyzed by 2−ΔΔCt method. U6 was the loading control of miR-27b-5p, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was that of LINC01234, SIRT5, CD44, Nanog and Oct4. The primer sequences shown in .
Table 1. Primer sequences for gene in our study
Western blot analysis
Total protein was extracted using ice-cold radio immunoprecipitation assay buffer (RIPA; Beyotime Institute of Biotechnology, Shanghai, China) containing 0.01% protease and phosphatase inhibitor (Sigma, Shanghai, China) and incubated on ice. The quantitative protein was isolated on 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis, electro-blotted onto the membrane, and sealed with 5% skim milk for 2 h. The membrane was incubated with primary antibodies SIRT5 (1:1000, Cell Signaling, Beverly, MA, USA) and GAPDH (1: 1000, Santa Cruz Biotechnology, CA, USA), as well as with secondary antibody. The bands were visualized in an enhanced chemiluminescence system. The gray value of protein bands was quantified by Image Lab software (National Institutes of Health, MD, USA).
Colony formation assay
Each well of the 6-well plate was seeded with 500 cells, with 3 wells in each group. After 12-d culture, the cells were stained with Giemsa solution (Sigma). The colony formation efficiency was calculated as (the number of colonies/the number of seeded cells) × 100% [Citation25].
Matrigel invasion assay
The Matrigel invasion assay was done using the BD Biocoat Matrigel Invasion Chamber (BD Biosciences, NJ, USA). From 5 randomly selected fields, the cells that had invaded through the membrane to the lower surface were counted under a light microscope [Citation25].
Wound-healing assay
Cells were wounded by dragging a 1 mL pipette tip across their monolayer. Images were taken using a DMI6000 inverted microscope (Leica Microsystems GmbH, Wetzlar, Germany) at 0 h and 24 h [Citation25].
Plasmids construction and 3ʹUntranslated regions (UTR) luciferase reporter assays
The targeting site sequence (wild type [WT]) of LINC01234 and SIRT5 mRNA 3’-UTR was artificially synthesized, and the WT targeting site was mutated to obtain the mutant type (MUT) sequence. The pmiR-RB-REPORTTM plasmid was digested using a restriction enzyme, and the artificially synthesized target gene segments WT and MUT were respectively inserted into pmiR-RB-REPORTTM vector (constructed by Guangzhou RiboBio Co., Ltd., Guangdong, China). Meanwhile, with empty plasmid used as the control, the sequenced luciferase reporter plasmids WT and MUT were used for subsequent transfection. Vectors containing WT and MUT were co-transfected with NC mimic or miR-27b-5p mimic into cells, respectively. Transfected for 48 h, ct he relative luciferase activity was determined using Renilla luciferase detection kits and analyzed using dual luciferase reporter analysis system (Promega, WI, USA) with firefly luciferase as the internal reference [Citation26].
RNA immunoprecipitation (RIP) assay
EZ-Magna RIP RNA-binding protein immunoprecipitation kit (Millipore Corporation, Billerica, Massachusetts, USA) was used for RIP assay. A2780 cells were lysed using the lysis buffer added with RNasin (Takara) and protease inhibitor (Roche Diagnostics GmbH, Mannheim, Germany). The lysate was centrifuged for 0.5 h and the supernatant was subjected to immunoprecipitation with anti-Ago2 or anti-IgG-coated magnetic beads [Citation27].
RNA pull-down assay
RNAs were labeled by biotin using the Pierce RNA 3ʹEnd Desthiobiotinylation Kit (Thermo Fisher Scientific). Briefly, A2780 cells were treated using 50 nmol/L biotinylated WT miR-27b-5p (WT-bio-miR-27b-5p) or MUT miR-27b-5p (MUT-bio-miR-27b-5p) and lysed for 0.5 h. Thereafter, the lysate was incubated with streptavidin magnetic beads, pre-coated using RNase-free BSA and yeast tRNA for 3 h. RT-qPCR was implemented to analyze LINC01234 content [Citation27].
In vivo xenograft experiment
OCSCs were transfected with LINC01234 low expression, miR-27b-5p high/low expression and SIRT5 high expression vector and the corresponding NCs . After a screening of stably transfected cell lines, approximately 1 × 105 logarithmically grown OCSCs were injected into BALB/c nude mice (5 mice/group). After a 7-week observation, the mice were euthanized and tumors were stripped [Citation28]. Tumor weight was measured and tumor volume was calculated [Citation23].
