ABSTRACT
There is no clear treatment guideline or individualized treatment plan for triple-negative breast cancer (TNBC). The aim of this study was to investigate more effective targets for TNBC-targeted therapy. MDA-MB-231 and BT549 cell lines were used to explore the function of LINC00649 on the proliferation, invasion, and migration of TNBC cells. A mice subcutaneous tumor model and a pulmonary metastasis model was established to identify the role of LINC00649 on the growth and metastasis of TNBC in vivo. LINC00649 was found to be a key molecule involved in the occurrence and development of TNBC by screening of public databases and detection of TNBC clinical samples. LINC00649 increased hypoxia-inducible factor 1α (HIF-1α) mRNA stability and protein expression by interacting with the nuclear factor 90 (NF90)/NF45 complex. In vitro, interference with LINC00649 inhibits MDA-MB-231 and BT549 cell proliferation, migration, and invasion, and the addition of HIF-1α revised this effect. In vivo experiments showed that LINC00649 promoted the growth and metastasis of TNBC. We demonstrated that LINC00649 interacts with the NF90/NF45 complex to increase the mRNA stability of HIF-1α and up-regulate HIF-1α expression, thereby inducing the proliferation, invasion, and migration of TNBC cells as well as tumor growth and metastasis.
Introduction
Breast cancer, a highly heterogeneous malignant tumor, is one of the most common malignant tumors in women [Citation1,Citation2]. As we all know, triple-negative breast cancer (TNBC) is a special type of breast cancer characterized by estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and human epidermal growth factor receptor-2 (HER-2)-negative [Citation3,Citation4]. Compared with other types of breast cancer, TNBC is more aggressive, the disease progresses quickly, and the incidence of distant metastasis is high [Citation5]. In a 5-year follow-up study, the rate of distant metastasis was significantly higher with TNBC than with non-TNBC (20.5% vs 11.7%), and the prognosis was poor [Citation6]. Notably, TNBC lacks therapeutic targets due to negative expression of ER, PR and HER-2, and there is no clear treatment guideline or individualized treatment plan at present. Therefore, in-depth study of TNBC-related genes and exploration of more effective targets for TNBC-targeted therapy will bring hope for treating patients with this type of breast cancer, which has important biological and clinical significance.
Table 1. DE lncRNAs in Stage II vs. I or Stage “III & IV” vs. I (p < 0.005)
Hypoxia is one of the most significant characteristics of tumor growth environment, and it is common in malignant tumors [Citation7,Citation8]. Hypoxia-inducible factor 1α (HIF-1α) is a signaling transcription factor commonly found in mammals and humans under hypoxia environment. It plays a key role in tumor progress, invasion, metastasis, and apoptosis, and is an independent prognostic factor for many solid tumors [Citation9,Citation10]. Studies have shown that HIF-1α is highly activated in TNBC, and high expression of HIF-1α plays a key role in the occurrence and development of TNBC [Citation11,Citation12]. It has been reported that the growth and metastasis of TNBC can be promoted by activating HIF-1α signaling [Citation13,Citation14]. In addition, Ganetespib inhibits HIF-1α activity by inducing HIF-1α degradation, thereby inhibiting the growth, invasion, and metastasis of TNBC [Citation15]. Therefore, it is very important to study the upstream molecular mechanism of HIF-1α affecting the growth and metastasis of TNBC.
The regulatory role of long non-coding RNAs (lncRNAs) in tumor progression has been widely confirmed. We first analyzed the expression matrix and clinical data of breast invasive carcinoma (BRCA) patients in the TCGA database in this study, and found that LINC00649 level was most significantly regulated in the TNBC tissues. It is suggested that LINC00649 may have an important effect on the growth and metastasis of TNBC, and may become a brand-new perspective for the treatment of TNBC. Recent studies have reported that LINC00649 has a high node degree and betweenness centrality (BC) value in the lncRNA-related competitive RNA expression network of prostate cancer, and can be used as a potential diagnostic marker [Citation16]. In addition, studies in colorectal cancer have shown similar results [Citation17].
Through bioinformatics prediction, we found binding sites between LINC00649 and NF90 proteins, suggesting an interaction between the two. Nuclear factor 90 (NF90), transcribed from the interleukin enhancer-binding factor 3 (ILF3) gene, is known to be a double-stranded RNA-binding protein that regulates mRNA stability and gene expression by forming a complex with NF45. Moreover, it has been confirmed that lncRNA DANCR increases the mRNA stability of HIF-1α through interaction with the NF90/NF45, and ultimately leads to metastasis and progression of nasopharyngeal carcinoma.
Therefore, we speculated that LINC00649 could enhance the mRNA stability of HIF-1α by interacting with the NF90/NF45 complex, and up-regulate the expression level of HIF-1α, thereby inducing the proliferation, invasion, migration, and tumor progression of TNBC cells.
Materials and methods
TCGA dataset analysis
The transcriptome expression matrix and clinical data of patients with Breast invasive carcinoma (BRCA) were analyzed in TCGA database. The three clinical information include “breast_carcinoma_estrogen_receptor_status” (ER), “breast_carcinoma_progesterone_receptor_status” (PR), “Lab_proc_her2_neu_chemistry_receptor_status” (HER2) is Negative. Meanwhile, the complete “pathologic_stage” (cancer stage) information was retained as the standard to extract RNA-Seq Raw Count data of TNBC, and a total of 112 samples were obtained. Among them, there are 19 Stage I, 72 Stage II, 19 Stage III and 2 Stage IV. Because there are only two samples for Stage IV, the sample size is too small, so the samples of Stage IV and Stage III are included as Stage “III & IV”. R packet DESeq2 (v.1.26.0) was used to compare the differential expression between groups (Stage II vs. Stage I or Stage “III & IV” vs. Stage I). Venn plot for differential expression lncRNAs in Stage II vs. I & Stage “III & IV” vs. I was made. Red/blue/yellow, respectively, represent high expression/low expression/differentiated expression inconsistencies in both groups. log2FC > 0 represents up-regulation, while log2FC < 0 represents down-regulation.
