ABSTRACT
SAMHD1 was reported to be related with the development of tumors, while its function in gastric cancer (GC) has not been elucidated yet. Here, we investigated the role and mechanism of SAMHD1 in regulating the proliferation of GC, as well as the mechanism of its expression regulation. Our results revealed that SAMHD1 was downregulated in GC tissues and cell lines, which was correlated with tumor size, depth of invasion and TNM stage. Overexpression of SAMHD1 inhibited the proliferation, clone formation, DNA synthesis and cell cycle progression, while knockdown of SAMHD1 promoted the proliferation of GC cells in vitro and vivo. Meanwhile, SAMHD1 inhibited the activation of MAPK p38 signaling pathway. Moreover, SB203580, as a MAPK p38 inhibitor, could reverse the proliferation and activation of MAPK p38 signaling pathway caused by knockdown of SAMHD1 in GC cells. Additionally, transcription factor Krüppel-like factor 4 (KLF4) bound to the core promoter of SAMHD1, increasing its transcriptional expression in GC cells. In conclusion, SAMHD1 suppressed the proliferation of GC through negatively regulating the activation of MAPK p38 signaling pathway and was upregulated by KLF4 in GC cells.
1. Introduction
Gastric cancer (GC) is the fifth malignant tumor worldwide, with more than 1 million new cases in 2020 [Citation1]. Since most of the patients with GC are in advanced stage after diagnosis, the mortality rate is high, with an estimated 769,000 deaths in 2020, making it the fourth most common cause of cancer-related deaths [Citation1]. Gastrectomy and D2 lymph node dissection combined with neoadjuvant or perioperative chemotherapy may benefit patients with GC at locally advanced stage [Citation2–4]. However, the 5-year survival rate is less than satisfactory [Citation5], and targeted therapy is one of the most promising ways to break through the treatment bottleneck. Currently, Trastuzumab, as a human epidermal growth factor receptor 2 (HER2) inhibitor, is the most common targeted drug in GC, which combining with first line chemotherapy could improve the overall survival for the patients with HER2+ [Citation2]. However, the percentage of patients with HER2+ was less than 20%, leading to limited application of Trastuzumab in treatment of GC [Citation6]. Thus, discovering novel molecules regulating development of GC may provide more options for future targeted therapies.
Human sterile alpha motif and HD-domain-containing protein 1 (SAMHD1) is a triphosphohydrolase that degrades deoxynucleoside triphosphates to the constituent deoxynucleoside and inorganic triphosphate, which blocks the replication of retrovirus [Citation7] and DNA virus [Citation8]. Additionally, SAMHD1 is also associated with a congenital inflammatory disease of the nervous system – Aicardi-Goutières syndrome [Citation9–11]. Recent studies have shown that SAMHD1 involves in regulating the development of lung cancer [Citation12,Citation13] and cutaneous T-cell lymphoma [Citation14]. SAMHD1 is downregulated in lung cancer [Citation12] and cutaneous T-cell lymphoma [Citation15,Citation16]. Furthermore, overexpression of exogenous SAMHD1 inhibits the proliferation of cell lines derived from lung cancer [Citation12] and cutaneous T-cell lymphoma [Citation14]. However, whether SAMHD1 functions to suppress GC remains unclear. The immunohistochemistry analysis showed that SAMHD1 was downregulated in GC tissues, which was associated with tumor size, depth of invasion and TNM stage, suggesting that low expression of SAMHD1 might be an important cause of GC proliferation.
In this work, we elucidated the mechanism of SAMHD1 inhibiting GC proliferation and its transcriptional regulation.
2. Methods
2.1 Patients and specimens
Paired GC and adjacent normal tissues were from 58 patients diagnosed with GC in The First Affiliated Hospital of Anhui Medical University between January 2019 and December 2020. None of the patients received preoperative adjuvant chemoradiotherapy. This study was conducted in accordance with the Declaration of Helsinki (2013) [Citation17].
2.2 Cell lines
MGC-803 and HGC-27 cells were generously gifted by Dr. Ping Wu (Anhui Medical University). AGS, MKN-45 and GES-1 cells were purchased from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences. All cell lines were authenticated using STR profiling (Biowing, China).
2.3 Plasmids construction and sources
Genomic DNA was extracted using the Cellular Genomic DNA Extraction Kit (Tiangen, China). And the promoters of SAMHD1 with different lengths were amplificated using the primers in Table S1. The purified DNA fragments and pGL3 vector were doubly digested with the Kpn I and Hind III restriction enzymes (Takara, Japan). Finally, the targeted fragments were inserted into the pGL3 vector using the DNA Insert Ligation (Thermofisher, USA). The pGL3-285 mutant was constructed using Rapid Site-specific Mutation kit (Tiangen) according to the manufacturer’s instruction.
The pcDNA3.1-empty vector and pcDNA3.1-SAMHD1 wild-type (WT) plasmids were obtained from Dr. Klaus Strebel (National Institutes of Health). In addition, the human pLVX-empty vector and pLVX-HA-SAMHD1 WT were from Wuhan MiaoLing Plasmid Sharing Platform (China). The pCDH-KLF4 (Krüppel-like factor 4), pCDH-empty vector, pLKO.1-shKLF4 and pLKO.1-empty vector plasmids were constructed by Tsingke Biotechnology Co., Ltd (China).