Statistical analysis
GraphPad Prism 6.0 (GraphPad Software Inc.) was applied to analyze the data. The measurement data were expressed as mean ± standard deviation. The t test was applied for two-group comparison, analysis of variance (ANOVA) for the comparison among multiple groups, and Tukey’s post hoc test for pairwise comparisons after ANOVA. Fisher's exact test was used to compare count data. Pearson`s correlation test was employed in the correlation analysis. Kaplan-Meier (K-M) curves was used for survival analyses. P < 0.05 was considered statistically significant.
Results
Bioinformatics analysis
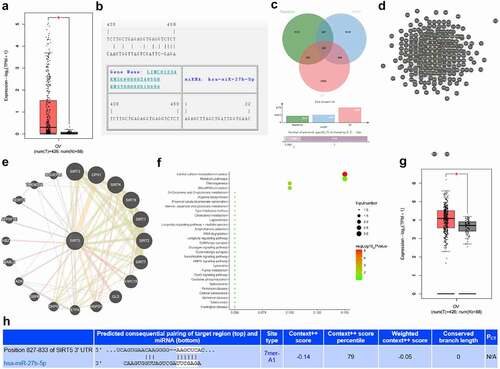
The search of the expression of LINC01234 in OC and normal samples included in TCGA and GTEx through the GEPIA database revealed that LINC01234 was highly expressed in OC ()). On the RNA22 database, there was a binding site between LINC01234 and miR-27b-5p ()). To further understand the downstream regulatory mechanism of miR-27b-5p, the TargetScan database and the mirDIP database were used to predict miRNA target genes. Through the GEPIA database, the genes that were significantly highly expressed in OC were obtained. The predicted downstream genes of miR-27b-5p and the highly expressed genes in OC were crossed, and finally, 282 candidate target genes were obtained ()). Further correlation analysis of these target genes revealed that among these target genes, SIRT5 is correlated with most genes ()). We further predicted SIRT5 related genes ()) and performed KEGG pathway enrichment analysis on the predicted genes ()). It was found that they were mainly enriched in signal pathways such as microRNAs in cancer, suggesting that the impact of SIRT5 on OC is likely to be affected by miRNAs. Further expression analysis found that SIRT5 was also highly expressed in OC ()). Targeting site prediction indicates that there was a binding site between SIRT5 and miR-27b-5p ()). These analysis results suggest that LINC01234 is very likely to influence SIRT5 and regulate the development of OC through miR-27b-5p.
Figure 1. LINC01234 downstream mechanism. A. The differential expression of LINC01234 in OC included in TCGA and GTEx. The abscissa indicated the sample type, the ordinate indicated LINC01234 expression, the red box plot indicated the tumor sample, and the gray box plot indicated the normal sample (* q < 0.01); B. LINC01234 and miR-27b-5p targeted binding site; C. miR-27b-5p downstream target genes, the three circles represented the three sets of prediction data, and the middle part represented the intersection; D. Candidate target gene correlation analysis, each circle indicated a gene, and the line between the circles indicated that there was a correlation between genes; E. SIRT5 related gene prediction, and the line indicated that there was a correlation between genes; F. KEGG pathway enrichment analysis of SIRT5 related gene, the abscissa represented GeneRatio, the ordinate represented the function item, the size of the circle represented the number of enriched genes in the pathway, the color represented the enrichment p value, and the histogram on the right indicated color scale; G. The differential expression of SIRT5 in OC included in TCGA and GTEx; H. The targeted binding site of miR-27b-5p and SIRT5.
Up-regulated LINC01234 exhibits in OC tissues and cells; identification of OCSCs
LINC01234 expression was detected in OC tissues and cells (). The results indicated that LINC01234 was highly expressed in OC tissues and cell lines. To study the relationship between LINC01234 expression and the clinicopathological features of patients with OCSCs, the patients were divided into two groups with low expression and high expression according to the median level of LINC01234 in cancer tissues. Then, study analysis revealed that a high level of LINC01234 was associated with poor survival of patients (Supplementary )) and was related to FIGO staging and lymph node metastasis, but not related to age, histological grade, and pathological subtype ().
Table 2. Association of LINC01234 expression and clinicopathological variables in OC patients
Figure 2. Up-regulated LINC01234 exhibits in OC tissues and cells and identification of OCSCs. A. LINC01234 expression in OC tissues detected via RT-qPCR; B. LINC01234 expression in OC cells detected via RT-qPCR; C. A2780 OCSCs marker CD133+ antigen flow detection, from left to right was the same type of control, CD133 antigen positive rate of A2780 cells, CD133 antigen positive rate of A2780 stem cells; D. Stem cell-related genes (CD44, Nanog, and Oct4) expression detected via RT-qPCR; * vs IOSE80 (normal ovarian cell line), P < 0.05. Panel A (n = 104), Panel B, D (N = 3). The measurement data were expressed as mean ± standard deviation.
