ABSTRACT
Long non-coding RNAs (lncRNAs) are relevant to the development of human cancers. Here, we aimed to investigate the role and mechanism of Linc00883 in the proliferation, invasion, and migration of colorectal cancer (CRC) cells. CRC cell lines SW480 and LoVo were applied as in vitro models in this study. Quantitative real-time PCR was applied to measure Linc00883, miR-577, and FKBP14 expressions. Cell Counting Kit-8, transwell, and wound-healing assays were carried out to confirm the function of Linc00883. Western blot was applied to detect the protein levels of the epithelial–mesenchymal transition-related proteins E-cadherin, vimentin, fibronectin, and α-SMA. RNA immunoprecipitation (RIP) and RNA pull-down experiments were performed to confirm the relationship between Linc00883 and miR-577. Linc00883 expression was elevated in CRC tissues and cells, and the patients with high expression of Linc00883 were related to a low survival rate and prone to distant metastasis. Moreover, we corroborated that Linc00883 and miR-577, miR-577 and FKBP14 are bound to each other. Linc00883 was negatively correlated with miR-577, and miR-577 was also negatively correlated with FKBP14. Furthermore, interference with Linc00883 restrained the proliferation, invasion, and migration of CRC cells through the miR-577/FKBP14 axis. In vivo studies also clarified that Linc00883 facilitated the growth of CRC tumors and the epithelial–mesenchymal transition (EMT) of CRC. Our results demonstrated that Linc00883 facilitated the proliferation, invasion, and migration of CRC cells by regulating the miR-577/FKBP14 axis.
Introduction
Colorectal cancer (CRC) has high mortality worldwide [Citation1,Citation2]. Although some progress has been made in CRC treatment, the prognosis of patients with CRC is still poor [Citation3]. Increasing evidence indicates that the proliferation, invasion, and migration of CRC are the main factors causing its high mortality rate [Citation4,Citation5]. Thus, an in-depth exploration of the underlying mechanisms of CRC cell proliferation, invasion, and migration is needed.
Long non-coding RNAs (lncRNAs) are a class of transcripts that are longer than 200 nucleotides and do not encode proteins [Citation6]. Various lncRNAs have momentous regulatory functions in the process of cell proliferation, metastasis, and oxidative stress response [Citation7–9]. As reported, the abnormally expressed lncRNAs are relevant to the occurrence and development of human cancers including CRC [Citation10,Citation11]. Recently, the competitive endogenous RNA (ceRNA) hypothesis has attracted widespread attention and lncRNAs interact with miRNAs, thereby restraining their expressions and functions [Citation12,Citation13]. For instance, Linc00707 facilitates CRC cell proliferation and metastasis by competitively binding miR-206, prompting that Linc00707 is a potential marker for CRC [Citation14]. Zhang et al. reported that SNHG14 expression is decreased in CRC tissues, and SNHG14 is related to the progression and transfer of CRC by competing with miR-92b-3p [Citation15]. Linc00883 (Ensemble gene ID: ENST00000495228), also known as DUBR, is abnormally expressed in human airway-related diseases [Citation16] and is elevated in neuroblastoma [Citation17]. Through the TCGA Colon Adenocarcinoma Datasets (COAD) database analysis (), we confirmed that Linc00883 was related to tumor typing and metastasis. However, whether Linc00883 functioned in the occurrence and development of CRC remained unclear.
Table 1. Linc00883 is related to tumor typing and metastasis.
MicroRNAs (miRNAs) are a class of short and conserved non-coding RNAs that regulate the expression and function of target mRNAs by binding to the 3’-untranslated region (3’-UTR) of their target mRNAs [Citation18,Citation19]. Previous studies confirmed that miR-144-3p decreases the BCL6 expression by targeting BCL6 3’-UTR, which in turn restrains CRC cell proliferation [Citation20]. Furthermore, Jiang et al. corroborated that miR-577 acts as a tumor suppressor in CRC by regulating HSP27 expression [Citation21]. FKBP14 is from the FKBP family, and the abnormal expression of FKBP14 participates in the progress of several malignant tumors [Citation22,Citation23]. That the bioinformatics online prediction software (starBase) predicted that there were several binding sites between miR-577 and FKBP14 attracted our attention, and Yang et al. indicated that the FKBP14 overexpression facilitates CRC cell proliferation and migration [Citation24]. However, whether miR-577 negatively regulates the FKBP14 expression and thus functions in CRC cell proliferation, invasion, and migration had not yet been fully clarified.