Clinical samples
In total, 46 pairs of TNBC tissues and para-cancerous normal tissues were collected from The First Affiliated Hospital of Zhengzhou University. The characteristic of patients was described in , including age, menopausal status, premenopausal, postmenopausal, tumor size, tumor node metastasis (TNM) stage, lymphnode metastasis. This study was approved by the ethics committee of The First Affiliated Hospital of Zhengzhou University, and all patients signed an informed consent form.
Table 2. Relationship between LINC00649 and clinical-pathological features in TNBC
Cell culture and treatment
Normal human mammary epithelial cell-line MCF-10A (ATCC: CRL-10317), non-TNBC cell lines MCF-7 (ATCC: CRL-3435) and BT-474 (ATCC: HTB-20), TNBC cell lines BT-549 (ATCC: HTB-122), BT-20 (ATCC: HTB-19), MDA-MB-231 (ATCC: HTB-26), MDA-MB-468 (ATCC: HTB-132) and MDA-MB-453 (ATCC: HTB-131) were all purchased from the American Type Culture Collection (ATCC). Cell lines used in this study were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with 10% (v/v) FBS at 37°C with 5% CO2. Hypoxic MDA-MB-231 and BT549 cells were maintained in a modular incubator and flushed with a gas mixture of 1% O2 and 5% CO2 for 10 h.
Cell transfection
The small interfering RNAs (siRNAs) targeting LINC00649, NF90 or NF45 was designed and synthesized by ApplyBio. The full-length HIF-1α cDNA were cloned into the pcDNA3.1 vector to generate pcDNA-HIF-1α constructs. Empty vector pcDNA 3.1 (Invitrogen) was used as the corresponding control. MDA-MB-231 or BT549 cell lines were transfected with si-control, si-LINC00649-1, si-LINC00649-2, si-NF90-1, si-NF90-2, si-NF45-1, si-NF45-2 using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Hypoxic MDA-MB-231 and BT549 cells were transfected with si-LINC00649, si-LINC00649+ pcDNA-HIF-1α using Lipofectamine 2000 (Invitrogen).
QRT-PCR
LINC01287, LINC00649, LINC00887, LINC01194 and LINC01518 mRNA levels in the TNBC tissues and para-cancerous normal tissues of TNBC patients were detected by quantitative real time-polymerase chain reaction (qRT-PCR) assay. The mRNA levels of LINC00649 in the all cells or the slices of mouse lung were detected by qRT-PCR assay. The mRNA levels of vascular endothelial growth factor A (VEGFA), angiopoietin-like protein 4 (ANGPTL4) and lysyloxidase (LOX) in the MDA-MB-231 and BT549 cells were detected by qRT-PCR assay. HIF-1α mRNA levels in the MDA-MB-231 and BT549 cells or the slices of mouse lung were detected. Total RNA was extracted from tissues or cells using the Trizol reagent (Thermo Fisher Scientific), and then reverse-transcribed into cDNA using cDNA Synthesis Kit (Takara). SYBR Green Real-time PCR Master mix (Takara) was used to perform the qRT-PCR reaction (20 μL) on the StepOne Real-Time PCR System (Applied Biosystems). Using the 2−ΔΔCt method to access the relative RNA levels. GAPDH and U6 were used as an internal reference.
Western blot
Western blot was used to detect the expressions of HIF-1α, NF90, NF45 in MDA-MB-231 and BT549 cell lines, and the protein expressions of HIF-1α in the lung slices. Cells or tissues were rinsed in phosphate buffer and dissociated using RIPA lysis buffer (EMD Millipore) on ice. Bradford method (Bio-Rad, Philadelphia) was used to determine the protein concentration. Then, protein from each sample was electrophoresed via SDS-PAGE and electrotransferred to a PVDF membrane (Millipore). The PVDF membranes were blocked in 5% nonfat milk, and then incubated with primary antibodies (anti-HIF-1α, ab179483, 1:1000; abcam; anti-NF90, ab92355, 1:10000; abcam; anti-NF45, ab154791, 1:1000; abcam) overnight at 4°C. Next, all membranes were incubated with HRP-labeled secondary antibody for 2 h. Finally, immunoblots were visualized by using an Enhanced Chemiluminescent (ECL) kit (Beyotime). The chemiluminescent signals of bands were recorded and β-actin was used as an internal control.
Cell proliferation and colony formation assay
Cell proliferation of MDA-MB-231 and BT549 cells was assessed by using a Cell Counting Kit-8 (CCK-8; Beyotime). MDA-MB-231 and BT549 cells were seeded into 96-well plates (1 × 103 cells/well) and cultured for 24 h. After that, CCK-8 regent (10 μL) was added and then incubated for 4 h. The absorbance was measured using Epoch microplate reader (BioTek), and then the percent cell viability was calculated.
For colony formation assay, MDA-MB-231 and BT549 cells were seeded in a 6-well plate at a density of 500 cells per well and cultured for 2 weeks. After that, colonies were fixed and stained with 1% crystal violet solution for 30 min. The visible colonies number was calculated and analyzed. PBS was used as control.