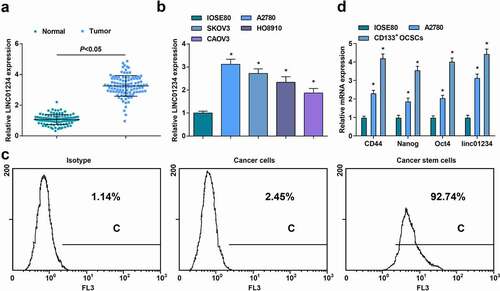
Tumor cell spheres were obtained by suspension culture in a serum-free medium containing growth factors. As manifested in flow cytometry ()), the CD133 antigen positive rate of the control cells was only 1.14%, and that of the A2780 cells was only 2.45%, but that of the A2780 tumor cell sphere (stem cells) was 92.74%.
Stem cell-related gene (CD44, Nanog, and Oct4) expression and linc01234 expression was also detected by RT-qPCR ()). It came out that CD44, Nanog, Oct4 and linc01234 expression in A2780 cells and CD133+ OCSCs were elevated, indicating the successful extraction of OCSCs.
Down-regulated LINC01234 or elevated miR-27b-5p represses OCSCs progression and tumorigenesis in vivo
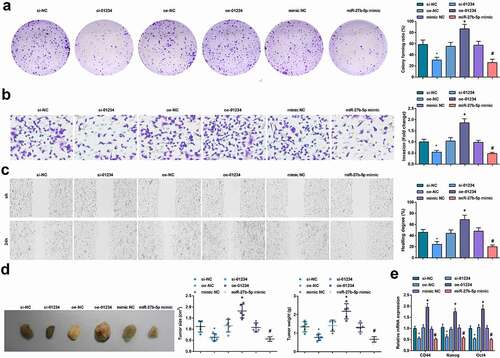
In CD133+ OCSCs isolated from A2780 cells, decreased LINC01234 or elevated miR-27b-5p could clearly reduce the abilities of colony formation, invasion and migration abilities, in vivo tumorigenicity and decrease expression of stem cell-related genes (CD44, Nanog, and Oct4). However, the effect of up-regulated LINC01234 on CD133+ OCSCs was contrary to those of decreased LINC01234. The above results suggested that down-regulation of LINC01234 or up-regulation of miR-27b-5p suppressed the proliferation, migration, invasion and self-renewal of OCSCs ().
Figure 3. Reduced LINC01234 or up-regulated miR-27b-5p restrains OCSCs progression and tumorigenesis in vivo. A. The colony formation ability examined via colony formation assay; B. The invasion ability of CD133+ OCSCs in each group detected via Transwell assay ; C. The migration ability of CD133+ OCSCs in each group detected via scratch test; D. In vivo xenograft experiment results; E. Stem cell-related genes (CD44, Nanog, and Oct4) expression detected via RT-qPCR; * vs the si-NC group, P < 0.05; + vs the oe-NC group, P < 0.05; # vs the mimic NC group, P < 0.05; Panel D (n = 5), Panel A-C, and E (N = 3).
LINC01234 was a sponge of miR-27a-5p
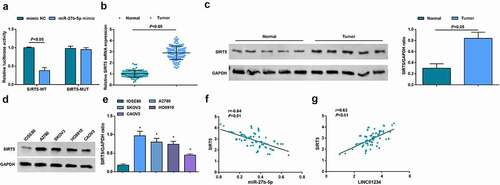
The relationship of miR-27b-5p with LINC01234 was analyzed via a bioinformatics website (https://cm.jefferson.edu/rna22/Precomputed/) ()). It came that overexpression of miR-27b-5p decreased the luciferase activity of the LINC01234-WT reporter gene. In contrast, miR-27b-5p exhibited little inhibitory effect on the luciferase activity of LINC01234-MUT reporter gene ()). RIP assay found that LINC01234 and miR-27b-5p were more enriched in A2780 cells ()) and RNA-pull down assay also confirmed binding relation between LINC01234 and miR-27b-5p ()).
Figure 4. MiR-27b-5p is a direct target of LINC01234. A. The effect of miR-27b-5p on LINC01234ʹ-WT and LINC01234ʹ-MUT luciferase reporters in A2780 cells; B. The enrichment of miR-27-5p and LINC01234 in A2780 cells indicated via RIP assay; C. The direct interaction between miR-27b-5p and LINC01234 verified via RNA pull-down assay; D. MiR-27b-5p expression in OC tissues and cells detected by RT-qPCR; E. The correlation analysis of miR-27b-5p and LINC01234 in human OC tissues; F. RT-qPCR was used to detect the expression of LINC01234 and miR-27b-5p after transfection; * vs IOSE80, P < 0.05; + vs si-NCP < 0.05; # vs oe-NC, P < 0.05; Panle B-D and F (N = 3), Panel E (n = 104).