2.4 Transient transfection and lentiviral transduction
Cells were transfected with plasmids using FuGENE® HD (Promega, USA) according to the manufacturer’s instruction. The lentiviruses containing the sequence targeting SAMHD1 (shRNA-1/-2) were packaged by Shanghai GeneChem Co., Ltd (China), and the sequences were: shRNA-1, CCGAATCATTGATACACCTCAATTT; shRNA-2, TGTACACGCATGCT GAAGC. After transfected with plasmids or infected with lentivirus, GC cells were cultured in puromycin (2 μg/ml) to maintain the stable overexpression or downregulation of SAMHD1.
2.5 RNA extraction and quantitative real‑time PCR (qRT‑PCR)
Total intracellular RNA was extracted using AxyPrep Total mRNA Mini kit (Corning, USA) and was reversely transcribed into cDNA using PrimeScriptTM RT Master Mix (Takara) according to the manufacturer’s instructions. The qRT-PCR was performed as previously described [Citation18] and the primers were listed in Table S1.
2.6 Cell proliferation assay
Cell proliferation was detected as previously described [Citation19]. Results were shown as a ratio relative to the control group, which was set as 1.
2.7 Clone formation assay
Cells were cultured in 6-well plates at a density of 200 cells/well. Following culture for 2 weeks, the cells were fixed with 4% paraformaldehyde for 15 min and then stained with crystal violet (Beyotime, China) for 15 min. Finally, the stained cells were photographed and counted.
2.8 Western blot
Western blot was performed as previously described [Citation19]. The information of primary antibodies is listed in Table S2. The gray values of protein bands were calculated using Image J software (version 1.53).
2.9 Cell cycle analysis
Cell cycle analysis was performed as previously described [Citation19]. The stained cells were detected by CytoFLEX flow cytometry (Beckman, USA) and CytExpert software (version 2.0, Beckman). The proportions of cells in G1/G0, S and G2/M phases were analyzed by Flow Jo software (version 7.6.1).
2.10 Ethynyldeoxyuridine (EdU) insert assay
EdU can replace thymidine and be inserted into the newly synthesized DNA during DNA replication. EdU acetylenyl and Azide Alexa Fluor 594 formed stable triazole rings, which could be detected at a 594-nm wavelength. In this study, we used BeyoClickTM EdU-594 kit (Beyotime) to detect DNA synthesis. Briefly, GC cells in the 12-well plates were cultured in medium containing 10 μM EdU for 2 h, and fixed with 4% paraformaldehyde for 15 min. After removing the supernatant, cells were treated with PBS with 0.3% Triton X-100 and then stained with the Click Additive Solution in 37°C away from the light for 30 min. Next, nucleus was labeled with Hoechst 33,342. Finally, the stained cells were analyzed by CytoFLEX flow cytometry (Beckman), and the percentages of EdU+ cells were calculated using CytExpert software (version 2.0, Beckman).
2.11 Immunohistochemistry (IHC)
IHC was performed as previously described [Citation19]. The expression levels of SAMHD1 protein in GC and adjacent normal tissues were scored by two pathologists and calculated using the following formula: staining intensity (0, negative; 1, weak; 2, middle; 3, strong) × positive area rate (1, 0%; 1, 1–25%; 2, 26–50%; 3, 51–75%; 4, 76–100%).
2.12 Animal experiment
Female nude mice aged 4 weeks were obtained from the Shanghai SLAC Laboratory Animal Co., Ltd. and kept under specific pathogen-free conditions at 22°C with 55% humidity and a 12-h light/12-h dark cycle. HGC-27 cells (shNC or shRNA-1, 106 cells/mouse) were injected subcutaneously into the middle-upper inguinal regions of the nude mice. The body weight of mice and the volume of tumors were measured every 7 days. After 4 weeks, the mice were euthanized by cervical dislocation and the tumors were resected for further detection.
2.13 RNA-seq and bioinformatics analysis
R software (version 3.6.2) was used for bioinformatics analysis in this study. RNA-seq was performed in the AGS and HGC-27 cells with different expression of SAMHD1 by Beijing Biomarker Technologies Co., Ltd. For analysis of RNA-seq data, the “org.Hs.eg.db” and “enrichplot” packages were used for screening out the differentially expressed genes and KEGG analysis respectively, and the heatmap was drew using the “heatmap” package. The data of 473 GC tissues and 41 adjacent normal tissues was downloaded from TCGA, which was normalized using log2(FPKM+0.01). Meanwhile, the data of 111 GC tissues and 21 adjacent normal tissues was also downloaded from GSE54129 database, which was normalized using “limma” package. And the differentially expressed transcriptional factors were identified using Wilcoxon test: P< 0.05, log2(fold change)>1.
2.14 Dual-luciferase reporter assay
The effect of KLF4 on the promoter of SAMHD1 was assessed by dual-luciferase reporter assay (Promega) according to the manufacturer’s instructions. Briefly, the cells were lysed with 1× PLB solution for 15 min. And 20 μl cell lysis was added into 100 μl LAR II reaction solution, and the firefly fluorescence was immediately detected by Luciferase activity detection instrument (GLOMAX MULTI, Promega). And then the reaction system was added with 100 μl Stop&Glo solution, and the renal fluorescence was also immediately detected. The ratio of firefly fluorescence vs renal fluorescence represented the transcriptional activity of pGL3 plasmids.
