ABSTRACT
Recent studies have unveiled that LINC00173 promotes small cell lung cancer progression. However, LINC00173 has not been studied in Wilms’ tumor (WT). N-glycosylation is a complex post-translational protein modification, and alterations of protein glycosylation have been identified to affect the development of multiple tumors, including WT. MGAT1, known as N-acetylglucosaminyltransferase I (GlcNAcT-1), could initiate synthesis of complex N-glycans, but it has never been related to LINC00173 in WT. This study aimed to explore if LINC00173 could impact WT progression via MGAT1. RT-qPCR and western blot were done to measure the expression and protein levels. Functional assays, as well as animal experiments were conducted to evaluate the function of genes in vivo and in vitro. Additionally, RNA pull-down, RIP, and dual-luciferase reporter assays were carried out to determine the molecular bindings. In vitro experiments proved that sh-LINC00173 inhibited WT cell invasion and promoted WT cell apoptosis, while in vivo experiments indicated sh-LINC00173 restrained WT progression. LINC00173 stabilized MGAT1 mRNA by recruiting HNRNPA2B1. Meanwhile, MGAT1 was verified to stabilize MUC3A protein by inducing N-glycosylation. In summary, our study first discovered that LINC00173 promoted WT progression through MGAT1-mediated MUC3A N-glycosylation, giving new clues to further understanding the mechanism underlying WT progression.
Introduction
Wilms’ tumor (WT) is the most common renal cancer in children, and every 1 out of 10,000 children suffers from WT [Citation1]. Recent years have witnessed a great improvement in the deepened understanding of the predisposition genes in WT. Many researchers have pointed out that WT is highly correlated with many genetic conditions, such as Bohring–Opitz syndromes [Citation2], DICER1 [Citation3] and PIK3CA-related overgrowth spectrum [Citation4]. The existing evidence supports that the underlying predisposition genes exhibit diverse functions and are engaged in multiple biological processes [Citation1]. Currently, although some predisposition genes of WT have been identified, more efforts still need to be made to enrich the knowledge of developmental and oncological mechanisms of WT. Further investigation into the roles of these genes should also be conducted to provide clinical reference for individuals with WT and their families.
The published research works have demonstrated that long non-coding RNAs (lncRNAs) play an indispensable role in the progression of malignancies, for instance, hepatocellular carcinoma [Citation5], cervical cancer [Citation6] as well as WT [Citation7]. To be specific, lncRNA myocardial infarction-associated transcript (MIAT) has been testified to propel WT cell proliferation and WT metastasis, which implies that MIAT is likely to be a potential biomarker for the diagnosis of WT [Citation7]. According to a recent report, long intergenic non-coding RNA 173 (LINC00173) has been identified to trigger chemo-resistance and growth of small cell lung cancer [Citation8]. Nevertheless, the role of LINC00173 in WT has not been explored yet. Accordingly, this study attempted to investigate whether LINC00173 was able to exert influences on the biological behaviors of WT cells, including apoptosis and migration, and on the progression and metastasis of WT.
Glycosylation represents the most complex protein modification. In recent years, alterations of protein glycosylation have been reported to remarkably impact tumor growth and metastasis [Citation9,Citation10]. In particular, protein N-glycosylation has been regarded as an essential modification at post-translational level and this process is proved to influence various biological processes [Citation11]. Notably, previous literature has indicated that mannosyl (alpha-1,3)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase (MGAT1) can cause the synthesis of complex N-glycans [Citation12]. However, the underlying mechanism of MGAT1 in WT remains unknown. In this study, we also intended to explore whether N-glycosylation mediated by MGAT1 played a part in modulating WT progression.
To sum up, this research mainly aimed to uncover the role played by LINC00173 in WT development and the related regulatory mechanism. Hopefully, the findings of this research could provide support for developing therapeutic target of WT.
Materials and methods
Cell culture
Normal human embryonic kidney HEK-293A cell line was procured from RIKEN BioResource (RCB0665, Japan). Three WT cell lines used in this study, namely HFWT, G-401 and SK-NEP-1, were purchased from American Type Culture Collection (ATCC; Manassas, VA, USA). Cells were cultured in RPMI 1640 medium (CD-02168-ML, Gibco, USA) added with 10% fetal bovine serum (FBS; Gibco) and 100 U/ml penicillin–streptomycin solution. The culture dishes were maintained in the humidified incubator with 5% CO2 at 37°C.
Quantitative reverse transcription polymerase chain reaction (RT-qPCR)
Trizol (abs60154, Absin, China) was used to extract total RNAs from targeted cells. Supplier’s guidance was strictly followed. Reverse transcription kit (11141ES10, Takara, Japan) was utilized to reversely transcribe the extracted RNAs (2 μg) into the corresponding cDNAs. Subsequently, RT-qPCR Kit (QR0100-1KT, Sigma-Aldrich, USA) was applied for RT-qPCR analysis. The expression levels of LINC00173, MGAT1, HNRNPA2B1 and MUC3A were calculated and demonstrated as 2−∆∆Ct, with GAPDH or U6 serving as internal control.
Cell transfection
The sh-RNAs (Ribobio, China) were used for knockdown of LINC00173 (sh-LINC00173-1/2/3), MGAT1 (sh-MGAT1-1/2/3), HNRNPA2B1 (sh-HNRNPA2B1-1/2/3) or MUC3A (sh-MUC3A-1/2/3). Sequences for sh-RNAs are provided in . The pcDNA3.1 vector (Ribobio, China) was utilized for overexpression of LINC00173, HNRNPA2B1, MGAT1 or MUC3A. Control vectors including sh-NC and pcDNA3.1 were also obtained. The above-mentioned plasmids were transfected into indicated cells respectively with Lipofectamine 2000 (XFSJ16444, Gibco, USA). Forty-eight hours later, the stably transfected cells were selected and involved in the following experiments. RT-qPCR was performed to test the knockdown or overexpression efficiency.
Table 1. Sequences of shRNAs, primers and MGAT1 3ʹUTR.
Terminal deoxynucleotidyl transferase (TdT) dUTP nck-end labeling (TUNEL) assay
TUNEL assay was carried out to evaluate apoptosis of G-401 and HFWT cells. After being fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100, indicated cells were incubated with 50 μL TdT reaction mix. Then, the nuclei were dyed by DAPI. Eventually, the fluorescence intensity was detected under a fluorescence microscope (XSP-63B, Shanghai Optical Instrument Factory).