In the present study, we quantified the Linc00883 expression in CRC tissues and cells and assessed the correlation between Linc00883 and the survival rate of CRC patients. Moreover, we sought to investigate the potential mechanism of Linc00883 in CRC toward providing novel potential biomarkers and targets for the prevention and treatment of CRC.
Materials and methods
Clinical specimens
The study was approved by the ethics committee of the First Affiliated Hospital of Zhengzhou University. Written informed consent was obtained from all of the patients involved. Sixty-four CRC patients were included in this study, and 64 pairs of CRC tissues and the matched adjacent normal tissues were collected from the First Affiliated Hospital of Zhengzhou University. All tissues were promptly frozen with liquid nitrogen and then stored in a low-temperature environment at −80°C.
Cell culture
Normal human colon epithelial cell lines NCM460 and CRC cell lines HCT-116, SW620, Caco2, HT29, SW480, and LoVo were taken from the Shanghai Institute of Biological Sciences (Shanghai, China). The above cells were grown in a modified Eagle medium with the addition of 10% fetal bovine serum (FBS, Hyclone, Logan, Utah, USA) and penicillin (100 U/ml, Thermo Fisher Scientific, Waltham, MA, USA), and streptomycin (100 μg/ml, Thermo Fisher Scientific). All cells were cultured in a damp environment at 37°C in 5% CO2.
Quantitative real-time PCR (qRT-PCR)
Total RNAs were isolated using TRIzol Regent (Invitrogen, Carlsbad, CA, USA) and a mirVanaTM PARISTM Kit (Life Technologies, Carlsbad, CA, USA) referring to the manufacturer’s instruction. Then, a PrimeScript™ RT-PCR kit (Takara, Dalian, China) or a TaqMan® MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific, USA) was applied to reverse transcribe RNA into cDNA using the manufacturer’s standard procedures. Subsequently, a real-time PCR assay was conducted on the 7,500 fluorescent quantitative PCR system using IQ SYBR Green SuperMix (Bio-Rad Foster, CA, USA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was applied for internal reference of Linc00883 and FKBP14; U6 was applied for internal reference of miR-577. The relative expressions of Linc00883, miR-577, and FKBP14 were measured using the 2−ΔΔCT method. All the primer sequences are presented in .
Table 2. The sequences of all primers used in qRT-PCR.
Cell transfection
Si-Linc00883 (si-DUBR, RiboBio, Guangdong, China, siB160509112439-1-5), si-FKBP14 (RiboBio, SIGS0011831-1), miR-577 inhibitor (RiboBio, #miR20003242-1-5), and miR-577 mimics (RiboBio, #miR10003242-1-5) were purchased from biological companies.
SW480 and LoVo CRC cells (1 × 105) were inoculated into six well-plates and cultivated for 24 h. Subsequently, the synthesized si-Linc00883 or/and miR-577 inhibitor, pcDNA-Linc00883 or/and miR-577 mimics, and si-FKBP14 were transfected into SW480 or LoVo cells using Lipofectamine 2000 Transfection Reagent (Invitrogen).
Cell counting kit-8 (CCK-8)
A CCK-8 assay was conducted to assess the proliferation of CRC cells as per the previously described methods [Citation25] with minor changes. CRC cells SW480 and LoVo (1 × 104 cells) were seeded into 96 well-plates and then incubated at 37°C in 5% CO2 for 24 h. To monitor the proliferation of CRC cells, the 10 μl CCK-8 reagent was added to each well at 1, 2, and 3 d, and the incubation continued for 4 h. A microplate reader (Bio-TEK, VT, USA) was applied to detect the optical density (OD) at 450 nm.
Transwell measurement
The invasion ability of CRC cells was analyzed by a transwell assay. SW480 and LoVo CRC cells (1 × 105/ml) were suspended in a 100 μl serum-free medium and plated in the upper chambers. Then, a 500 μl modified Eagle medium with 10% FBS was added to the lower chambers. Twenty-four hours later, the stained cells were counted using a microscope (Olympus, Tokyo, Japan). We repeated the transwell measurement at least three times.
Wound-healing experiment
SW480 and LoVo CRC cells (1 × 105) were inoculated into 6 well-plates. After the confluence of CRC cells reached approximately 80%, a 200 μl pipette tip was used to produce three separate wounds (as parallel groups). Then, the floating cells were washed away. A representative image of cells migrating into the wound was taken using a microscope (Olympus). We repeated the wound-healing experiment at least three times.