2.15 Immunochromatin precipitation (ChIP)
SimpleChIP® Plus Enzymatic Chromatin IP Kit (Magnetic Beads, CST, USA) was used for the ChIP assay according to the manufacturer’s instructions. Briefly, cells were crosslinked with methanal and then the nuclear were extracted. Chromatin digestion was performed using ultrasonic cell disruptor. The anti-KLF4 antibodies were added into the DNA samples, which were rotated and incubated overnight. Next, 30 μl Magnetic Beads were used to bind the DNA-KLF4 complex, which was then purified through centrifugal column. The purified DNA was used for further PCR and the primers were listed in Table S1. Finally, the PCR products were separated in agarose gels and photographed under ultraviolet light.
2.16 Statistical analysis
The quantitative data obtained from three independent and repeated experiments are presented as the mean ± standard error, and the qualitative differences between two groups were analyzed by unpaired Student’s t-test. The relationships between expression levels of SAMHD1 and clinicopathological features were determined by chi-square test, Fisher’s exact test or Student’s t-test. * P< 0.05 and ** P< 0.01 were considered to indicate a statistically significant difference.
3. Results
3.1 SAMHD1 was downregulated in GC tissues and cell lines
SAMHD1 was down-regulated in GC tissues compared with adjacent normal tissues ()). Moreover, the expression levels of SAMHD1 protein were associated with the tumor size, depth of invasion (T stage) and TNM stage in patients with GC (). Meanwhile, the expression levels of SAMHD1 protein and mRNA in GC cells were also lower than that in GES-1 cells ()).
Figure 1. Expression of SAMHD1 in gastric cancer (GC) tissues, adjacent normal (Adjacent) tissues, GC cells and gastric normal epithelial mucosal cells (GES-1). (a) Immunohistochemical staining of SAMHD1 protein in GC and Adjacent tissues. (b) The immunostaining score is presented. The expression levels of SAMHD1 protein (c, d) and mRNA (e) in GC cells (MKN-45, AGS, MGC-803 and HGC-27) and GES-1 cells.
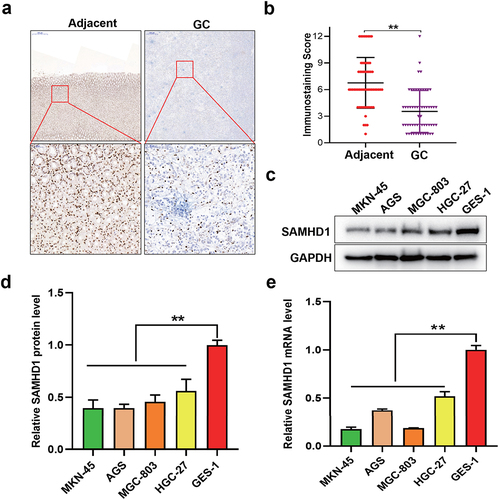
Table 1. The relationship between clinicopathologic factors and expression levels of SAMHD1 in patients with gastric cancer.
3.2 Knockdown of SAMHD1 promoted the proliferation and clone formation of GC cells
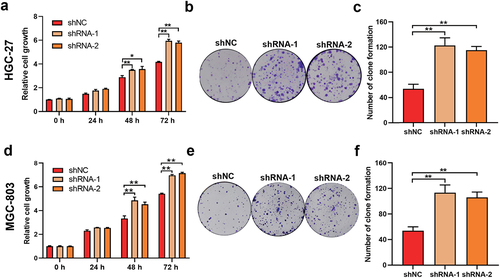
To determine the effect of lower expression of SAMHD1 on the proliferation of GC, we established the HGC-27 and MGC-803 cells with stable knockdown of SAMHD1 (Fig. S1). As shown in ), the proliferation of HGC-27 cells in shRNA-1 and −2 groups was faster than that in shNC group. Additionally, compared with shNC group, the abilities of clone formation of HGC-27 cells were also higher in shRNA-1 and −2 groups ()). Meanwhile, knockdown of SAMHD1 also promoted the proliferation and clone formation of MGC-803 cells ()).
Figure 2. Knockdown of SAMHD1 enhanced the proliferation and clone formation of HGC-27 and MGC-803 cells. (a, b) CCK-8 and clone formation assays were used to detect the proliferation and clone formation of HGC-27 cells in shNC, shRNA-1 and shRNA-2 groups respectively, and the number of clone formation is presented in histogram (c). The proliferation (d) and clone formation (e, f) of MGC-803 cells in shNC, shRNA-1 and shRNA-2 groups were also detected.
3.3 Overexpression of exogenous SAMHD1 suppressed the proliferation, clone formation, DNA synthesis and cell cycle progression of MKN-45 cells
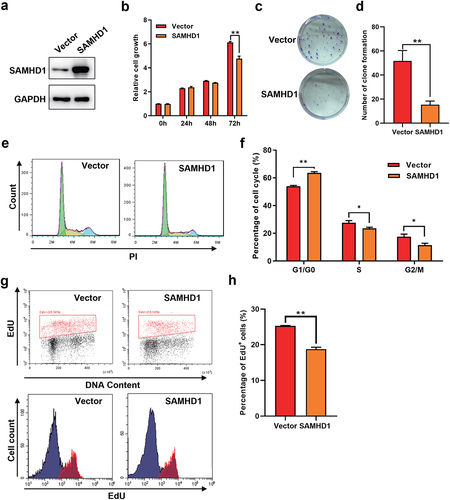
To verify the role of SAMHD1 in inhibiting the development of GC, we overexpressed the SAMHD1 protein in MKN-45 cells ()), which inhibited the cell proliferation ()). Moreover, overexpression of SAMHD1 also suppressed the clone formation of MKN-45 cells ()). Dysregulation of cell cycles is a key cause in the development of GC. However, whether SAMHD1 influenced the cell cycles of GC cells was unclear. As shown in ), compared with empty vector group, the MKN-45 cells with overexpression of SAMHD1 had lower percentages of S and G2/M phases and a higher percentage of G1/G0 phase, suggesting that SAMHD1 suppressed the cell cycle progression of GC cells. DNA synthesis occurs in S phase and is important for tumor cell division. Overexpression of exogenous SAMHD1 also slowed the DNA synthesis of MKN-45 cells ()). Collectively, overexpression of exogenous SAMHD1 suppressed the proliferation, clone formation, DNA synthesis and cell cycle progression of GC cells.