Flow cytometry analysis
Annexin V-FITC Apoptosis Kit was used in this assay to assess the apoptosis of HFWT and G-401 cells. Manufacturer’s instruction was strictly followed. After being washed with phosphate buffer saline (PBS), the transfected cells were stained by Annexin V-FITC for 10 min in a dark room. The result was analyzed via flow cytometry (DxFLEX, Thermo Fisher).
Wound healing assay
The transfected G-401 and HFWT cells were planted into 6-well plates. Then, the cells with 80% confluence were scratched with the 200-μL sterile pipette tip. After the scratched cells were washed off, 2% FBS was added into the medium. The culture dishes were placed into the incubator containing 5% CO2 at 37°C. The scratches at 0 h and 24 h were imaged under a microscope and further analyzed by MShot Image Analysis System.
Transwell assay
Transwell filter chambers were used based on the manufacturer’s guidance. The stably transfected G-401 and HFWT cells were suspended in the apical chamber, where the serum-free RPMI 1640 medium (Gibco, Gaithersburg, MD, USA) was added. The basolateral chamber was added with 10% FBS (Gibco, USA). Twenty-four hours later, the migrated cells were fixed with methanol for 10 min. Subsequently, 0.5% crystal violet was used for staining nuclei. The number of migrated cells was counted under a light microscope.
RNA binding protein immunoprecipitation (RIP)
Imprint® RNA Immunoprecipitation Kit (RIP-12RXN, Sigma-Aldrich, MA, USA) was used to perform RIP assay with G-401 and HFWT cells, following the user manual. Cell lysates were incubated with anti-immunoglobulin G (IgG), Anti-U2AF2 (ab37530, Abcam, Cambridge, UK), Anti-TAF15 (# 28409S, Cell Signaling Technology, USA), Anti-SRSF1 (32–4500, Invitrogen), Anti-HNRNPA2B1 (ab31645, Abcam), Anti-MGAT1 (sc-376079, Santa Cruz Biotechnology, CA, USA) or Anti-MUC3A (sc-7315, Santa Cruz Biotechnology) antibodies coated with magnetic beads at 4°C overnight. After purification, the immunoprecipitated RNAs were further subjected to RT-qPCR.
Western blot
Proteins from G-401 and HFWT were isolated by using protein lysis buffer (RIPA20110527, TBD, China) and then extracted with Total Protein Extraction Kit (PROTTOT-1KT, Sigma-Aldrich, USA). After that, SDS Quick Match Gel Kit (P0670-250 ml, Beyotime Biotechnology, Shanghai, China) was applied and the extracted proteins (60 μg) were transferred onto polyvinylidene fluoride (PVDF) membranes. Subsequently, the membranes were blocked with 5% skimmed milk and primary antibodies were added for co-culture at 4°C overnight. The primary antibodies used in this study included anti-GAPDH, anti-cleaved caspase-3, anti-Bcl-2, anti-Bax, anti-β-actin, anti-MGAT1, anti-HNRNPA2B1, anti-MUC1, anti-MUC13 and anti-MUC3A. Afterward, secondary antibodies were added for incubation. The expressions of tested proteins were analyzed by J software. β-Actin and GAPDH acted as the internal references in this study.
Dual-luciferase reporter assay
pcDNA3.1 or pcDNA3.1-LINC00173 was co-transfected with pGL3 or pGL3+ MGAT1 promoter into HEK293T cells. Likewise, pcDNA3.1 or pc DNA3.1-LINC00173 was also co-transfected with pmirGLO or pmirGLO+MGAT1-3ʹUTR into HEK293T cells. In addition, pcDNA3.1 or pcDNA3. 1-HNRNPA2B1 was co-transfected with pmirGLO, pmirGLO+MGAT1-3ʹUTR or pmirGLO-MGAT1-3ʹUTR (MUT) into HFWT and G-401 cells. Then, the luciferase activity was detected under different conditions and renilla luciferase activity served as the internal reference.
RNA pull-down assay
Pierce™ RNA 3’ End Desthiobiotinylation Kit was utilized for obtaining biotinylated MGAT1 3ʹUTR probe (Bio- MGAT1-3ʹUTR), biotinylated LINC00173 probe (Bio-LINC00173) and negative control probes (Bio-NC). The supplier’s instructions were strictly followed. Then, the biotinylated RNAs were cultivated with streptavidin beads. After obtaining cell lysates, streptavidin-coated magnetic beads were added for incubation overnight. RT-qPCR or western blot was subsequently performed to detect the expressions of relevant RNAs or proteins.
Concanavalin A (ConA) pull-down assay
Con A antibody was incubated with G-401 and HFWT cell lysates at 4°C. Con A Sepharose beads were added to precipitate lectin-binding proteins. After elution, the mannoproteins were separated from the total proteins and then the protein levels were measured through western blot.
Tumor xenograft model
Male BALB/c mice at 6–8 wk of age were utilized in our study. All mice were obtained from Jiangsu Jicui Yaokang Biological Technology Co. Ltd. HFWT cells transfected with sh-NC, sh-LINC00173-1 or sh-MGAT1-1 in 100 μL PBS were implanted subcutaneously in the right flank of BALB/c mice. Tumor size and volume was observed and recorded every 3 d since d 7. Tumor volume was expressed in mm3 using the formula: V = 0.5 × (length × width [Citation2]). At d 28 after injection, mice were sacrificed, and the tumor tissues were obtained for further analysis. Animal studies were reviewed and approved Jiangdu People’s Hospital of Yangzhou.
Hematoxylin–eosin (HE) staining
HE staining was conducted as previously described [Citation13]. The paracancerous lymph nodes of the tumor tissues from nude mice were fixed in paraformaldehyde (PFA) overnight, embedded in paraffin, and then cut into sections with 4 μm thickness. The sections were then dewaxed with xylene, hydrated with gradient ethanol, and stained with 5% Hematoxylin for 5 min, followed by soaking in tap water for 15 min or warm water for 5 min at about 50°C. The samples were then counterstained with eosin solution for 2 min. After washing and dehydration, an inverted microscope was used for photography and observation of the sections.
Statistical analysis
Statistical analysis was done with the help of SPSS statistical software package and GraphPad Prism 7.0. Each experiment was performed at least three times. All data were presented as mean ± standard deviation (SD). To compare the differences between groups, Student’s t-test, one-way analysis of variance (ANOVA) or two-way ANOVA were used. P < 0.05 was viewed with statistically significant difference.