Cell migration and invasion assay
The transwell assay was implemented for studying the invasion and migration ability of MDA-MB-231 and BT549 cell lines. After cells were trypsinized and resuspended in serum-free medium, 1 × 106 cells/mL MDA-MB-231 or BT549 cells (100 μL) were seeded in the upper Transwell chambers (Millipore). In the cell invasion assay, the upper Transwell chambers need to coat with Matrigel (Corning). The lower chamber was added with complete medium containing with 10% FBS as a chemoattractant and incubated for 24 h. Then, cells that invaded through the Matrigel were fixed with 4% paraformaldehyde and stained with 1% crystal violet solution. The invasive cells in lower chamber were imaged (magnification, ×200) and the number of counted under the microscope (Olympus).
RNA pull-down
An RNA pull-down assay used in this study was performed using a Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher). MDA-MB-231 cells (1 × 107 cells) were lysed and incubated with biotinylated-LINC00649 probe or the antisense fragment. Next, the cell lysates were then mixed with streptavidin agarose beads (Sigma) at 4°C overnight, and then the streptavidin beads were incubated at 37°C for 2 h. Finally, proteins bound to LINC00649 were examined by Western blotting.
RIP
RNA immunoprecipitation (RIP) assays were performed with EZ-Magna RIP Kit (Millipore) according to the guidelines. MDA-MB-231 cells (1 × 107 cells) were lysed in RIP lysis buffer (100 μL), and then resuspended with 900 μL RIP buffer. Then, magnetic beads conjugated to the NF90 or NF45 antibody were incubated with the supernatant overnight at 4°C. Next, the beads were pelleted at 2500 rpm for 30 sec, and then washed using RIP wash buffer. Finally, the immunoprecipitates were incubated with Proteinase K at 55°C for 30 min. QRT-PCR was performed to detect LINC00649 or HIF-1α level after the isolated RNA was extracted using TRIzol regent (Invitrogen). Total RNA was used as input controls.
Animal studies
A total of 24 BALB/c nude mice (6 weeks old) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. The short hairpin RNAs (shRNA) targeting TIPE3 (shTIPE3) was designed and synthesized by ApplyBio. MDA-MB-231 cells transfected with sh-control or sh-LINC00649 according to the manufacturer’s instructions. Mice were randomly selected, and a mice subcutaneous tumor model was established by subcutaneously injecting MDA-MB-231 cells (5 × 106 cells/200 μl PBS) transfected with sh-control or sh-LINC00649 into nude mice (n = 6/group). Tumor volume in each group was measured weekly. Four weeks later, tumors were collected after the mice were euthanized, and the weight was measured after the mice were sacrificed. Pulmonary metastasis model was established by injecting MDA-MB-231 cells (2 × 106 cells) transfected with sh-control or sh-LINC00649 were injected into nude mice via tail vein (n = 6/group). Lungs were harvested after 4 weeks and stained with hematoxylin and eosin. All animal experiments in this study were approved by the Animal Care and Use Committee of The First Affiliated Hospital of Zhengzhou University.
Statistical analysis
Statistical analyses of all data were carried out by SPSS 20.0 software (IBM Corp.). The measurement data are presented as the mean ± SD. An unpaired Student’s t-test and One-way ANOVA were used to analyze the difference between groups. P < 0.05 was considered a statistically significant difference.
Results
LINC00649 was up-regulated in TNBC
To investigate the lncRNAs related to the growth and metastasis of TNBC, we analyzed the expression matrix and clinical data of breast invasive carcinoma (BRCA) patients in the TCGA database. Since Stage IV only has two samples, the samples of Stage IV and Stage III are included in the same group and named Stage “III & IV”. We found 44 differentially expressed lncRNAs (Stage II vs I) and 35 differentially expressed lncRNAs (Stage III & IV vs I) (). Using Venn-Diagrams, we built a list of six overexpressed genes and three underexpressed genes in common between the two experimental conditions ( and ). TNBC tissues and para-cancerous normal tissues were from 46 TNBC patients, and qRT-PCR was used to detect the levels of some lncRNAs that have been reported in tumors, including LINC01287, LINC00649, LINC00887, LINC01194 and LINC01518 [Citation16–21]. As shown in the , the levels of LINC01287, LINC00649, LINC00887, LINC01194 and LINC01518 were up-regulated in the TNBC tissues when compared to para-cancerous normal tissues. Next, LINC00649 with large differential expression was selected as the study object to analyze its correlation with clinical parameters in patients with TNBC. The results showed that LINC00649 level was correlated with TNM stage and lymphnode metastasis ().
Figure 1. LINC00649 was up-regulated in TNBC and was associated with tumor prognosis (a) Venn plot for DE lncRNAs in Stage II vs. I & Stage “III&IV” vs. I. (b) QRT-PCR was used to detected the mRNA levels of LINC01287, LINC00649, LINC00887, LINC01194 and LINC01518 in the TNBC tissues and para-cancerous normal tissues from 46 TNBC patients. *P < 0.05 vs. Adjacent. (c) QRT-PCR was used to detected LINC00649 level in the MCF-10A, MCF-7, BT474, BT-549, BT-20, MDA-MB-231, MDA-MB-468 and MDA-MB-453 cells. *P < 0.05 vs. MCF-10A. (d) The LINC00649 levels in the nuclear and cytosolic fractions derived from MDA-MB-231 and BT549 cells.