Figure 3. Overexpression of exogenous SAMHD1 inhibited the proliferation, clone formation, cell cycle progression and DNA synthesis of MKN-45 cells. MKN-45 cells were transfected with pcDNA3.1-SAMHD1 or -empty vector plasmids for 72 h. (a) The expression levels of SAMHD1 were detected. (b) The proliferation of MKN-45 cells was assessed. (c, d) The effect of SAMHD1 on the clone formation of MKN-45 cells was determined. (e, f) The percentages of cell cycles were detected using flow cytometry assay. (g, h) EdU insert assay was applied to detect the DNA synthesis of MKN-45 cells.
3.4 Knockdown of SAMHD1 accelerated the growth of subcutaneous transplantation tumors
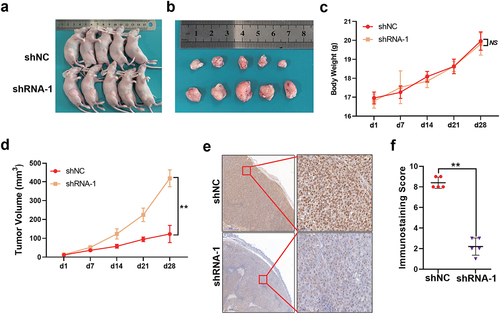
To further explore the effect of SAMHD1 on the GC proliferation in vivo, we constructed the subcutaneous tumor model of nude mice ()). As shown in ), the resected tumors in shRNA-1 group were larger than those in shNC group. There was no difference in the body weight of mice among shNC and shRNA-1 group ()). As shown in ), the tumors in shRNA-1 group grew faster than those in shNC group. Additionally, compared with shNC group, the tumors in shRNA-1 group maintained extremely lower expression of SAMHD1 ()). To sum up, knockdown of SAMHD1 promoted the growth of subcutaneous transplantation tumors.
Figure 4. Knockdown of SAMHD1 accelerated the growth of subcutaneous transplantation tumors in nude mice. HGC-27 cells from shNC or shRNA-1 groups were injected subcutaneously into the middle-upper inguinal regions of the nude mice. (a) The mice with tumors were photographed under white light. (b) The tumors were resected from mice and photographed. (c) The body weight of mice in two groups. (d) The length and width of tumors were measured, and the tumor volume was calculated as: (length × width 2)/2. (e, f) Immunohistochemical staining of SAMHD1 protein in tumors and the staining score is presented. NS, no significance.
3.5 SAMHD1 inhibited the proliferation of GC cells via suppressing the MAPK p38 signaling pathway
Kodigepalli et al [Citation20] found that SAMHD1 suppresses the proliferation of THP-1 cells through inhibiting the PI3K/AKT signaling pathway [Citation20]. However, the mechanism of SAMHD1 inhibiting GC proliferation has not yet been elucidated. Thus, we performed the RNA-seq in HGC-27 and AGS cells with different expression of SAMHD1. After stable overexpression of SAMHD1 in AGS cells (FigureS2), 197 genes were upregulated and 596 genes were downregulated (FigureS3a), which were mainly enriched in PI3K/AKT and MAPK signaling pathway ()). Meanwhile, after stable knockdown of SAMHD1 in HGC-27 cells, 140 genes were upregulated and 294 genes were downregulated (Figure S3b), which were mainly enriched in MAPK signaling pathway ()). Taken together, the MAPK was the common signaling pathway related to the expression levels of SAMHD1 in AGS and HGC-27 cells.
Figure 5. SAMHD1 inhibited the proliferation of gastric cancer cells via suppressing MAPK p38 signaling pathway. KEGG analysis of enriched signaling pathways in AGS cells (a) and HGC-27 cells (b) with different expression of SAMHD1. (c) Fifty-five common differently expressed genes are presented in heatmap. (d) The expression levels of MAP2K6 mRNA were detected. (e) The expression of MAP2K6 protein and phosphorylation levels of p38 at Thr180/Tyr182 were also detected. After treated with SB203580 for 48 h, the proliferation of HGC-27 cells (f) and the activation of MAPK p38 were assessed (g).