Results
LINC00173 promotes WT progression
First, to explore the role of LINC00173 in WT in vitro, LINC00173 expression in WT cell lines was assessed. RT-qPCR data depicted that LINC00173 expression was higher in WT cell lines (HFWT, G-401, SK-NEP-1) than in normal human embryonic kidney HEK-293A cell line, and HFWT and G-401 displayed the highest LINC00173 level ()). Thus, HFWT and G-401 cell lines were selected for subsequent loss-of-function assays to probe into the impacts of LINC00173 knockdown on WT cell proliferation and apoptosis. The knockdown efficiency of sh-RNAs targeting LINC00173 (sh-LINC00173-1/2/3) in HFWT and G-401 cells was tested through RT-qPCR. As shown in ), sh-LINC00173-1 and sh-LINC00173-2 knocked down LINC00173 expression more efficiently than sh-LINC00173-3, so sh-LINC00173-1/2 was applied in the following experiments. Subsequently, to evaluate apoptosis of WT cells, we conducted TUNEL assay and flow cytometry analyses with HFWT and G-401 cells. Results illustrated that WT cell apoptosis was increased by sh-LINC00173-1/2 (). In addition, western blot was used to detect the levels of apoptosis-related proteins including cleaved caspase-3, Bcl-2 and Bax in WT cells, the result of which showed that cleaved-caspase 3 and Bax levels were elevated, while Bcl-2 levels were decreased in WT cells transfected with sh-LINC00173-1/2, consistently showing that WT cell apoptosis was facilitated by the knockdown of LINC00173 (Figure S1A). Thereafter, we examined the impact of LINC00173 deficiency on WT cell migration via wound healing and transwell assays. First, to rule out the possibility that the mitigated migration of WT cells was attributed to the strengthened apoptosis caused by LINC00173 knockdown, flow cytometry was done to detect the apoptosis of WT cells within 24 h, which was the duration of migration assay. We confirmed by flow cytometry analysis that at 24 h, apoptosis of LINC00173 depleted WT cells had no significant change in comparison with sh-NC transfected WT cells (Figure S1B). Later, results of wound healing and transwell assays evidenced that downregulated LINC00173 hindered the migration of WT cells ().
Figure 1. LINC00173 inhibits apoptosis of WT cells and promotes cell migration.(a) RT-qPCR was performed to quantify LINC00173 expression in HEK-293A cells and three WT cell lines, namely HFWT, G-401 and SK-NEP-1. (b) The knockdown efficiency of sh-LINC00173-1/2/3 was tested in HFWT and G-401 cells through RT-qPCR. (c-d) Apoptosis of HFWT and G-401 cells were evaluated by TUNEL assay and flow cytometry analysis. (e-f) Migration capabilities of WT cells were assessed through wound healing assay and transwell assay under LINC00173 knockdown. *P<0.05, **P<0.01.
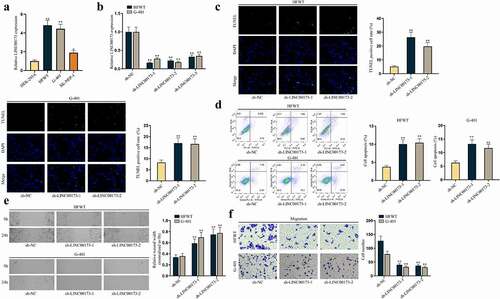
Moreover, to verify the effect of LINC00173 on WT in vivo, we conducted animal experiments. HFWT cells transfected with sh-LINC00173-1 or sh-NC were injected into mice to generate xenografts. The results revealed that the inhibition of LINC00173 could repress WT tumor growth (Figure S1C-D). HE staining of the tumor samples from mice showed that metastasis nodes in sh-LINC00173-1 group were significantly fewer than in the sh-NC group, indicating that LINC00173 depletion suppressed tumor metastasis in vivo (Figure S1E). In short, LINC00173 knockdown could impede WT development.
MGAT1 hampers cell apoptosis and propels cell migration in WT
It was discovered that N-glycosylation could regulate multiple functional genes in TW [Citation14]. MGAT1 is known as a crucial gene to synthesize processed mannose cores into complex N-glycans or hybrid N-linked oligosaccharide structures [Citation12,Citation15]. More importantly, a former report has pointed out MGAT1 is associated with tumor metastasis in hepatocellular carcinoma (HCC) [Citation16]. Thus, we wondered whether MGAT1 participated in WT progression. Expectedly, RT-qPCR revealed that MGAT1 expression was higher in three WT cell lines than in HEK-293A cells, especially in HFWT and G-401 cells ()). The following RT-qPCR outcomes confirmed the successful knockdown of MGAT1 by sh-MGAT1-1/2/3, and sh-MGAT1-1/2 with higher knockdown efficiency was used in the following assays ()). Subsequently, data from TUNEL assay and flow cytometry analyses demonstrated that MGAT1 knockdown accelerated apoptosis of WT cells (). Similarly, protein levels of cleaved caspase-3 and Bax increased and Bcl-2 level decreased under MGAT1 inhibition, according to western blot data in WT cells (Figure S2A). Meanwhile, we verified via flow cytometry analysis that apoptosis of WT cells at 24 h was not altered after MGAT1 knockdown (Figure S2B). Then, wound healing and transwell assays verified that the migration of WT cells was hampered by MGAT1 knockdown ().
Figure 2. MGAT1 hampers cell apoptosis and propels cell migration in WT.(a) MGAT1 expression in HEK-293A cells and three WT cells (HFWT, G-401 and SK-NEP-1) was detected through RT-qPCR. (b) The knockdown efficiency of sh-MGAT1-1/2/3 was examined in HFWT and G-401 cells by RT-qPCR. (c-d) TUNEL assay and flow cytometry analysis were performed to assess apoptosis capabilities of HFWT and G-401 cells with MGAT1 knockdown. (e-f) Wound healing assay and transwell assay were carried out to evaluate migration of WT cells with MGAT1 knockdown. *P<0.05, **P<0.01.