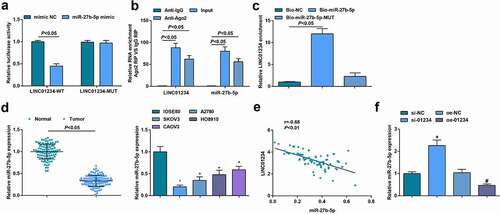
Low expression of miR-27b-5p was present in OC tissues and cells (, )). Based on the median level of miR-27b-5p in cancer tissues, patients were divided into miR-27b-5p low expression and miR-27b-5p high expression group, and study analysis suggested that patients with miR-27b-5p low expression presented poor survival (Supplementary )). In addition, the level of miR-27b-5p was related to FIGO staging and lymph node metastasis, but not related to age, histological grade, and pathological subtype (). Moreover, through Pearson analysis, it was observed that LINC01234 expression was negatively correlated with miR-27b-5p expression in OC tissues ()).
Table 3. Association of miR-27b-5p expression and clinicopathological variables in OC patients
After transfection of si-NC, si-LINC01234, oe-NC, or oe-LINC01234, RT-qPCR was used to detect the expression of miR-27b-5p. It was demonstrated that CD133+ OCSCs transfected with si-LINC01234 showed elevated miR-27b-5p expression while those transfected with oe-LINC01234 exhibited reduced miR-27b-5p expression ()). According to the above results, miR-27b-5p was a direct target of LINC01234.
Down-regulated miR-27b-5p reverses the inhibition of reduced LINC01234 on OCSCs development and tumorigenesis in vivo
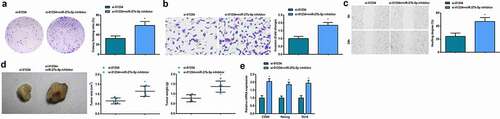
In CD133+ OCSCs isolated from A2780 cells, transfection of miR-27b-5p inhibitor based on si-LINC01234 promoted the abilities of colony formation, invasion and migration abilities, enhanced in vivo tumorigenicity and increased expression of stem cell-related genes (CD44, Nanog, and Oct4). Evidently, down-regulation of miR-27b-5p could reverse the inhibitory effect of reduced LINC01234 on OCSCs and tumorigenesis in vivo ().
Figure 5. Reduced miR-27b-5p reverses the suppression of decreased LINC01234 on OCSCs progression and tumorigenesis in vivo. A. The colony formation ability examined via colony formation assay; B. The invasion ability of CD133+ OCSCs in each group detected via Transwell assay; C. The migration ability of CD133+ OCSCs in each group detected via scratch test; D. In vivo xenograft experiment results; E. Stem cell-related genes (CD44, Nanog, and Oct4) expression detected via RT-qPCR; * vs the si-01234, P < 0.05; Panel D (n = 5), Panel A-C, and E (N = 3).
MiR-27b-5p directly targets SIRT5
The targeting relationship of miR-27-5p and SIRT5 was predicted via a bioinformatics website (http://www.targetscan.org/vert-72/)()). It turned out that miR-27b-5p decreased the luciferase activity of theSIRT5-3ʹUTR-WT reporter in A2780 cells. In contrast, miR-27b-5p had little inhibitory effect on luciferase activity of the SIRT5-3ʹUTR-MT reporter ()).
Figure 6. SIRT5 is a direct target of miR-27b-5p. A. The effect of miR-27b-5p on SIRT5-3ʹUTR-WT and SIRT5-3ʹUTR-MUT luciferase reporters in A2780 cells; B. SIRT5 expression in OC tissues detected via RT-qPCR; C. SIRT5 expression in OC tissues detected via Western blot analysis; D/E. SIRT5 expression in OC cells detected via Western blot analysis; F/G. The correlation analysis of miR-27b-5p and SIRT5, and of LINC01234 and SIRT5 in human OC tissues; * vs IOSE80, P < 0.05; Panel A, D, E (N = 3), Panel B, C, F, G (n = 104).
Moreover, lowly expressed miR-27b-5p was present in OC tissues and cells ()), while elevated SIRT5 was exhibited in OC tissues and cells (). Patients were allocated into two groups with low expression and high expression of SIRT5 (median level of SIRT5 in cancer tissues as the point). The survival analysis showed that high levels of SIRT5 indicated poor survival (Supplementary )) and SIRT5 levels were related to FIGO staging and lymph node metastasis, but not related to age, histological grade, and pathological subtype (). Through Pearson analysis of the correlation between miR-27b-5p and SIRT5, and between LINC01234 and SIRT5 in OC tissues, we found that SIRT5 was negatively correlated with miR-27b-5p , while was positively correlated with LINC01234 ().