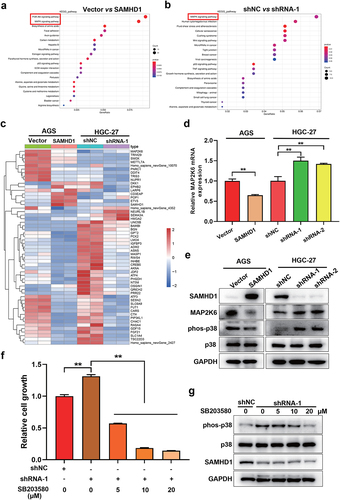
Extracellular regulated protein kinases (ERK) 1/2, c-Jun N-terminal kinase, p38 and ERK5 are the classical MAPK signaling pathways [Citation21]. But, which one of MAPK signaling pathways was regulated by SAMHD1? Next, we discovered 55 common differentially expressed genes in HGC-27 and AGS cells (FigureS3c), which are presented in heatmap ()). Among them, MAP2K6 was the significant one related to expression of SAMHD1 ()). Additionally, overexpression of SAMHD1 decreased the expression of MAP2K6 mRNA and protein, while knockdown of SAMHD1 increased the expression of MAP2K6 ()). MAP2K6, also named as MKK6, is the critical one of upstream kinases in MAPK p38 signaling pathway [Citation22,Citation23]. As expected, SAMHD1 inhibited the activation of MAPK p38 signaling pathway, and the opposite results happened after knockdown of SAMHD1()).
To determine the role of MAPK p38 signaling pathway in SAMHD1 inhibiting the GC proliferation, the HGC-27 cells with stable knockdown of SAMHD1 were treated with MAPK p38 inhibitor (SB203580). As shown in ), downregulation of SAMHD1 promoted the proliferation of HGC-27 cells, which was reversed by SB203580. Besides, SB203580 also suppressed the knockdown of SAMHD1 caused activation of MAPK p38 signaling pathway ()). Taken together, SAMHD1 inhibited the proliferation of GC via negatively regulating the MAPK p38 signaling pathway.
3.6 KLF4 bound to the core promoter of SAMHD1, increasing its transcriptional activity
To illuminate the mechanism of expression regulation of SAMHD1 in GC, we tried to find out transcriptional factors regulating expression of SAMHD1. Schematic diagrams of pGL3 plasmids containing of SAMHD1 promoter are presented in FigureS4a. Dual luciferase reporting assay showed that the core promoter region was within the first 285 bp before translation start site of SAMHD1 gene (FigureS4b). In TCGA and GSE54129 databases, there were 43 differentially expressed transcription factors in GC tissues ()). Online analysis in UCSC and JASPAR websites showed that among those differentially expressed transcription factors, KLF4 was the only one that could bind to the core promoter of SAMHD1. KLF4 was downregulated in GC cells compared with GES-1 cells ()). Additionally, the expression levels of SAMHD1 mRNA and protein were positively correlated with the expression levels of KLF4 protein in GC cells ()). The binding sequence of KLF4 is presented in , and the sequence potentially bound by KLF4 in the core promoter of SAMHD1 is presented in . Moreover, we constructed the pGL3-285 mutant plasmid ()). As shown in (g, h), KLF4 increased the relative fluorescence activity of pGL3-285 wild type, but not pGL3-285 mutant in AGS and MKN-45 cells. ChIP assay proved the binding of KLF4 protein to the core promoter of SAMHD1 ()). To sum up, KLF4 increased the transcriptional activity of SAMHD1 via binding to its core promoter.
Figure 6. KLF4 binding to the core promoter of SAMHD1 increased its transcriptional activity in gastric cancer (GC) cells. (a) Venn diagram of differentially expressed transcriptional factors in GC tissues from TCGA and GSE54129 databases. (b) The expression levels of KLF4 protein in GC and GES-1 cells were detected. (c) Correlation analysis between expression levels of KLF4 protein and SAMHD1 mRNA and protein in GC cells (GES-1 cell was regarded as control). (d) The DNA sequence bound by KLF4 protein. (e) The potential sequence in the core promoter of SAMHD1 bound by KLF4 protein. (f) The sequence of pGL3-285 wild type and the mutant in binding region. AGS (g) and MKN-45 (h) cells were co-transfected with pCDH-KLF4 (or pCDH-empty vector) and pGL3-285 wild type (or pGL3-285 Mut or -Basic) plasmids for 48 h, and the relative luciferase activities were detected. (i) ChIP assay verified the binding of KLF4 to the core promoter of SAMHD1.
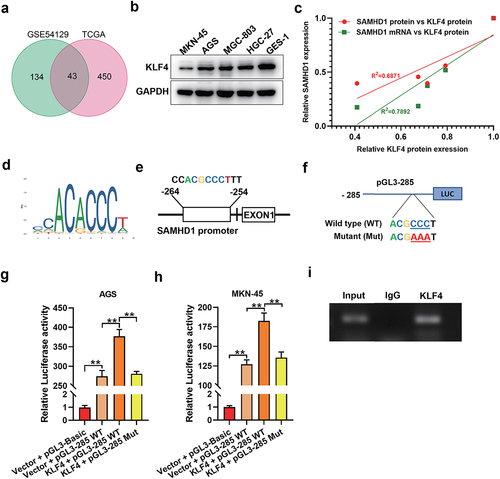
Figure 7. Overexpression of SAMHD1 inhibited the proliferation of GC cells caused by knockdown of KLF4. After transfected with pCDH-KLF4 or -empty vector plasmids for 48 h, qRT-PCR was applied to detect the expression levels of SAMHD1 and KLF4 mRNAs in AGS (a) and MKN-45 (b) cells, and western blot was used to detect the expression levels of SAMHD1 and KLF4 protein in AGS and MKN-45 cells (c). After co-transfected with shKLF4 (or shNC) and pLVX-SAMHD1 (or pLVX-empty vector) plasmids, the expression of SAMHD1 and KLF4 proteins (d) and the proliferation (e) in AGS cells were detected.