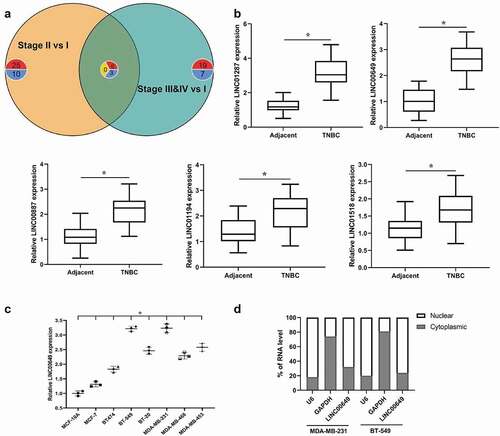
We also performed qRT-PCR to detect the mRNA level of LINC00649 in normal human mammary epithelial cell line (MCF-10A), non-TNBC cell line (MCF-7 and BT-474), TNBC cell line (BT-549, BT-20, MDA-MB-231, MDA-MB-468 and MDA-MB-453). When compared with MCF-10A cells, LINC00649 level was increased in other cells, and the differential expressions of LINC00649 in MDA-MB-231 and BT549 cells was the most obvious (). Therefore, we selected MDA-MB-231 and BT549 cells to study the regulatory mechanism of LINC00649 on the growth and metastasis of TNBC. As we all know, the subcellular localization of lncRNA determines its function [Citation22]. Our data showed that LINC00649 was located in the nuclear instead of cytoplasm by using qRT-PCR (). Taken together, these findings imply that LINC00649 was up-regulated in the nuclear of TNBC cell and was closely associated with tumor prognosis.
Interference with LINC00649 inhibits TNBC cell proliferation, migration, and invasion
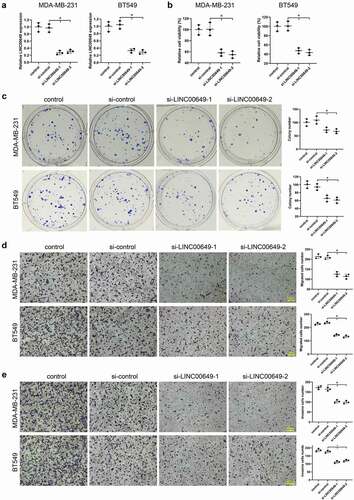
In order to study the effect of LINC00649 on proliferation, migration, and invasion of TNBC cells, we transfected si-LINC00649-1 or si-LINC00649-2 into MDA-MB-231 and BT549 cells, and divided them into control group, si-control group, si-LINC00649-1 group and si-LINC00649-2 group. The results of qRT-PCR showed that when compared to control group and si-control group, LINC00649 levels were down-regulated after MDA-MB-231 and BT549 cells were transfected with si-LINC00649-1 or si-LINC00649-2 (). The proliferation of MDA-MB-231 and BT549 cells in each group was detected by CCK-8 assay, and the results showed that the cell activity in the LINC00649 interference group (si-LINC00649-1 and si-LINC00649-2) was significantly reduced (). In addition, clone formation experiments showed that the colony number in the INC00649 interference group was dramatically lower than that in the si-control and control group (). We also found that interference with LINC00649 expression significantly reduced the number of migrated cells compared with the si-control group (). Notably, the statistical results of cell invasion showed that down-regulation of LINC00649 level could reduce the number of invaded cells (). Overall, these results indicate that interference with LINC00649 inhibits TNBC cell proliferation, migration, and invasion.
Figure 2. Interference with LINC00649 inhibits TNBC cell proliferation, migration and invasion. si-control, si-LINC00649-1, si-LINC00649-2 were transfected into MDA-MB-231 and BT549 cells, and divided them into control group, si-control group, si-LINC00649-1 group and si-LINC00649-2 group. (a) QRT-PCR was used to detected LINC00649 level in the MDA-MB-231 and BT549 cells. (b) CCK-8 assay was performed to detect the cell viability. (c) Colony formation assay was used to detect cell clone formation ability. Transwell assay detected migration (d) and invasion abilities (e). Scale bar = 200 μm. *P < 0.05 vs. si-control.
LINC00649 interacts with NF90 and NF45
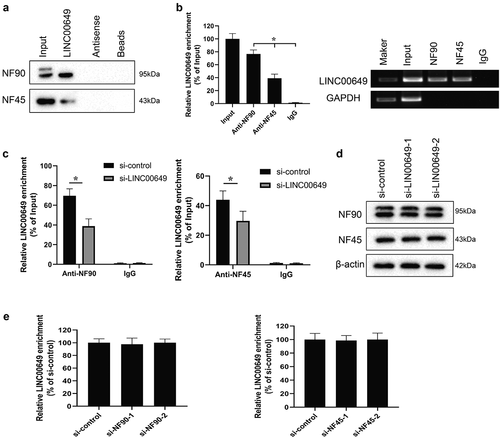
We first used RNAInter (http://www.rna-society.org/rnainter/) to predict the protein binding to LINC00649, and found that LINC00649 might bind to NF90 (Supplementary figure 1). Notably, NF90 can form a complex with NF45 to regulate mRNA stability and gene expression. Therefore, we verified the interaction between LINC00649 and NF90, LINC00649, and NF45 by RNA pull-down and RIP experiments in the MDA-MB-231 cell. The Western blot and qRT-PCR results were in agreement with our guess and showed that LINC00649 interacts with NF90 and NF45 proteins (). After transfected with si-LINC00649 in the MDA-MB-231 cells, the interaction between LINC00649 and NF90, LINC00649, and NF45 was decreased (). Notably, the protein levels of NF90 and NF45 remained unchanged after down-regulation of LINC00649 (). Furthermore, MDA-MB-231 cells were transfected with si-NF90-1, si-NF90-2, si-NF45-1, si-NF45-2. We found that LINC00649 mRNA level did not change after down-regulation of NF90 and NF45 (). Furthermore, the same effect was seen in BT549 cells (Supplementary figure 2). Thus, our data suggest that LINC00649 can bind to NF90 and NF45 without affecting their protein expression.