Table 4. Association of SIRT5 expression and clinicopathological variables in OC patients
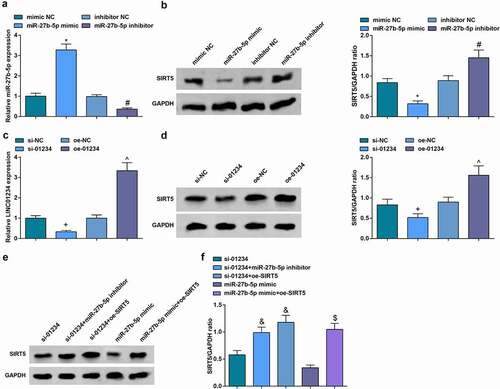
After transfection of mimic NC, miR-27b-5p mimic, inhibitor NC or miR-27b-5p inhibitor to CD133+ OCSCs isolated from A2780 cells, RT-qPCR and Western blot analysis detected the expression of miR-27b-5p and SIRT5. It was indicated that upregulated miR-27b-5p suppressed SIRT5 expression, and inhibited miR-27b-5p elevated SIRT5 expression (). To further study the relationship between LINC01234 and miR-27b-5p and SIRT5, we also tested SIRT5 expression after overexpression or inhibition of LINC01234, and found that the level of SIRT5 increased after overexpression of LINC01234 while decreased after inhibition of LINC01234 (). In addition, we combined LINC01234 with miR-27b-5p and SIRT5 to measure SIRT5 expression and found that inhibiting miR-27b-5p or up-regulating SIRT5 can reverse SIRT5 expression regulated by silenced LINC01234. Elevating SIRT5 reversed the down-regulation of SIRT5 mediated by restored miR-27b-5p ().
Figure 7. SIRT5 is a direct target of miR-27b-5p. A.RT-qPCR was used to detect miR-27b-5p expression after transfection; B. Western blot analysis was performed to determine SIRT5 protein expression after transfection; C. RT-qPCR was used to detect LINC01234 expression after transfection; D. Western blot analysis was performed to determine SIRT5 protein expression after transfection; E/F. Western blot analysis was performed to determine SIRT5 expression after transfection; N = 3; * vs mimic NC, P < 0.05; # vs inhibitor NC, P < 0.05;+ vs si-NC, P < 0.05; ^ vs oe-NC, P < 0.05; & vs si-01234, P < 0.05; $ vs miR-27b-5p mimic, P < 0.05.
The above results manifested that SIRT5 was a direct target of miR-27b-5p and LINC01234 can regulate SIRT5 via miR-27b-5p.
Up-regulation of SIRT5 transforms the repression of elevated miR-27b-5p on OCSCs progression and tumorigenesis in vivo
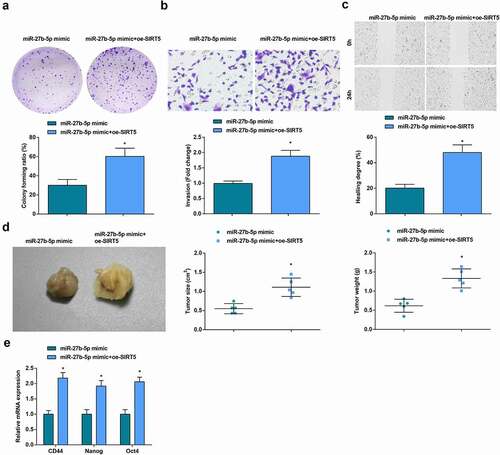
In CD133+ OCSCs isolated from A2780 cells, transfection of miR-27b-5p mimic and SIRT5 overexpression vector promoted the abilities of colony formation, invasion and migration abilities, in vivo tumorigenicity and stem cell-related gene expression, demonstrating that elevation of SIRT5 could reverse the inhibitory effect of up-regulated miR-27b-5p on OCSCs and tumorigenesis in vivo ().
Figure 8. Elevated SIRT5 reverses the inhibitory influence of up-regulated miR-27b-5p on OCSCs progression and tumorigenesis in vivo. A. The colony formation ability examined via colony formation assay; B. The invasion ability of CD133+ OCSCs in each group detected via Transwell assay; C. The migration ability of CD133+ OCSCs in each group detected via scratch test; D. In vivo xenograft experiment results; E. Stem cell-related genes (CD44, Nanog, and Oct4) expression detected via RT-qPCR; * vs the miR-27b-5p mimic, P < 0.05; Panel D (n = 5), Panel A-C, and E (N = 3).