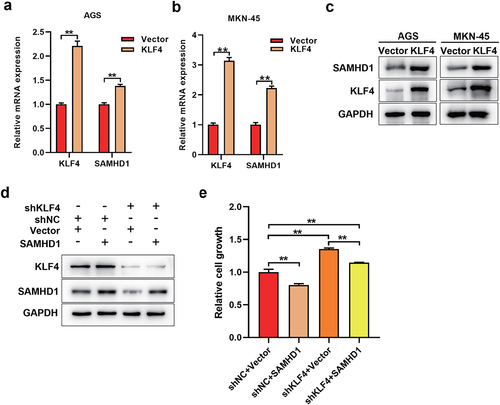
3.7 Overexpression of SAMHD1 inhibited the proliferation of GC cells caused by knockdown of KLF4
As shown in ), overexpression of KLF4 increased the expression levels of SAMHD1 mRNA and protein in AGS and MKN-45 cells. On the contrary, knockdown of KLF4 decreased the expression levels of SAMHD1 protein in AGS cells ()). Additionally, overexpression of SAMHD1 rescued the knockdown of KLF4 induced decrease of SAMHD1 ()). Moreover, knockdown of KLF4 promoted the proliferation of AGS cells, which was reversed partially by overexpression of SAMHD1 ()).
4. Discussion
Studies show that SAMHD1 was downregulated in lung cancer tissues [Citation12,Citation13]. In this study, we firstly found that SAMHD1 expressed less in GC tissues, which was associated with tumor size, depth of invasion and TNM stage. SAMHD1 inhibited the proliferation of Hela (cervical cancer) [Citation24], A549 (lung cancer) [Citation12,Citation13] and HuT78 (cutaneous T-cell lymphoma) [Citation14] cells, consistent with our results in GC cells. Moreover, we found that knockdown of SAMHD1 promoted the growth of subcutaneous transplantation of GC cells. Gao et al [Citation13] proved that overexpression of SAMHD1 inhibited the proliferation of A549 cells in vivo [Citation13]. However, Kodigepalli et al [Citation25] established the leukemia models of nude mice by injecting intravenously with THP-1 cells, and the result shows no significant difference in the tumor growth between the knockout of SAMHD1 group and control group [Citation25]. The different role of SAMHD1 in solid and hematologic tumors may be due to the different modeling approaches and complex environments in vivo.
Karthik et al [Citation20] found that SAMHD1 regulated the PI3K/AKT signaling pathway in THP-1 cells using the microarray analysis [Citation20]. Besides, they also discovered other signaling pathways, which may be related to the expression of SAMHD1, including interferon, NF-κB, MAPK p38 and death receptor in THP-1 cells [Citation20]. In this study, we chose the RNA-seq method because of its higher sensitivity. Besides, we not only selected the HGC-27 cells with stable knockdown of SAMHD1, but also AGS cells with stable overexpression of SAMHD1 to screen out the enriched signaling pathways. Further experiment determined that SAMHD1 inhibited the activation of MAPK p38 signaling pathway in GC cells. MAPK p38 is the critical one of signaling pathways regulating the proliferation of GC [Citation26,Citation27]. As expected, we discovered that SAMHD1 suppressed the GC proliferation via negatively regulating the MAPK p38 signaling pathway.
Methylation regulates expression of SAMHD1 in lung cancer [Citation12], cutaneous T-cell lymphoma [Citation15] and skin cutaneous melanoma [Citation28]. The methylation levels of SAMHD1 promoter are negatively correlated with its expression in skin cutaneous melanoma [Citation28]. DNA methylation inhibitor increases the expression of SAMHD1 mRNA and protein in lung cancer and cutaneous T-cell lymphoma [Citation12,Citation15]. However, based on the online analysis of the EWAS database (https://ngdc.cncb.ac.cn/ewas/datahub/index), the methylation levels of SAMHD1 promoter are positively correlated with its expression in GC, thus methylation can’t explain the phenomenon about its less expression. Valverde et al [Citation29] found that interferon regulatory factor 1 increases the expression of SAMHD1 through binding to its promoter [Citation29]. In this study, we found that KLF4, as the one of differentially expressed genes in GC, upregulated the expression of SAMHD1. Additionally, KLF4 was downregulated in GC cells and knockdown of KLF4 decreased the expression of SAMHD1, suggesting that KLF4 at a low expression level was insufficient to positively regulate the transcription of SAMHD1 gene, leading to the decline of its expression in GC.
KLF4, as one member of KLF family, owns zinc fingers binding to specific DNA sequence [Citation30]. KLF4 expresses widely in epithelial cells from the skin and gastrointestinal tract mucosa, but was downregulated in GC [Citation31] and colorectal cancer [Citation32]. It is well known that KLF4 inhibits the development of GC [Citation33,Citation34] and colorectal cancer [Citation32]. Studies have shown that KLF4 suppresses the development of GC through regulating the expression of oncogenes or tumor suppressor genes. In detail, KLF4 upregulates the expression of caudal type homeobox 2 [Citation35] and ghrelin [Citation36], and downregulates the expression of C-X-C Motif Chemokine Ligand 8 [Citation34], miR-106a [Citation37,Citation38], inhibitor of apoptosis-stimulating protein of p53 [Citation39] and PODXL [Citation30] in GC. In this study, SAMHD1 was another downstream gene upregulated by KLF4.