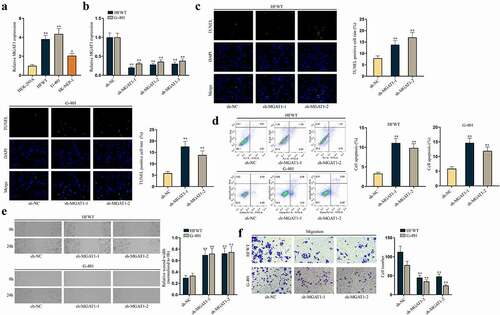
Similarly, in vivo experiments verified that MGAT1 depletion could slow down WT tumor growth (Figure S2C-D). HE staining also verified that metastasis node number in mice was reduced by sh-MGAT1-1 (Figure S2E). In summary, MGAT1 knockdown could accelerate WT cell apoptosis and restrain cell migration in vitro and retard WT tumor growth and metastasis in vivo.
LINC00173 upregulates MGAT1 in WT cells
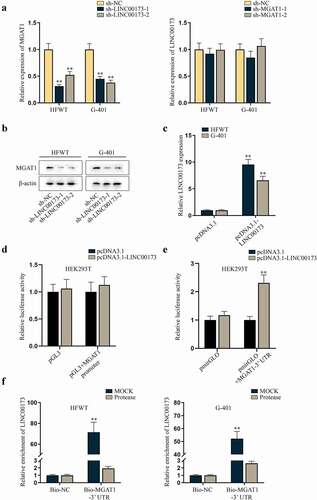
Since lncRNAs are widely acknowledged as regulators of functional genes, including in WT [Citation7,Citation17], we interrogated whether MGAT1 was a target of LINC00173 in WT cells. First, we investigated the correlation between LINC00173 and MGAT1 expression levels in HFWT and G-401 cells. According to RT-qPCR data, MGAT1 expression decreased with LINC00173 knockdown in HFWT and G-401 cells, whereas LINC00173 expression was not altered with MGAT1 knockdown ()). This finding suggested that MGAT1 was a down-stream target of LINC00173 in WT cells. Accor-dingly, we verified that the protein level of MGAT1 was decreased in HFWT and G-401 cells transfected with sh-LINC00173-1/2 ()). To figure out the specific regulatory mechanism, a series of mechanism experiments were carried out. Prelim-inarily, pcDNA3.1-LINC00173 was utilized to overexpress LINC00173 in HFWT and G-401 cells ()). In dual-luciferase reporter assay, we found that luciferase activity of pmirGLO+MGAT1-3ʹUTR, rather than pGL3+ MGAT1 promoter, increased after cells were co-transfected with pcDNA3.1-LINC00173 (). This finding suggested that LINC00173 targeted MGAT1 at post-transcriptional level. In subsequent RNA pull-down assay, we analyzed LINC00173 enrichment in Bio-MGAT1-3ʹUTR before and after cells were treated with Protease to hydrolyze proteins [Citation18]. As a result, the enrichment of LINC00173 was abundant in the Mock group, which that was little in Protease-treated cells, which indicated that the interaction between LINC00173 and MGAT1-3ʹUTR was mediated by certain proteins ()). Therefore, it was concluded that LINC00173 could upregulate MGAT1 expression in WT cells.
Figure 3. LINC00173 positively modulates MGAT1 expression in WT cells.(a) MGAT1 or LINC00173 expression was measured in HFWT and G-401 cells transfected with sh-LINC00173-1/2 or sh-MGAT1-1/2 relative to sh-NC via RT-qPCR. (b) Western blot was performed to examine the protein level of MGAT1 in HFWT and G-401 cells transfected with sh-NC or sh-LINC00173-1/2. (c) The overexpression efficiency of pcDNA3.1-LINC00173 was tested in WT cells through RT-qPCR. (d-e) Dual luciferase reporter experiments were carried out in HEK293T cells with co-transfection of pcDNA3.1-LINC00173 and indicated luciferase reporters. (f) After RNA pull-down assay, RT-qPCR was performed to quantify the enrichment of LINC00173 in the pulldowns of Bio-NC or Bio-MGAT1-3’UTR in HFWT and G-401 cells treated with or without Protease. **P<0.01.
LINC00173 recruits HNRNPA2B1 to stabilize MGAT1 mRNA
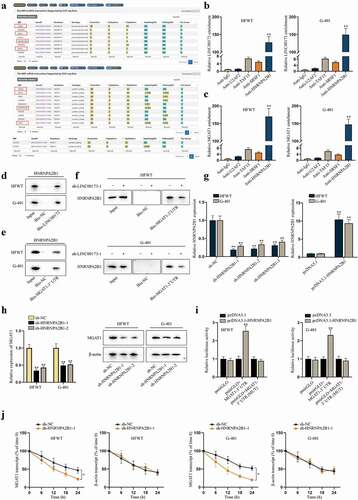
Reviewing previous literature, RNA-binding proteins (RBPs) have been identified as vital mediators in the post-transcriptional regulation of genes by lncRNAs [Citation19]. Based on the findings that LINC00173 interacted with MGAT1-3ʹUTR indirectly, we tried to explore whether regulation of LINC00173 on MGAT1 was mediated by a certain RBP. We searched on starBase (http://starbase.sysu.edu.cn/) to find potential RBPs potentially binding with LINC00173 (under the condition of CLIP-Data ≥3) and MGAT1 (under the condition of CLIP-Data ≥6) at the same time. After comparing the two sets of predicted RBPs, we identified U2AF2, TAF15, SRSF1 and HNRNPA2B1 as candidate RBPs ()). Subsequently, RIP assay was performed with HFWT and G-401 cells. Supported with RT-qPCR analysis, enrichment of LINC00173 and MGAT1 was discovered to be substantial only in anti-HNRNPA2B1 (). Then, western blot following RNA pull-down assay confirmed that HNRNPA2B1 protein was obviously pulled down by Bio-LINC00173 and Bio-MGAT1-3ʹUTR in two WT cell lines (). Moreover, we found that the binding between MGAT1-3ʹUTR and HNRNPA2B1 was inhibited after LINC00173 knockdown ()). These data confirmed that LINC00173 influenced the binding between HNRNPA2B1 and MGAT1 3ʹUTR in WT cells. Later, the knockdown and overexpression efficiency of sh-HNRNPA2B1-1/2/3 and pcDNA3.1-HNRNPA2B1 were examined in HFWT and G-401 cells by RT-qPCR ()). Two interference sequences (sh-HNRNPA2B1-1/2) were chosen for the following experiments for their higher efficiency. Then, RT-qPCR and western blot data represented that MGAT1 expression at both RNA level and protein level was decreased as a result of HNRNPA2B1 knockdown in WT cells ()). To further examine whether HNRNPA2B1 targeted MGAT1 at 3ʹUTR region, we carried out dual-luciferase reporter experiments in HFWT and G-401 cells. Consequently, we verified that HNRNPA2B1 overexpression enhanced the luciferase activity of pmirGLO+MGAT1-3ʹUTR rather than that of pmirGLO+MGAT1-3ʹUTR (MUT), confirming that HNRNPA2B1 targeted MGAT1 at 3ʹUTR region ()). Then, we detected whether HNRNPA2B1 could stabilize MGAT1 mRNA in WT cells. HFWT and G-401 cells were treated with 50 mM α-amanitin to block the transcription of MGAT1. RT-qPCR revealed that HNRNPA2B1 knockdown facilitated the degradation of MGAT1 mRNA in WT cells ()). Altogether, we summarized that LINC00173 could stabilize the mRNA of MGAT1 by recruiting HNRNPA2B1.