Figure 3. LINC00649 interacts with NF90 and NF45 in the MDA-MB-231 cells. The interaction between LINC00649 and NF90, LINC00649 and NF45 by RNA pull-down (a) and RIP experiments (b) in the MDA-MB-231 cells. MDA-MB-231 cells were grouped into si-control group and si-LINC00649 group. *P < 0.05 vs. IgG. (c) RIP assay was performed to detect the interaction between LINC00649 and NF90, LINC00649 and NF45 by qRT-PCR. *P < 0.05 vs. si-control. (d) Western blot was used to detect the protein levels of NF90 and NF45. MDA-MB-231 cells were transfected with si-NF90-1, si-NF90-2, si-NF45-1, si-NF45-2. (e) QRT-PCR was used to detected LINC00649 level.
LINC00649 regulates HIF-1α mRNA stability by binding with NF90/NF45
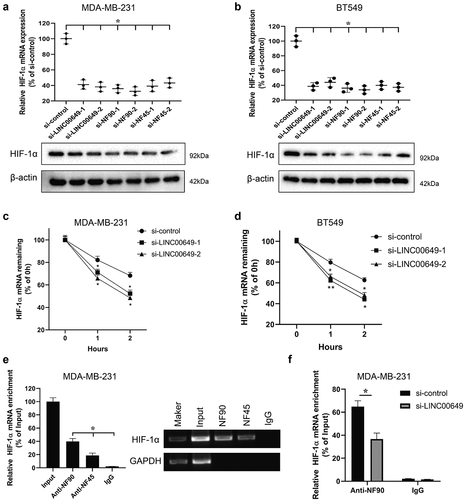
Studies have shown that lncRNA DANCR increases the stability of HIF-1α mRNA by interacting with the NF90/NF45 and affects the metastasis and progression of nasopharyn geal carcinoma [Citation23]. Therefore, we will investigate whether LINC00649 can participate in regulating HIF-1α mRNA stability by binding with NF90/NF45. MDA-MB-231 and BT549 cells were transfected with si-control, si-LINC00649-1 (or si-LINC00649-2), si-NF90-1 (or si-NF90-2), si-NF45-1 (or si-NF45-2), and si-NF45-1 (or si-NF45-2), respectively. The HIF-1α expressions were detected by qRT-PCR and Western blot, and the results showed that interference with LINC00649, NF90 and NF45 could significantly reduce the expression of HIF-1α in the MDA-MB-231 and BT549 cells (). In order to further analyze the molecular mechanism by which LINC00649 regulates HIF-1α gene expression, MDA-MB-231 and BT549 cells transfected with si-LINC00649-1 or si-LINC00649-2 were treated with actinomycin D. After HIF-1α mRNA level was detected, the results showed that down-regulation of LINC00649 reduced the stability of HIF-1α mRNA (). Furthermore, RIP assay showed that HIF-1α mRNA was highly enriched in the protein samples precipitated by NF90 and NF45 antibodies, and was higher in the protein samples precipitated by NF90 antibodies in the MDA-MB-231 cells (). Therefore, we examined the interaction between HIF-1α and NF90 was verified by RIP assay after MDA-MB-231 transfected with si-LINC00649 (). Altogether, these results suggest that LINC00649 can regulate HIF-1α mRNA stability by binding with NF90/NF45.
Figure 4. LINC00649 regulates HIF-1α mRNA stability by binding with NF90/NF45. MDA-MB-231 and BT549 cells were transfected with si-control, si-LINC00649-1, or si-LINC00649-2, si-NF90-1, si-NF90-2, si-NF45-1, si-NF45-2, si-NF45-1, si-NF45-2, respectively. The HIF-1α mRNA level and protein expression were detected by qRT-PCR (a) and Western blot (b). MDA-MB-231 and BT549 cells transfected with si-LINC00649-1 or si-LINC00649-2 were treated with actinomycin D. *P < 0.05 vs. si-control. HIF-1α mRNA levels in the MDA-MB-231 (c) and BT549 cells (d) were detected by qRT-PCR. *P < 0.05, **P < 0.01 vs. si-control. (e) RIP assay was performed to detect the interaction between NF90 and HIF-1α, NF45 and HIF-1α. *P < 0.05 vs. IgG. (f) The interaction between HIF-1α and NF90 was verified by RIP assay. *P < 0.05 vs. si-control.