The chronic infection of Helicobacter pylori (HP) leads to gastric atrophic gastritis and intestinal metaplasia, which is closely related to the incidence of gastric antrum cancer. Cytotoxin-associated proteins A (CagA) is one of typical pathogenic factors of HP, and involves in the regulation of GC cell proliferation and metastasis. Zhao et al found that the expression levels of KLF4 in HP infected GC tissues are lower than that in GC tissues without HP infection [Citation40]. HP infection suppresses the expression of KLF4 through increasing the methylation of its promoter [Citation40]. Besides, CagA also inhibits the expression of KLF4 via upregulating its upstream miR-155 [Citation41]. However, whether CagA regulates the expression of SAMHD1 directly or indirectly remains unknown. In chronic infection, virus evolve a mechanism to antagonize the antiviral ability of SAMHD1 in host. For example, the Vpx or Vpr of HIV recruits SAMHD1 protein to Cullin ring-finger ubiquitin ligase 4 E3 ubiquitin ligase complex, and then the ubiquitinated SAMHD1 protein is degraded by proteasome pathway [Citation42,Citation43]. Epstein-Barr Virus-Encoded Protein Kinase 4 phosphorylates SAMHD1, resulting in loss of its ability to block the replication of EBV [Citation44]. Thus, it is unknown whether SAMHD1 involves in chronic infection of HP, which will be identified in the follow-up study.
In conclusion, our study revealed that SAMHD1 inhibited proliferation of GC cells via negatively regulating the MAPK p38 signaling pathway and KLF4 promoted the transcription of SAMHD1 in GC cells.
Ethical statement
The study was approved by the Ethics Committee of the First Affiliated Hospital of Anhui Medical University. The protocol for animal experiment was approved by Animal Use and Care Committee of Anhui Medical University.
Informed consent
All patients signed the informed consent for this study.
Supplemental Material
Download PDF (324.4 KB)Supplemental Material
Download PDF (324.4 KB)Acknowledgements
We thank Dr. Dandan Zang in the Center for Scientific Research of Anhui Medical University for her assistance with the flow cytometry.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384101.2022.2085356
Additional information
Funding
References
- Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249.
- Japanese Gastric Cancer A. Japanese gastric cancer treatment guidelines 2018 (5th edition). Gastric Cancer. 2021;24(1):1–21.
- Al-Batran SE, Homann N, Pauligk C, et al. Perioperative chemotherapy with fluorouracil plus leucovorin, oxaliplatin, and docetaxel versus fluorouracil or capecitabine plus cisplatin and epirubicin for locally advanced, resectable gastric or gastro-oesophageal junction adenocarcinoma (FLOT4): a randomised, phase 2/3 trial. Lancet. 2019;393(10184):1948–1957.
- Schuhmacher C, Gretschel S, Lordick F, et al. Neoadjuvant chemotherapy compared with surgery alone for locally advanced cancer of the stomach and cardia: European organisation for research and treatment of cancer randomized trial 40954. J Clin Oncol. 2010;28(35):5210–5218.
- Zeng H, Chen W, Zheng R, et al. Changing cancer survival in China during 2003-15: a pooled analysis of 17 population-based cancer registries. Lancet Glob Health. 2018;6(5):e555–e67.
- Van Cutsem E, Bang YJ, Feng-Yi F, et al. HER2 screening data from ToGA: targeting HER2 in gastric and gastroesophageal junction cancer. Gastric Cancer. 2015;18(3):476–484.
- Goldstone DC, Ennis-Adeniran V, Hedden JJ, et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480(7377):379–382.
- Chen Z, Zhu M, Pan X, et al. Inhibition of hepatitis B virus replication by SAMHD1. Biochem Biophys Res Commun. 2014;450(4):1462–1468.
- Park K, Ryoo J, Jeong H, et al. Aicardi-Goutieres syndrome-associated gene SAMHD1 preserves genome integrity by preventing R-loop formation at transcription-replication conflict regions. PLoS Genet. 2021;17(4):e1009523.
- Franzolin E, Coletta S, Ferraro P, et al. SAMHD1-deficient fibroblasts from aicardi-goutieres syndrome patients can escape senescence and accumulate mutations. FASEB J. 2020;34(1):631–647.
- Rice GI, Bond J, Asipu A, et al. Mutations involved in aicardi-goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nat Genet. 2009;41(7):829–832.
- Wang JL, Lu FZ, Shen XY, et al. SAMHD1 is down regulated in lung cancer by methylation and inhibits tumor cell proliferation. Biochem Biophys Res Commun. 2014;455(3–4):229–233.
- Wu Y, Niu Y, Wu Y, et al. SAMHD1 can suppress lung adenocarcinoma progression through the negative regulation of STING. J Thorac Dis. 2021;13(1):189–201.
- Kodigepalli KM, Li M, Liu SL, et al. Exogenous expression of SAMHD1 inhibits proliferation and induces apoptosis in cutaneous T-cell lymphoma-derived HuT78 cells. Cell Cycle. 2017;16(2):179–188.
- de Silva S, Wang F, Hake TS, et al. Downregulation of SAMHD1 expression correlates with promoter DNA methylation in sezary syndrome patients. J Invest Dermatol. 2014;134(2):562–565.
- Kohnken R, Kodigepalli KM, Mishra A, et al. MicroRNA-181 contributes to downregulation of SAMHD1 expression in CD4+ T-cells derived from sezary syndrome patients. Leuk Res. 2017;52:58–66.
- World Medical A. World medical association declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310(20):2191–2194.
- Meng L, Chen Z, Jiang Z, et al. MiR-122-5p suppresses the proliferation, migration, and invasion of gastric cancer cells by targeting LYN. Acta Biochim Biophys Sin (Shanghai). 2020;52(1):49–57.