Figure 4. LINC00173 stabilizes MGAT1 mRNA by recruiting HNRNPA2B1.(a) Through starBase, candidate RBPs that might interact with LINC00173 were selected under the condition of CLIP-Data >= 3, and potential RBPs that might interact with MGAT1 were screened out under the condition of CLIP-Data >= 6. Overlapped results were highlighted in box. (b-c) In RIP assay, the level of LINC00173 and MGAT1 precipitated by indicated antibodies was detected through RT-qPCR. (d-e) After RNA pull-down assay, western blot was performed to measure the protein level of HNRNPA2B1 pulled down by Bio-LINC00173, Bio-MGAT1-3’UTR or Bio-NC. (f) RNA pull-down assay and western blot detected the level of HNRNPA2B1 pulled down by Bio-NC or Bio-MGAT1-3’UTR before and after WT cells were transfected with sh-LINC00173-1. (g) By RT-qPCR, knockdown efficiency of sh-HNRNPA2B1-1/2/3 and overexpression efficiency of pcDNA3.1-HNRNPA2B1 were tested in HFWT and G-401 cells. (h) RT-qPCR and western blot were conducted to measure the mRNA and protein levels of MGAT1 in HFWT and G-401 cells transfected with sh-NC, sh-HNRNPA2B1-1 or sh-HNRNPA2B1-2. (i) Dual luciferase reporter assay assessed the putative affinity between HNRNPA2B1 and MGAT1 3’UTR in HFWT and G-401 cells. (j) MGAT1 mRNA stability was detected by RT-qPCR at different time points under the influence of HNRNPA2B1 knockdown, with β-actin as negative control. *P<0.05, **P<0.01.
MGAT1 stabilizes MUC3A protein through N-glycosylation in WT cells
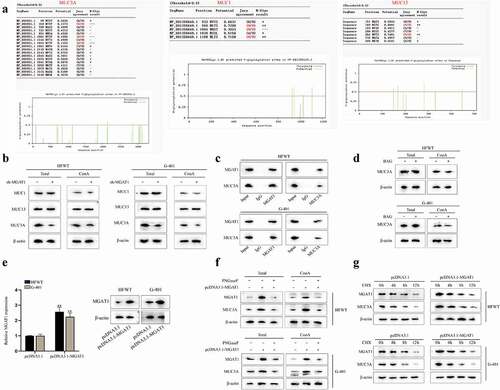
Later, we proceeded to investigate the target downstream of MGAT1. MGAT1 has been reported to initiate synthesis of complex N-glycans and to facilitate cancer progression [Citation12,Citation20]. According to published studies, mucin (MUC) family genes have been found to be dysregulated in multiple cancers [Citation21,Citation22] and most members of MUC family genes can be glycosylated [Citation23,Citation24]. Thus, we tried to determine whether MGAT1 could mediate N-glycosylation of MUC family in WT cells. First, we searched for the MUCs related to WT. Based on previous study, MUC1 [Citation25], MUC3A [Citation26] and MUC13 [Citation27,Citation28] were correlated with poor prognosis in renal tumors. Then, with the help of NetPhos 3.1 Server (https://services.healthtech.dtu.dk/service.php?NetPhos-3.1), the N-glycosylation sites of the above MUC family genes were predicted. The potential N-glycosylation sites with the values over 0.5 were chosen ()). Thereafter, western blot was performed to measure the protein levels of MUC1, MUC13 and MUC3A in whole cell and in the pulldown of Con A. It turned out that both total protein and isolated mannoprotein level of MUC3A obviously declined due to MGAT1 knockdown, but levels of total MUC1 and MUC13 and their enrichment in ConA pulldown presented no variation after MGAT1 knockdown, indicating that MGAT1 only regulated the N-glycosylation of MUC3A ()). Subsequently, through CoIP assay and western blot, we confirmed that MGAT1 could interact with MUC3A ()). Benzyl-α-N-acetylgalactosamine (BAG; abs42014051, Absin, China), a synthetic analogue of N-acetylgalactosamine, served as an inhibitor of O-glycosylation [Citation29]. Based on the results of ), we noticed that neither total protein nor mannoprotein level of MUC3A had significant change on account of BAG treatment, which meant that MGAT1 did not mediate O-glycosylation of MUC3A in these WT cells. Later, as shown in ), it was confirmed that MGAT1 expression was enhanced after pcDNA3.1-MGAT1 was transfected into HFWT and G-401 cells. Furthermore, PNGaseF, an inhibitor of N-glycosylation, was utilized for deglycosylation treatment [Citation30]. According to the results of western blot, we found that total protein level and mannoprotein level of MUC3A and MGAT1 increased in response to MGAT1 overexpression, which was reversed by PNGaseF treatment in WT cells, implying that MGAT1 regulated MUC3A through N-glycosylation ()). Referring to published report, N-glysosylation may affect protein stability [Citation31]. Therefore, MUC3A protein stability was examined in HFWT and G-401 cells. Chlorhexidine (CHX; S0083, BioVision, China) was used to inhibit protein synthesis. WT cells were treated with CHX and the relevant data were gleaned every 4 h. Based on the results of western blot, we concluded that MGAT1 overexpression could strengthen MUC3A protein stability ()). In summary, MGAT1 stabilized MUC3A protein through N-glycosylation in WT cells.