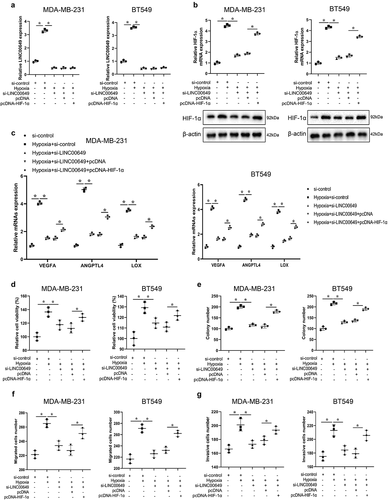
LINC00649 promotes the proliferation, migration, and invasion of TNBC cells through regulating HIF-1α level
Based on the interaction of LINC00649 and NF90/NF45, NF90/NF45 and HIF-1α, we examined whether LINC00649 affects the proliferation, migration and invasion of TNBC cells by regulating HIF-1α level. Hypoxic MDA-MB-231 and BT549 cells were transfected with si-LINC00649 and/or pcDNA-HIF-1α, respectively, and then grouped into si-control, hypoxia+si-control, hypoxia+si-LINC00649, hypoxia+si-LINC00649+ pcDNA, hypoxia+si-LINC00649+ pcDNA-HIF-1α groups. Obviously, compared to nonhypoxic MDA-MB-231 and BT549 cells, the expression levels of LINC00649 and HIF-1α in the hypoxic MDA-MB-231 and BT549 cells were significantly up-regulated (). The silence of LINC00649 in the hypoxic MDA-MB-231 and BT549 cells could revised this effect, indicating that LINC00649 could regulate HIF-1α level (). Furthermore, the expressions of HIF-1α were increased after hypoxic MDA-MB-231 or BT549 cells were co-transfected with si-LINC00649 and pcDNA-HIF-1α, while the level of LINC00649 was unchanged (). We also performed qRT-PCR to detect the levels of the HIF-1α target genes VEGFA, ANGPTL4 and LOX in the five groups, and the results showed that VEGFA, ANGPTL4 and LOX levels had the same trend of HIF-1α ().
Next, the results from CCK-8 assay showed that the cell activity in the hypoxic MDA-MB-231 and BT549 cells was significantly increased (). Cell activity was decreased after the silence of LINC00649 in the hypoxia+si-LINC00649 group, which could be revised by the overexpression of HIF-1α in the hypoxia+si-LINC00649+ pcDNA-HIF-1α group (). The colony number of MDA-MB-231 and BT549 cells had the same trend of cell activity (). The above results suggested that interference with LINC00649 could inhibit TNBC cell proliferation via HIF-1α. We also found that interference with LINC00649 expression significantly reduced the number of migrated cells compared with the hypoxia+si-control group, and then the addition of HIF-1α revised this effect (). Furthermore, the trend of cell invasion in MDA-MB-231 or BT549 cells was consistent with that of cell migration (). Altogether, the above data suggest that LINC00649 promotes the proliferation, migration, and invasion of TNBC cells through regulating HIF-1α level.
Figure 5. LINC00649 promotes the proliferation, migration and invasion of TNBC cells through regulating HIF-1α level. Hypoxic MDA-MB-231 and BT549 cells were grouped into si-control, hypoxia+si-control, hypoxia+si-LINC00649, hypoxia+si-LINC00649+ pcDNA, hypoxia+si-LINC00649+ pcDNA-HIF-1α groups. (a) QRT-PCR was used to detected LINC00649 level. (b) The HIF-1α mRNA level and protein expression were detected by qRT-PCR and Western blot. (c) Western blot was used to detect the protein levels of VEGFA, ANGPTL4 and LOX. (d) CCK-8 assay was performed to detect the cell viability. (e) Colony formation assay was used to detect cell clone formation ability. Transwell assay detected migration (f) and invasion abilities (g). *P < 0.05 vs. si-control/hypoxia+si-control/hypoxia+si-LINC00649+ pcDNA.
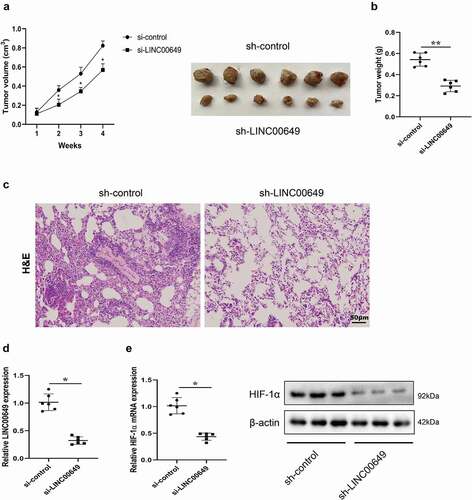
LINC00649 promoted the growth and metastasis of TNBC in vivo
In order to verify the specific role of LINC00649 on the growth of TNBC, we established a mice subcutaneous tumor model. MDA-MB-231 cells transfected with sh-control or sh-LINC00649 were injected subcutaneously into nude mice. Tumor volume was measured weekly in both groups of mice, and the results showed that interfering with LINC00649 could significantly inhibit tumor growth (). The mice were sacrificed 4 weeks later, the tumor was removed and the weight was measured, and the results showed that the tumor weight was reduced after MDA-MB-231 cells were transfected with sh-LINC00649 (). Pulmonary metastasis model was established to identify the function of LINC00649 on the metastasis of TNBC in vivo. MDA-MB-231 cells transfected with sh-control or sh-LINC00649 were injected into nude mice via tail vein. The tumor tissue cells in model group were disordered and the nuclear staining was deep after LINC00649 intervention compared with the control group by HE staining (). The mRNA level of LINC00649 was down-regulated in the tumors of sh-LINC00649 group mice (). Moreover, we found that interfering with LINC00649 significantly reduced the expression levels of HIF-1α mRNA and protein in the tumors (). These findings reveal that LINC00649 promoted the growth and metastasis of TNBC in vivo through HIF-1α.