Dual-luciferase reporter gene assay
SW480 and LoVo CRC cells (1 × 104) were grown in 24-well plates. Linc00883-WT (WT) represented the recombinant plasmid obtained by inserting the WT-Linc00883 sequence containing the miR-577 binding sites (UUUAUCU) into the dual-luciferase reporter vector pmirGLO. Linc00883-Mut (Mut) represented the recombinant plasmid obtained by inserting the mutated Mut-Linc00883 sequence containing the miR-577 binding sites (AAAUAGA) into the dual-luciferase reporter vector pmirGLO.
Next, Linc00883-WT or Linc00883-Mut was constructed and co-transfected into SW480 and LoVo cells with mimics-NC or miR-577 mimics. In addition, FKBP14-WT, FKBP14-Mut luciferase reporter plasmids were structured and co-transfected into SW480 and LoVo cells with miR-577 mimics or/and pcDNA-Linc00883, and their corresponding controls. Forty-eight hours after transfection, we analyzed the luciferase activity using the Dual-Luciferase Reporter Gene Assay Kit (Promega, Madison, WI, USA) given the standard procedures of the reagent manufacturer.
RNA immunoprecipitation (RIP) assay
RIP assays were performed using the RNA-Binding Protein Immunoprecipitation Kit (Millipore, USA) according to its instructions. Cells were incubated with AGO2 antibody or Anti-IgG antibody (Abcam, UK) overnight. The next day, the samples were washed and incubated with 0.1% SDS/Proteinase K. The expression levels of miR-577 and Linc00883 were detected using qRT-PCR [Citation26].
RNA pull-down assay
RNA pull-down assay was performed using the Pierce™ Magnetic RNA-Protein Pull-Down Kit (Thermo, USA) according to its instructions. Biotin-labeled Linc00883 was transfected into cells and incubated with streptavidin magnetic beads. The expression levels of miR-577 were detected by qRT-PCR.
Western blot analysis
After harvesting the SW480 and LoVo CRC cells, the cells were lysed with RIPA buffer (150 mM NaCl, 0.5% EDTA, 50 mM Tris, 0.5% NP40). Then, the protein concentrations were quantified using the BCA kit (Pierce, Rockford, IL, USA); the proteins of different molecular weights were separated on a 10% sodium dodecyl sulfate-polyacrylamide gradient gel (SDS-PAGE) and then transferred into the PVDF membranes (Millipore, Massachusetts, USA). After blocking the membranes with 5% BSA at room temperature for 2 h, the membranes were incubated with the primary antibodies including anti-FKBP14 (Abcam, 0.1 µg/ml, ab251703), anti-α-SMA (Abcam, 0.341 µg/ml, ab7817), anti-vimentin (Abcam, 1:1000, ab137321), anti-fibronectin (Abcam, 1:1000, ab45688), anti-E-cadherin (Abcam, 1 µg/ml, ab231303), and anti-β-actin (Abcam, 1 µg/ml, ab8226) overnight at 4°C. After that, a horseradish peroxidase (HRP) labeled secondary antibody (Proteintech Group, Inc., IL, USA) was incubated with the membranes at room temperature for 1 h. An ECL chemiluminescence system (Pierce) was applied to quantify the protein bands [Citation27].
Xenograft CRC tumor model
Twelve BALB/c female nude mice (SPF) obtained from the Experimental Animal Center of Zhengzhou University were used to construct a mouse model of CRC. After lenti-NC and lenti-Linc00883 were transfected into SW480 CRC cells, the cells (2.5 × 106) were injected subcutaneously into the dorsal side of nude mice [Citation28,Citation29]. The tumor volume was measured once a week, and the tumor volume was calculated. After 5 weeks, the nude mice were sacrificed, and the tumors were weighed. Xenograft CRC tumor models were successfully induced in all nude mice. Our research was approved by the Medical Ethics Committee of the First Affiliated Hospital of Zhengzhou University, and the animal experimental protocol was approved by Zhengzhou University Animal Care and Use Committee.
Statistical analysis
SPSS 21.0 software was applied to assess all experimental data. The data were presented as mean ± standard deviation. When comparing the two groups, we conducted the Student’s t-test; when comparing among more than two groups, we conducted one-way ANOVA followed by Tukey’s posttest. A P value of less than 0.05 was considered a significant difference.