Figure 5. MGAT1 stabilizes MUC3A protein stability via N-glycosylation in WT cells.(a) N-glycosylation sites of MUC3A, MUC1 and MUC13 were predicted via NetPhos 3.1 Server. (b) Western blot measured the total protein of MUC1, MUC13 and MUC3A in HFWT and G-401 cells. After ConA pull-down, mannoprotein levels of MUC1, MUC13 and MUC3A were also detected through western blot in WT cells with or without MGAT1 knockdown. (c) In ChIP assay, western blot analysis was applied for measuring the enrichment of MGAT1 and MUC3A in indicated groups. (d) By western blot, the total protein of MUC3A in HFWT and G-401 cells was examined before and after BAG addition. After ConA pull-down assay, mannoprotein level of MUC3A was also detected via western blot under different conditions. (e) The overexpression efficiency of pcDNA3.1-MGAT1 in HFWT and G-401 cells was tested by RT-qPCR and western blot. (f) Western blot measured total protein levels of MGAT1 and MUC3A in HFWT and G-401 cells in pcDNA3.1, pcDNA3.1-MGAT1 or pcDNA3.1-MGAT1+PNGaseF groups. After ConA pull-down assay, mannoprotein level of MGAT1 and MUC3A in WT cells was also examined under different conditions. (g) After addition of CHX, western blot was conducted to detect protein levels of MGAT1 and MUC3A in HFWT and G-401 cells with or without MGAT1 overexpression every 4 hours. **P<0.01.
MGAT1 restrains WT cell apoptosis and facilitates cell migration via MUC3A
Moreover, we investigated the effects of MUC3A on apoptosis and migration of WT cells. First, RT-qPCR and western blot data showed that the expression of MUC3A was knocked down by sh-MUC3A-1/2/3 and was upregulated by pcDNA3.1-MUC3A in HFWT and G-401 cells ()). Sh-MUC3A-1/2 with higher knockdown efficiency was applied in the following assays. No obvious alterations in MGAT1 were discovered in response to MUC3A silence (Figure S3A). Subsequently, we found that with MUC3A knockdown, the apoptosis of WT cells was promoted ()). The results of western blot depicted that MUC3A knockdown elevated cleaved-caspase-3 and Bax levels and diminished Bcl-2 level, further confirming the promoting influences of MUC3A silence on cell apoptosis (Figure S3B). Additionally, we confirmed that knocking down MUC3A could not affect the apoptosis rate of WT cells within 24 h (Figure S3C). Based on this finding, we conducted transwell assay and confirmed that MUC3A knockdown retarded WT cell migration ()).
Figure 6. MUC3A restrains cell apoptosis and facilitates cell migration in WT.(a) RT-qPCR and western blot were conducted to test knockdown efficiency of sh-MUC3A-1/2/3 and overexpression efficiency of pcDNA3.1-MUC3A in HFWT and G-401 cells. (b-c) Flow cytometry and transwell assays evaluated the effects of MUC3A knockdown on apoptosis and migration of WT cells. (d-e) Flow cytometry and transwell assays were done to determine apoptosis and migration of HFWT and G-401 cells transfected with sh-NC, sh-MGAT1-1, sh-MGAT1-1+pcDNA3.1, or sh-MGAT1-1+pcDNA3.1-MUC3A. **P<0.01.
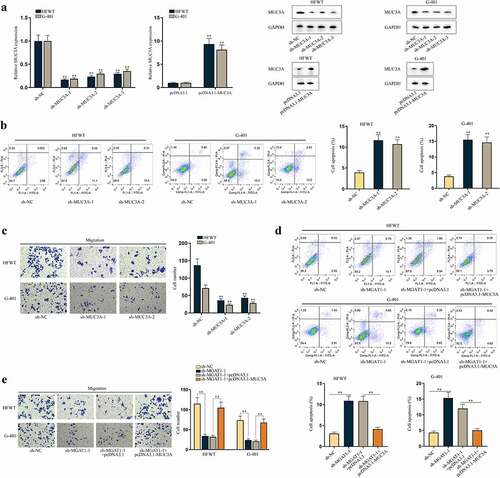
Thereafter, we examined whether MGAT1 regulated apoptosis and migration of WT cells via MUC3A. As depicted in Figure S3D, MGAT1 knockdown resulted in a reduction in MUC3A and MGAT1 levels, and the decline in MUC3A level was reversed by pcDNA3.1-MUC3A co-transfection, while the decrease in MGAT1 expression was hardly altered under the same condition. Flow cytometry analysis and western blot assays validated that the stimulating effects of MGAT1 downregulation on WT cell apoptosis could be offset by MUC3A upregulation () and S3E). Moreover, we confirmed by flow cytometry analysis that apoptosis of WT cells of the abovementioned groups was not altered within 24 h (Figure S3F). Then, we verified that WT cell migration retarded by MGAT1 knockdown was recovered by MUC3A upregulation ()). In conclusion, MGAT1 was able to affect apoptosis and migration of WT cells via modulation on MUC3A.
Discussion
Renal tumor occupies 7% of all neoplasm cases in children and the most common subtype is WT [Citation32]. Recently, lncRNAs are emerging as a new focus in cancer research as they have been discovered to participate in the progression of diverse malignant tumors, including WT [Citation33]. According to the latest report, lncRNA SNHG16 has been identified to play an oncogenic role in WT by sponging miR-200a-3p [Citation17]. Referring to the published work, it was revealed that LINC00173 functioned as an oncogene to facilitate the malignancy of melanoma [Citation34], and LINC00173 silencing weakened CRC cell growth and metastasis via miR-765/PLP2 axis [Citation35]. This study was the first to concentrate on the effects of LINC00173 in WT and tried to unveil its related mechanism. At the very beginning, we confirmed the relation between LINC00173 and WT by validating the upregulation of LINC00173 in WT cell lines. Then, functional assay results evidenced that LINC00173 knockdown accelerated apoptosis and retarded migration of WT cells. The results of animal experiments along with HE staining manifested the suppressive impacts of LINC00173 silence on tumor progression and metastasis. The findings together suggested that LINC00173 could restrain WT cell apoptosis and promote WT cell migration in vitro and accelerate WT tumor growth and metastasis in vivo.