Discussion
LncRNAs demonstrate key influences in the process of malignancies and LINC01234 has been manifested to be a pro-oncogene in all kinds of tumors. For this reason, we aim to investigate the influence of LINC01234 on the differentiation and self-renewal of OCSCs through the targeted modulation of SIRT5 and adsorption of miR-27b-5p.
The observation of the study was that up-regulated LINC01234 was exhibited in OC tissues and cells. In addition, down-regulated LINC01234 suppressed OCSCs progression and tumorigenesis of OC. Many studies are applied for the verification of the results. For instance, LINC01234 is apparently elevated in gastric cancer (GC) tissues, and knockout of LINC01234 stimulates apoptosis and restrains tumorigenesis in mouse xenografts [Citation29]. In the meantime, LINC01234 is up-regulated in non-small cell lung cancer (NSCLC) in contrast with normal lung tissue and linked positively with poor prognosis, and depression of LINC01234 reduces cell growth [Citation30]. This is consonant with the fact that LINC01234 silence leads to reduced proliferation and colony formation, and elevated apoptosis of EC cells and inhibited in vivo EC tumor growth [Citation8]. Moreover, it suits well with the previously defined role that LINC01234 decrease reduces cell proliferation, migration, and invasion of colorectal cancer cells, while represses cell cycle entry and stimulates cell apoptosis [Citation31]. An interesting finding of the study was that LINC01234 was a direct target of miR-27b-5p. In fact, lncRNAs can act as miRNA sponges, reducing their regulatory effect on mRNAs [Citation32]. A study has found that down-regulated LINC01234 exhibits as a competing endogenous RNA for miR-27b-3p [Citation6].
In this work, we have exhibited evidence manifesting that elevated miR-27b-5p suppressed OCSCs progression and tumorigenesis of OC. According to a former research, miR-27b-5p is identified as a tumor suppressor for OC, and it could restrain the viability, migration ability and invasion capacity of tumor cells [Citation14]. Also, the anti-tumor effect of overexpression of miR-27b-5p has been proved in oral squamous cell carcinoma by suppressing cell proliferation [Citation13]. miR-27b is considered as the reduced miRNA in DGCR8 depression cells and accelerates cell proliferation in OC cells [Citation33]. Meanwhile, a study has indicated that down-regulation of miR-27b-5p could contribute to the promotion of the growth and progression of OSCC [Citation13]. Besides, changed miR-27b expression influences the cell migration and proliferation of NSCLC [Citation34]. A study has manifested that enhanced miR-27b represses CSC proliferation and migration in colorectal cancer [Citation35]. Additionally, elevated miR-27b-3p depresses cell proliferation and colony formation in GC cells, in contrast, miR-27b-3p reduction strengthens these malignant behaviors [Citation36]. The study also expressed that miR-27b-5p had a direct targeting relationship with SIRT5. This is consistent with our study that SIRT5 is targeted by some miRNAs, including miR-27b in sporadic colorectal cancer [Citation37]. However, the relationship of miR-27b-5p with SIRT5 remains to be further verified.
The most outstanding results of the study revealed that up-regulation of SIRT5 transformed the repression of elevated miR-27b-5p on OCSCs progression and tumorigenesis in vivo of OC. The results have been demonstrated via plenty of studies. For example, SIRT5 is elevated in OC tissues, forecasting a poor reaction to chemotherapy [Citation16,Citation17]. Moreover, SIRT5 is increased in cellular transformation, promotes proliferation and tumorigenesis in human breast tumors, and is implicated with poor prognosis [Citation38]. This finding is also reported by Zhang et al. that depression of SIRT5 lessens hepatocellular carcinomas (HCC) cell proliferation and SIRT5 up-regulation elevates HCC cell proliferation, while SIRT5 decrease induces HCC cell apoptosis [Citation39]. This also accords with our earlier observations, which manifests that SIRT5 is also reported to be up-regulated in clear cell renal cell carcinoma (ccRCC) tissues, and its silence represses ccRCC cell proliferation, indicating that SIRT5 facilitates ccRCC tumorigenesis [Citation40].In summary, the study concludes that LINC01234 silence or miR-27b-5p elevation represses OCSCs progression and tumorigenesis of OC in vivo via regulation of SIRT5 expression. These findings may help us to understand the mechanism of OC deeply. However, the specific mutation sites that cause the progress of the disease have not been further studied, which is the future study direction. In addition, the further mechanism of SIRT5 on OC tumorigenesis and progression, as well as the underlying association between LINC01234, miR-27b-5p, and SIRT5 was not fully studied in this study due to limited time and fund. Thus, further research should be undertaken to investigate the mechanisms.