Figure 6. LINC00649 promoted the growth and metastasis of TNBC in vivo. A mice subcutaneous tumor model was established by subcutaneously injecting MDA-MB-231 cells transfected with sh-control or sh-LINC00649 into nude mice (n = 6/group). (a) Tumor volume was measured weekly. (b) Tumor weight was measured after 4 weeks. Pulmonary metastasis model was established by injecting MDA-MB-231 cells transfected with sh-control or sh-LINC00649 were injected into nude mice via tail vein (n = 6/group). (c) After 4 weeks, HE staining was performed to detect the degree of tumor metastasis. Scale bar = 50 µm. (d) QRT-PCR was used to detected LINC00649 level in the tumors. (e) The HIF-1α mRNA level and protein expression in the tumors were detected by qRT-PCR and Western blot. *P < 0.05, **P < 0.01 vs. si-control.
Discussion
In the present study, we identified six overexpressed genes and three underexpressed genes by analyzing the expression matrix and clinical data of breast invasive carcinoma (BRCA) patients in the TCGA database. And we then detected the expression levels of some lncRNAs that have been reported in tumors, and found that LINC00649 was most significantly changed in the TNBC tissues when compared to normal tissues. The limitation of this study lies in the small clinical sample size, which will be expanded in subsequent studies. Furthermore, LINC00649 is correlated with TNM stage and lymphnode metastasis, indicating that LINC00649 have an important effect on the growth and metastasis of TNBC. In addition, we found that the differential expressions of LINC00649 in MDA-MB-231 and BT549 cells was the most obvious when compared to other TNBC cell lines, normal human mammary epithelial cell line and non-TNBC cell lines. Therefore, we selected MDA-MB-231 and BT549 cells to study the regulatory mechanism of LINC00649 on the growth and metastasis of TNBC, and identified that interference with LINC00649 inhibits TNBC cell proliferation, migration and invasion in vitro by performing CCK-8 assay, colony formation assay and transwell assay.
NF90 binds to nuclear factor 45 (NF45), the product of interleukin enhancer-binding factor-2 (ILF2), to form a NF90/NF45 heterodimer complex [Citation24]. The role of NF90 in a variety of malignant tumors has been widely studied, which also triggered us to explore the relationship between NF90/NF45 and TNBC, as well as the potential molecular mechanism, in order to provide new ideas for the treatment of TNBC generation [Citation25–29]. We also demonstrated that LINC00649 can bind to NF90 and NF45. Notably, NF90 and NF45 protein levels remained unchanged after down-regulation of LINC00649, and LINC00649 mRNA level did not change after down-regulation of NF90 and NF45. These results support that lncRNA LINC00649 affects TNBC by interacting with the NF90/NF45.
Under hypoxia condition, HIF-1α protein exists stably and is transferred into the nuclear. After forming heterodimer complex with HIF-1β in the nuclear, HIF-1α protein participates in cellular processes by activating hypoxia-induced gene transcription and regulating downstream gene expressions [Citation30]. Recent studies have shown that HIF-1α has not only intracellular but also intercellular regulatory functions [Citation31,Citation32]. In this study, our data showed that LINC00649 was located in the nuclear instead of cytoplasm. Next, we also found that the down-regulation of LINC00649 reduced the HIF-1α mRNA stability. and HIF-1α could interact with NF90 and NF45. Furthermore, the interaction between HIF-1α and NF90 was declined after MDA-MB-231 cells transfected with si-LINC00649. These findings show that LINC00649 can regulate HIF-1α mRNA stability by binding with NF90/NF45. In vitro experiments showed that the proliferation, migration and invasion of TNBC cells was enhanced under hypoxic condition, and then inhibited after the silence of LINC00649. The addition of HIF-1α revised this effect, suggesting that LINC00649 promotes the proliferation, migration and invasion of TNBC cells through regulating HIF-1α level. Moreover, we established a mice subcutaneous tumor model and a pulmonary metastasis model to identify the role of LINC00649 on the growth and metastasis of TNBC. Our data suggest that silence of LINC00649 inhibited the growth and metastasis of TNBC in vivo through HIF-1α.
In summary, lncRNA LINC00649 was first found to be upregulated in TNBC and promote the growth and metastasis of TNBC. By interacting with the NF90/NF45 complex, lncRNA LINC00649 increased the HIF-1α mRNA stability and up-regulated the expression of HIF-1α.
Availability of data and material
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Supplemental Material
Download Zip (2.2 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Groza M, Zimta AA, Irimie A, et al. Recent advancements in the study of breast cancer exosomes as mediators of intratumoral communication. J Cell Physiol. 2020;235(2):691–705.
- Guan XF, Liu Z, and Zhao Z, et al. Emerging roles of low-density lipoprotein in the development and treatment of breast cancer. Lipids Health Dis. 2019;18(1) ;137-+.
- Yin L, Duan -J-J, and Bian X-W, et al. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020;22(1):61-+.
- Dass SA, Tan KL, and Selva Rajan R, et al. Triple negative breast cancer: a review of present and future diagnostic modalities. Medicina-Lithuania. 2021;57(1):62-+.
- Qavi Q, Alkistawi F, and Kumar S, et al. Male triple-negative breast cancer. Cureus. 2021;13(4):e14542.
- Qiu J, Xue X, Hu C, et al. Comparison of clinicopathological features and prognosis in triple-negative and non-triple negative breast cancer. J Cancer. 2016;7(2):167–173.
- Vaupel P. The role of hypoxia-induced factors in tumor progression. Oncologist. 2004;9:10–17.
- Reiterer M, Colaco R, and Emrouznejad P, et al. Acute and chronic hypoxia differentially predispose lungs for metastases. Sci Rep. 2019;9(1): 10246-+.
- Hajizadeh F, Okoye I, and Esmaily M, et al. Hypoxia inducible factors in the tumor microenvironment as therapeutic targets of cancer stem cells. Life Sci. 2019; 237;116952-+.