Moreover, based on previous evidence, protein glycosylation alterations have been investigated in multiple tumors and proven to stimulate tumor development [Citation9,Citation36]. Notably, N-glycosylation is suggested to regulate multiple functional genes in TW according to a former study [Citation14]. Further, a previous research has suggested that MGAT1 is able to initiate the synthesis of N-glycans [Citation12], but our study first linked MGAT1 with WT. Our data first showed the upregulation of MGAT1 in three WT cell lines. Similarly, based on loss-of-function assays, we confirmed that MGAT1 also served as an oncogene to inhibit apoptosis and propel migration of WT cells in vitro and positively regulate WT tumor growth and metastasis in vivo.
Subsequently, we first discovered that LINC00173 positively regulated MGAT1 expression but MGAT1 failed to regulate LINC00173 expression in WT cells, suggesting that MGAT1 was downstream of LINC00173. Then, luciferase reporter assay suggested that LINC00173 might affect MGAT1 expression post-transcriptionally via indirect binding to MGAT1 3ʹUTR. Recent reports have substantially revealed the affinity between lncRNAs and RBPs in tumor cells [Citation37,Citation38]. Herein, HNRNPA2B1 was validated to be recruited by LINC00173 in WT cells. The involvement of HNRNPA2B1 in different malignancies has been unveiled. For example, miR503HG bound to HNRNPA2B1 to regulate NF-κB pathway and aggravate hepatocellular cancer progression [Citation39]. LINC01234 recruited HNRNPA2B1 to aggravate the malignancy of lung cancer [Citation40]. In this study, it was confirmed that LINC00173 could modulate MGAT1 mRNA stability via the interaction with HNRNPA2B1.
Furthermore, we explored the targets for MGAT1 in WT cells. Considering that MUC family genes could be glycosylated and further affect cancer progression [Citation21,Citation22,Citation24,Citation41], we speculated that MGAT1 could mediate N-glycosylation of MUC family genes in WT cells. Supported by bioinformatics analysis, MUC1, MUC3A and MUC13 with potential N-glycosylation sites were picked out as the candidates. Interestingly, by RNA pull-down and Con A pull-down assays, we found that only MUC3A interacted with MGAT1 and N-glycosylation of MUC3A could be mediated by MGAT1. Previously, MUC3A was suggested to predict unfavorable prognosis in colorectal cancer [Citation42]. Herein, we uncovered the upregulation of MUC3A in WT cells and confirmed that its knockdown could hinder migration and aggravate apoptosis of WT cells. In addition, a previous study put forward that N-glycosylation could impact protein stability [Citation31]. Accordingly, this study evidenced that MGAT1 overexpression could enhance the protein stability of MUC3A via affecting MUC3A N-glycosylation. Finally, it was reflected in rescue experiments that overexpression of MUC3A could restore the effects of MGAT1 knockdown on WT cell apoptosis and migration.
To summarize, the oncogenic role of LINC00173 and MGAT1 was ascertained via in vitro and in vivo experiments. Moreover, LINC00173 was proved to enhance MGAT1 mRNA stability via interaction with HNRNPA2B1, and MGAT1 was verified to positively affect MUC3A protein stability via modulation on MUC3A N-glycosylation. The limitation of study lies in that the relation between LINC00173 and WT needs to be further confirmed with involvement of clinical samples. Nevertheless, the novel findings of this study might provide an innovative perspective to a better understanding of mechanisms in WT and help develop potential biomarkers for WT diagnosis and treatment.
Supplemental Material
Download Zip (8.5 MB)Acknowledgments
We are grateful to the help provided by all lab personnel in this research.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384101.2022.2070399
Additional information
Funding
References
- Mahamdallie S, Yost S, Poyastro-Pearson E, et al. Identification of new Wilms tumour predisposition genes: an exome sequencing study. Lancet Child Adolesc Health. 2019;3:322–331.
- Russell B, Johnston JJ, Biesecker LG, et al. Clinical management of patients with ASXL1 mutations and Bohring-Opitz syndrome, emphasizing the need for Wilms tumor surveillance. Am J Med Genet Part A. 2015;167a:2122–2131.
- Schultz KAP, Williams GM, Kamihara J, et al. DICER1 and associated conditions: Identification of *gies. Clin Cancer Res off J Am Assoc Cancer Res. 2018;24:2251–2261.
- Gripp KW, Baker L, Kandula V, et al. Nephroblastomatosis or Wilms tumor in a fourth patient with a somatic PIK3CA mutation. Am J Med Genet Part A. 2016;170:2559–2569.
- Chen J, Huang X, Wang W, et al. LncRNA CDKN2BAS predicts poor prognosis in patients with hepatocellular carcinoma and promotes metastasis via the miR-153-5p/ARHGAP18 signaling axis. Aging (Albany NY). 2018;10:3371–3381.
- Zeng J, Ma YX, Liu ZH, et al. LncRNA SNHG7 contributes to cell proliferation, invasion and prognosis of cervical cancer. Eur Rev Med Pharmacol Sci. 2019;23:9277–9285.
- Zhao XS, Tao N, Zhang C, et al. Long noncoding RNA MIAT acts as an oncogene in Wilms’ tumor through regulation of DGCR8. Eur Rev Med Pharmacol Sci. 2019;23:10257–10263.
- Zeng F, Wang Q, Wang S, et al. Linc00173 promotes chemoresistance and progression of small cell lung cancer by sponging miR-218 to regulate Etk expression. Oncogene. 2020;39:293–307.
- Liu D, Shriver Z, Venkataraman G, et al. Tumor cell surface heparan sulfate as cryptic promoters or inhibitors of tumor growth and metastasis. Proc Natl Acad Sci USA. 2002; 99:568–573.
- Zhou R, Wu Y, Wang W, et al. Circular RNAs (circRNAs) in cancer. Cancer Lett. 2018;425:134–142.
- Xiong Y, Karuppanan K, Bernardi A, et al. Effects of N-Glycosylation on the structure, function, and stability of a plant-made Fc-fusion anthrax decoy protein. Front Plant Sci. 2019;10:768.
- Biswas B, Batista F, Sundaram S, et al. MGAT1 and complex N-Glycans regulate ERK signaling during spermatogenesis. Sci Rep. 2018;8:2022.
- Hu YP, Jin YP, Wu XS, et al. LncRNA-HGBC stabilized by HuR promotes gallbladder cancer progression by regulating miR-502-3p/SET/AKT axis. Mol Cancer. 2019;18:167.