Disclosure Conflict-of-interest
The authors declare that they have no conflicts of interest.
Supplemental Material
Download Zip (308.3 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here
Additional information
Funding
References
- Guo J-Y, Wang X-Q, Sun L-F. MicroRNA-488 inhibits ovarian cancer cell metastasis through regulating CCNG1 and p53 expression. Eur Rev Med Pharmacol Sci. 2020;24(6):2902–2910.
- Lu J, Chen H, He F, et al. Ginsenoside 20(S)-Rg3 upregulates HIF-1alpha-targeting miR-519a-5p to inhibit the Warburg effect in ovarian cancer cells. Clin Exp Pharmacol Physiol. 2020;47(8):1455–1463.
- Hoogstad-van Evert JS, Bekkers R, Ottevanger N, et al. Harnessing natural killer cells for the treatment of ovarian cancer. Gynecol Oncol. 2020;157(3):810–816.
- Murillo-Maldonado JM, Riesgo-Escovar JR. The various and shared roles of lncRNAs during development. Dev Dyn. 2019;248(11):1059–1069.
- Kong S, Tao M, Shen X, et al. Translatable circRNAs and lncRNAs: driving mechanisms and functions of their translation products. Cancer Lett. 2020;483:59–65.
- Chen Z, Chen X, Lu B, et al. Up-regulated LINC01234 promotes non-small-cell lung cancer cell metastasis by activating VAV3 and repressing BTG2 expression. J Hematol Oncol. 2020;13(1):7.
- Chen Y, Zhao H, Li H, et al. LINC01234/MicroRNA-31-5p/MAGEA3 Axis Mediates the Proliferation and Chemoresistance of Hepatocellular Carcinoma Cells. Mol Ther Nucleic Acids. 2020;19:168–178.
- Ma J, Han L-N, Song J-R, et al. Long noncoding RNA LINC01234 silencing exerts an anti-oncogenic effect in esophageal cancer cells through microRNA-193a-5p-mediated CCNE1 downregulation. Cell Oncol (Dordr). 2020;43(3):377–394.
- Liu D, Jian X, Xu P, et al. Linc01234 promotes cell proliferation and metastasis in oral squamous cell carcinoma via miR-433/PAK4 axis. BMC Cancer. 2020;20(1):107.
- Guo L, Peng Y, Meng Y, et al. Expression profiles analysis reveals an integrated miRNA-lncRNA signature to predict survival in ovarian cancer patients with wild-type BRCA1/2. Oncotarget. 2017;8(40):68483–68492.
- Jiang Y, Hu Z, Zuo Z, et al. Identification of circulating MicroRNAs as a promising diagnostic biomarker for cervical intraepithelial neoplasia and early cancer: a meta-analysis. Biomed Res Int. 2020;2020:4947381.
- Liu W, Lv C, Zhang B, et al. MicroRNA-27b functions as a new inhibitor of ovarian cancer-mediated vasculogenic mimicry through suppression of VE-cadherin expression. RNA. 2017;23(7):1019–1027.
- Wang M, Qiu Y, Zhang R, et al. MEHP promotes the proliferation of oral cancer cells via down regulation of miR-27b-5p and miR-372-5p. Toxicol In Vitro. 2019;58:35–41.
- Liu CH, Jing XN, Liu XL, et al. Tumor-suppressor miRNA-27b-5p regulates the growth and metastatic behaviors of ovarian carcinoma cells by targeting CXCL1. J Ovarian Res. 2020;13(1):92.
- Hong J, Mei C, Raza SHA, et al. SIRT5 inhibits bovine preadipocyte differentiation and lipid deposition by activating AMPK and repressing MAPK signal pathways. Genomics. 2020;112(2):1065–1076.
- Sun X, Wang S, Gai J, et al. SIRT5 promotes cisplatin resistance in ovarian cancer by suppressing DNA damage in a ROS-Dependent Manner Via Regulation of the Nrf2/HO-1 Pathway. Front Oncol. 2019;9:754.
- Sun X, Wang S, Li Q. Comprehensive analysis of expression and prognostic value of sirtuins in ovarian cancer. Front Genet. 2019;10:879.
- Terraneo N, Jacob F, Dubrovska A, et al. Novel therapeutic strategies for ovarian cancer stem cells. Front Oncol. 2020;10:319.
- Munoz-Galvan S, Carnero A. Targeting cancer stem cells to overcome therapy resistance in ovarian cancer. Cells. 2020;9(6):1402.