- Chen ZM, Huang L, Li MM, et al. Inhibitory effects of isocryptotanshinone on gastric cancer. Sci Rep. 2018;8(1):9307.
- Kodigepalli KM, Bonifati S, Tirumuru N, et al. SAMHD1 modulates in vitro proliferation of acute myeloid leukemia-derived THP-1 cells through the PI3K-Akt-p27 axis. Cell Cycle. 2018;17(9):1124–1137.
- Yue J, Lopez JM. Understanding MAPK signaling pathways in apoptosis. Int J Mol Sci. 2020;21(7):2346.
- Kumar GS, Page R, Peti W. The interaction of p38 with its upstream kinase MKK6. Protein Sci. 2021;30(4):908–913.
- He D, Wu H, Xiang J, et al. Gut stem cell aging is driven by mTORC1 via a p38 MAPK-p53 pathway. Nat Commun. 2020;11(1):37.
- Clifford R, Louis T, Robbe P, et al. SAMHD1 is mutated recurrently in chronic lymphocytic leukemia and is involved in response to DNA damage. Blood. 2014;123(7):1021–1031.
- Kodigepalli KM, Li M, Bonifati S, et al. SAMHD1 inhibits epithelial cell transformation in vitro and affects leukemia development in xenograft mice. Cell Cycle. 2018;17(23):2564–2576.
- Fang Y, Yang J, Zu G, et al. Junctional adhesion molecule-like protein promotes tumor progression and metastasis via p38 signaling pathway in gastric cancer. Front Oncol. 2021;11:565676.
- Huang N, Cui X, Li W, et al. IL33/ST2 promotes the malignant progression of gastric cancer via the MAPK pathway. Mol Med Rep. 2021;23(5):361.
- Chen W, Cheng P, Jiang J, et al. Epigenomic and genomic analysis of transcriptome modulation in skin cutaneous melanoma. Aging (Albany NY). 2020;12(13):12703–12725.
- Valverde-Estrella L, Lopez-Serrat M, Sanchez-Sanchez G, et al. Induction of Samhd1 by interferon gamma and lipopolysaccharide in murine macrophages requires IRF1. Eur J Immunol. 2020;50(9):1321–1334.
- Zhang J, Zhu Z, Wu H, et al. PODXL, negatively regulated by KLF4, promotes the EMT and metastasis and serves as a novel prognostic indicator of gastric cancer. Gastric Cancer. 2019;22(1):48–59.
- Li JC, Chen QH, Jian R, et al. The partial role of KLF4 and KLF5 in gastrointestinal tumors. Gastroenterol Res Pract. 2021;2021:2425356.
- Taracha-Wisniewska A, Kotarba G, Dworkin S, et al. Recent discoveries on the involvement of kruppel-like factor 4 in the most common cancer types. Int J Mol Sci. 2020;21(22):8843.
- Kong F, Sun T, Kong X, et al. Kruppel-like factor 4 suppresses serine/threonine kinase 33 activation and metastasis of gastric cancer through reversing epithelial-mesenchymal transition. Clin Cancer Res. 2018;24(10):2440–2451.
- Liu Z, Wu X, Tian Y, et al. H. pylori infection induces CXCL8 expression and promotes gastric cancer progress through downregulating KLF4. Mol Carcinog. 2021;60(8):524–537.
- Chen BJ, Zeng S, Xie R, et al. hTERT promotes gastric intestinal metaplasia by upregulating CDX2 via NF-kappaB signaling pathway. Oncotarget. 2017;8(16):26969–26978.
- Lee HJ, Kang YM, Moon CS, et al. KLF4 positively regulates human ghrelin expression. Biochem J. 2009;420(3):403–411.
- Zhu M, Zhang N, He S. Transcription factor KLF4 modulates microRNA-106a that targets Smad7 in gastric cancer. Pathol Res Pract. 2019;215(8):152467.
- Zhu M, Zhang N, Lu X, et al. Negative regulation of kruppel-like factor 4 on microRNA-106a at upstream transcriptional level and the role in gastric cancer metastasis. Dig Dis Sci. 2018;63(10):2604–2616.
- Wang L, Li Y, Li L, et al. Role of Kruppel-like factor 4 in regulating inhibitor of apoptosis-stimulating protein of p53 in the progression of gastric cancer. Oncol Lett. 2018;15(5):6865–6872.
- Zhao R, Liu Z, Xu W, et al. Helicobacter pylori infection leads to KLF4 inactivation in gastric cancer through a TET1-mediated DNA methylation mechanism. Cancer Med. 2020;9(7):2551–2563.
- Ou Y, Ren H, Zhao R, et al. Helicobacter pylori CagA promotes the malignant transformation of gastric mucosal epithelial cells through the dysregulation of the miR-155/KLF4 signaling pathway. Mol Carcinog. 2019;58(8):1427–1437.
- Banchenko S, Krupp F, Gotthold C, et al. Structural insights into Cullin4-RING ubiquitin ligase remodelling by Vpr from simian immunodeficiency viruses. PLoS Pathog. 2021;17(8):e1009775.
- Cingoz O, Arnow ND, Puig Torrents M, et al. Vpx enhances innate immune responses independently of SAMHD1 during HIV-1 infection. Retrovirology. 2021;18(1):4.
- Zhang K, Dongwen L, Li R. Conserved herpesvirus protein kinases target SAMHD1 to facilitate virus replication. Cell Rep. 2019;28(2):449–59 e5.