- Martinerie C, Gicquel C, Louvel A, et al. Altered expression of novH is associated with human adrenocortical tumorigenesis. J Clin Endocrinol Metab. 2001;86:3929–3940.
- Li Y, Liu Y, Zhu H, et al. N-acetylglucosaminyltransferase I promotes glioma cell proliferation and migration through increasing the stability of the glucose transporter GLUT1. FEBS Lett. 2020;594:358–366.
- Takayama H, Ohta M, Iwashita Y, et al. Altered glycosylation associated with dedifferentiation of hepatocellular carcinoma: a lectin microarray-based study. BMC Cancer. 2020;20:192.
- Zhao XS, Tao N, Zhang C, et al. Long noncoding RNA SNHG16 acts as an oncogene in Wilms’ tumor through sponging miR-200a-3p. Eur Rev Med Pharmacol Sci. 2020;24:4145–4151.
- Ferrall-Fairbanks MC, Barry ZT, Affer M, et al. PACMANS: a bioinformatically informed algorithm to predict, design, and disrupt protease-on-protease hydrolysis. Protein Sci. 2017;26:880–890. Platt MO.
- Yang S, Wang J, Ng RT. Inferring RNA sequence preferences for poorly studied RNA-binding proteins based on co-evolution. BMC Bioinformatics. 2018;19:96.
- Beheshti Zavareh R, Sukhai MA, Hurren R, et al. Suppression of cancer progression by MGAT1 shRNA knockdown. PloS one. 2012;7:e43721.
- Tian Y, Denda-Nagai K, Kamata-Sakurai M, et al. Mucin 21 in esophageal squamous epithelia and carcinomas: analysis with glycoform-specific monoclonal antibodies. Glycobiology. 2012;22:1218–1226.
- Kai Y, Amatya VJ, Kushitani K, et al. Mucin 21 is a novel, negative immunohistochemical marker for epithelioid mesothelioma for its differentiation from lung adenocarcinoma. Histopathology. 2019;74:545–554.
- Chen H, Liu T, Liu J, et al. Circ-ANAPC7 is upregulated in acute myeloid leukemia and appears to target the MiR-181 family. Cell Physiol Biochem. 2018;47:1998–2007.
- Piyush T, Rhodes JM, Yu LG. MUC1 O-glycosylation contributes to anoikis resistance in epithelial cancer cells. Cell Death Discov. 2017;3:17044.
- Leroy X, Buisine MP, Leteurtre E, et al. [MUC1 (EMA): a key molecule of carcinogenesis?]. Ann Pathol. 2006;26:257–266.
- Niu T, Liu Y, Zhang Y, et al. Increased expression of MUC3A is associated with poor prognosis in localized clear-cell renal cell carcinoma. Oncotarget. 2016;7:50017–50026.
- Sheng Y, Ng CP, Lourie R, et al. MUC13 overexpression in renal cell carcinoma plays a central role in tumor progression and drug resistance. Int J Cancer. 2017;140:2351–2363.
- Xu Z, Liu Y, Yang Y, et al. High expression of Mucin13 associates with grimmer postoperative prognosis of patients with non-metastatic clear-cell renal cell carcinoma. Oncotarget. 2017;8:7548–7558.
- Chen L, Sundbäck J, Olofsson S, et al. Interference with O-glycosylation in RMA lymphoma cells leads to a reduced in vivo growth of the tumor. Int J Cancer. 2006;119:1495–1500.
- Walpole C, McGrane A, Al-Mousawi H, et al. Investigation of facilitative urea transporters in the human gastrointestinal tract. Physiol Rep. 2018;6:e13826.
- Li SF, Zhu CS, Wang YM, et al. Downregulation of β1,4-galactosyltransferase 5 improves insulin resistance by promoting adipocyte commitment and reducing inflammation. Cell Death Dis. 2018;9:196.
- Goyal S, Mishra K, Sarkar U, et al. Diagnostic utility of Wilms’ tumour-1 protein (WT-1) immunostaining in paediatric renal tumours. Indian J Med Res. 2016;143:S59–s67.
- Zhu KR, Sun QF, Zhang YQ. Long non-coding RNA LINP1 induces tumorigenesis of Wilms’ tumor by affecting Wnt/β-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23:5691–5698.
- Yang F, Lei P, Zeng W, et al. Long noncoding RNA LINC00173 promotes the malignancy of melanoma by promoting the expression of IRS4 through competitive binding to microRNA-493. Cancer Manag Res. 2020;12:3131–3144.
- Yu Y, Lu X, Yang C, et al. Long noncoding RNA LINC00173 contributes to the growth, invasiveness and chemo-resistance of colorectal cancer through regulating miR-765/PLP2 axis. Cancer Manag Res. 2020;12:3363–3369.
- Liu W, Yan X, Liu W, et al. Alterations of protein glycosylation in embryonic stem cells during adipogenesis. Int J Mol Med. 2018;41:293–301.
- Ren P, Xing L, Hong X, et al. LncRNA PITPNA-AS1 boosts the proliferation and migration of lung squamous cell carcinoma cells by recruiting TAF15 to stabilize HMGB3 mRNA. Cancer Med. 2020;9:7706–7716.
- Zhu P, He F, Hou Y, et al. A novel hypoxic long noncoding RNA KB-1980E6.3 maintains breast cancer stem cell stemness via interacting with IGF2BP1 to facilitate c-Myc mRNA stability. Oncogene. 2021;40:1609–1627.
- Wang H, Liang L, Dong Q, et al. Long noncoding RNA miR503HG, a prognostic indicator, inhibits tumor metastasis by regulating the HNRNPA2B1/NF-κB pathway in hepatocellular carcinoma. Theranostics. 2018;8:2814–2829.
- Chen Z, Chen X, Lei T, et al. Integrative analysis of NSCLC Identifies LINC01234 as an oncogenic lncRNA that interacts with HNRNPA2B1 and regulates miR-106b biogenesis. Mol ther. 2020;28:1479–1493.
- Wang Y, Liao X, Ye Q, et al. Clinic implication of MUC1 O-glycosylation and C1GALT1 in esophagus squamous cell carcinoma. Sci China Life Sci. 2018;61:1389–1395.
- Niu T, Liu Y, Zhang Y, et al. Increased expression of MUC3A is associated with poor prognosis in localized clear-cell renal cell carcinoma. Oncotarget. 2016;7:50017–50026.