- Zhu KY, Tian Y, Li Y-X, et al. The functions and prognostic value of Kruppel-like factors in breast cancer. Cancer Cell Int. 2022;22(1):23.
- Song Y-J, Tan J, Gao X-H, et al. Integrated analysis reveals key genes with prognostic value in lung adenocarcinoma. Cancer Manag Res. 2018;10:6097–6108.
- Wang WY, Cao Y-X, Zhou X, et al. Stimulative role of ST6GALNAC1 in proliferation, migration and invasion of ovarian cancer stem cells via the Akt signaling pathway. Cancer Cell Int. 2019;19(1):86.
- Chen Q, LIU X, XU L, et al. Long non-coding RNA BACE1-AS is a novel target for anisomycin-mediated suppression of ovarian cancer stem cell proliferation and invasion. Oncol Rep. 2016;35(4):1916–1924.
- Chen MZ, Zhong L, Wei D-M, et al. [Effects of Cellular Density on the Induction of Suspension Globe of Ovarian Cancer Stem Cells]. Sichuan Da Xue Xue Bao Yi Xue Ban. 2017;48(5):758–762.
- Chen D, Zhang Y, Wang J, et al. MicroRNA-200c overexpression inhibits tumorigenicity and metastasis of CD117+CD44+ ovarian cancer stem cells by regulating epithelial-mesenchymal transition. J Ovarian Res. 2013;6(1):50.
- Lin B, Zhao H, Li L, et al. Sirt1 improves heart failure through modulating the NF-kappaB p65/microRNA-155/BNDF signaling cascade. Aging (Albany NY). 2020;13(10):14482–14498.
- Wang S, Jiang W, Zhang X, et al. LINC-PINT alleviates lung cancer progression via sponging miR-543 and inducing PTEN. Cancer Med. 2020;9(6):1999–2009.
- Qin W, Ren Q, Liu T, et al. MicroRNA-155 is a novel suppressor of ovarian cancer-initiating cells that targets CLDN1. FEBS Lett. 2013;587(9):1434–1439.
- Chen X, Chen Z, Yu S, et al. Long Noncoding RNA LINC01234 Functions as a Competing Endogenous RNA to Regulate CBFB Expression by Sponging miR-204-5p in Gastric Cancer. Clin Cancer Res. 2018;24(8):2002–2014.
- Chen Z, Chen X, Lei T, et al. Integrative Analysis of NSCLC Identifies LINC01234 as an Oncogenic lncRNA that Interacts with HNRNPA2B1 and Regulates miR-106b Biogenesis. Mol Ther. 2020;28(6):1479–1493.
- Liao X, Zhan W, Zhang J, et al. Long noncoding RNA LINC01234 promoted cell proliferation and invasion via miR-1284/TRAF6 axis in colorectal cancer. J Cell Biochem. 2020;121(10): 4295–4309.
- Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA-LncRNA Interactions. Methods Mol Biol. 2016;1402:271–286.
- Guo Y, Tian P, Yang C, et al. Silencing the double-stranded RNA binding protein DGCR8 inhibits ovarian cancer cell proliferation, migration, and invasion. Pharm Res. 2015;32(3):769–778.
- Zhang J, Hua X, Qi N, et al. MiR-27b suppresses epithelial-mesenchymal transition and chemoresistance in lung cancer by targeting Snail1. Life Sci. 2019;254:117238.
- Chen Y, Zhang B, Jin Y, et al. MiR-27b targets PI3K p110alpha to inhibit proliferation and migration in colorectal cancer stem cell. Am J Transl Res. 2019;11(9):5988–5997.
- Tao J, Zhi X, Zhang X, et al. miR-27b-3p suppresses cell proliferation through targeting receptor tyrosine kinase like orphan receptor 1 in gastric cancer. J Exp Clin Cancer Res. 2015;34(1):139.
- Slaby O, Sachlova M, Brezkova V, et al. Identification of microRNAs regulated by isothiocyanates and association of polymorphisms inside their target sites with risk of sporadic colorectal cancer. Nutr Cancer. 2013;65(2):247–254.
- Greene KS, Lukey MJ, Wang X, et al. SIRT5 stabilizes mitochondrial glutaminase and supports breast cancer tumorigenesis. Proc Natl Acad Sci U S A. 2019;116(52):26625–26632.
- Zhang R, Wang C, Tian Y, et al. SIRT5 Promotes Hepatocellular Carcinoma Progression by Regulating Mitochondrial Apoptosis. J Cancer. 2019;10(16):3871–3882.
- Potvin C, Leclerc D, Tremblay G, et al. Cloning, sequencing and expression of a Bacillus bacteriolytic enzyme in Escherichia coli. Mol Gen Genet. 1988;214(2):241–248.