- de Heer Ec, Jalving M, Harris AL, et al. HIFs, angiogenesis, and metabolism: elusive enemies in breast cancer. J Clin Investig. 2020;130(10):5074–5087.
- Lin A, Li C, Xing Z, et al. The LINK-A lncRNA activates normoxic HIF1 alpha signalling in triple-negative breast cancer. Nat Cell Biol. 2016;18(2):213-+.
- Chen X, Iliopoulos D, Zhang Q, et al. XBP1 promotes triple-negative breast cancer by controlling the HIF1 alpha pathway. Nature. 2014;508(7494):103-+.
- Tosatto A, Sommaggio R, Kummerow C, et al. The mitochondrial calcium uniporter regulates breast cancer progression via HIF-1. EMBO Mol Med. 2016;8(5):569–585.
- Chen Y, Zhang B, Bao L, et al. ZMYND8 acetylation mediates HIF-dependent breast cancer progression and metastasis. J Clin Investig. 2018;128(5):1937–1955.
- Xiang L, Gilkes DM, Chaturvedi P, et al. Ganetespib blocks HIF-1 activity and inhibits tumor growth, vascularization, stem cell maintenance, invasion, and metastasis in orthotopic mouse models of triple-negative breast cancer. J Mol Med Jmm. 2014;92(2):151–164.
- Ye G, Guo L, Xing Y, et al. Identification of prognostic biomarkers of prostate cancer with long non-coding RNA-mediated competitive endogenous RNA network. Exp Ther Med. 2019;17(4):3035–3040.
- He M, Lin Y, Xu Y. Identification of prognostic biomarkers in colorectal cancer using a long non-coding RNA-mediated competitive endogenous RNA network. Oncol Lett. 2019;17(3):2687–2694.
- Song C, Sun P, He Q, et al. Long non-coding RNA LINC01287 promotes breast cancer cells proliferation and metastasis by activating Wnt/beta-catenin signaling. Eur Rev Med Pharmacol Sci. 2019;23(10):4234–4242.
- Tian Y, Yu M, Sun L, et al. Long non-coding RNA00887 reduces the invasion and metastasis of non-small cell lung cancer by causing the degradation of miRNAs. Oncol Rep. 2019;42(3):1173–1182.
- Xing Z, Zhang Z, Gao Y, et al. The lncRNA LINC01194/miR-486-5p axis facilitates malignancy in non-small cell lung cancer via regulating CDK4. Onco Targets Ther. 2020;13:151–163.
- Zhang D, Zhang H, Wang X, et al. LINC01518 knockdown inhibits tumorigenicity by suppression of PIK3CA/Akt pathway in oesophageal squamous cell carcinoma. Artif Cells Nanomed Biotechnol. 2019;47(1):4284–4292.
- Mao C, Wang X, Liu Y, et al. A G3BP1-Interacting lncRNA promotes ferroptosis and apoptosis in cancer via nuclear sequestration of p53. Cancer Res. 2018;78(13):3484–3496.
- Wen X, Liu X, Mao Y-P, et al. Long non-coding RNA DANCR stabilizes HIF-1 alpha and promotes metastasis by interacting with NF90/NF45 complex in nasopharyngeal carcinoma. Theranostics. 2018;8(20):5676–5689.
- Guan D, Altan-Bonnet N, Parrott AM, et al. Nuclear factor 45 (NF45) is a regulatory subunit of complexes with NF90/110 involved in mitotic control. Mol Cell Biol. 2008;28(14):4629–4641.
- Fung LF, Lo AKF, Yuen PW, et al. Differential gene expression in nasopharyngeal carcinoma cells. Life Sci. 2000;67(8):923–936.
- Jiang W, Huang H, Ding L, et al. Regulation of cell cycle of hepatocellular carcinoma by NF90 through modulation of cyclin E1 mRNA stability. Oncogene. 2015;34(34):4460–4470.
- Vumbaca F, Phoenix KN, Rodriguez-Pinto D, et al. Double-stranded RNA-binding protein regulates vascular endothelial growth factor mRNA stability, translation, and breast cancer angiogenesis. Mol Cell Biol. 2008;28(2):772–783.
- Guo NL, Wan Y-W, Tosun K, et al. Confirmation of gene expression - based prediction of survival in non-small cell lung cancer. Clin Cancer Res. 2008;14(24):8213–8220.
- Guo Y, Fu P, Zhu H, et al. Correlations among ERCC1, XPB, UBE2I, EGF, TAL2 and ILF3 revealed by gene signatures of histological subtypes of patients with epithelial ovarian cancer. Oncol Rep. 2012;27(1):286–292.
- Chanmee T, Ontong P, Izumikawa T, et al. Hyaluronan production regulates metabolic and cancer stem-like properties of breast cancer cells via hexosamine biosynthetic pathway-coupled HIF-1 signaling. J Biol Chem. 2016;291(46):24105–24120.
- Zhang W, Zhou X, Yao Q, et al. HIF-1-mediated production of exosomes during hypoxia is protective in renal tubular cells. Am J Physiol Renal Physiol. 2017;313(4):F906–F913.
- Wang Q-L, Huang W-X, and Zhang P-J, et al. Colorimetric determination of the early biomarker hypoxia-inducible factor-1 alpha (HIF-1 alpha) in circulating exosomes by using a gold seed-coated with aptamer-functionalized Au@Au core-shell peroxidase mimic. Mikrochim Acta. 2020;187(1):61-+.