Results
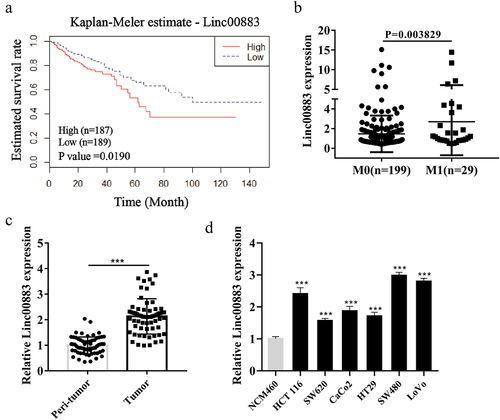
Linc00883 expression is elevated in tissue samples and cells of colorectal cancer
Increasing evidence shows that lncRNAs are closely related to the evolution of various human tumors [Citation30,Citation31]. However, lncRNAs relevant to CRC have not been fully elucidated. Here, we first assessed the correlation between lncRNA Linc00883 and the survival rate of CRC patients. The results clarified that CRC patients with low expression (n = 189) had a higher survival rate than those with high expression (n = 187; p = 0.019; )), and the Linc00883 in CRC patients with distant metastasis (M1: n = 29) was higher than that in patients with non-metastatic CRC (M0: n = 199) (p = 0.003829; )), suggesting that Linc00883 might be a potential target for CRC. Next, we analyzed the Linc00883 expression in CRC tissue samples (n = 64) and CRC cell lines, and the results demonstrated that compared with the matched normal tissues adjacent to cancer and colon epithelial cell line NCM460, Linc00883 expression was elevated in CRC tissue samples (p < 0.001; )) and CRC cell lines (p < 0.001; )). The above experimental data corroborated that the elevated expression of Linc00883 might be related to the poor prognosis and metastasis of CRC.
Figure 1. Expression of Linc00883 in colorectal cancer tissue samples and cells. (a) The patient cohort was obtained from the gene information of the TCGA Colon Adenocarcinoma Datasets (COAD) database that was downloaded from the cBioPortal website. The expression of Linc00883 greater than 0.87 was defined as a high expression, while the expression of Linc00883 lower than 0.87 was defined as a low expression. Kaplan-Meier analysis of the correlation between Linc00883 and survival rate of colorectal cancer (CRC) patients with low expression of Linc00883 (n = 189) and with high expression of Linc00883 (n = 187). (b) Quantitative Real-Time PCR (qRT-PCR) was applied to detect the expression of Linc00883 in patients with distant metastasis of CRC (M1: n = 29) or non-metastatic CRC (M0: n = 199). The expression of Linc00883 in CRC tissues (n = 64) (c) and CRC cell lines (d) was assessed using qRT-PCR. ***P < 0.001 vs. Peri-tumor or NCM460 (colon epithelial cell line). All experiments were performed at least in triplicate.
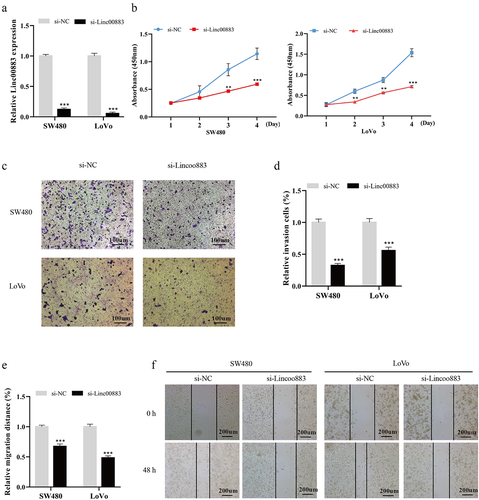
Knockdown of Linc00883 restrains CRC cell proliferation, invasion, and migration
Immediately after, to further probe into the carcinogenic function of Linc00883 in CRC, we knocked down Linc00883 expression in SW480 and LoVo CRC cell lines. The qRT-PCR results corroborated that Linc00883 was successfully knocked down in SW480 and LoVo CRC cells (p < 0.001; )). From the results of CCK-8, we expounded that the Linc00883 knockdown weakened the CRC cell proliferation (p < 0.01; )). A transwell assay corroborated that the Linc00883 knockdown repressed the invasion of CRC cells (p < 0.001; ) (c–2). Moreover, the wound-healing experiment also showed that the Linc00883 knockdown restrained the CRC cell migration (p < 0.001; )(e–2). Overall, the interference with Linc00883 inhibited CRC cell proliferation, invasion, and migration.
Figure 2. Influence of Linc00883 on CRC cell proliferation, invasion, and migration. Si-Linc00883 and its corresponding control were transfected into CRC cell lines SW480 and LoVo. (a) Analysis of the Linc00883 expression in CRC cells by qRT-PCR. (b) Cell Counting Kit-8 (CCK-8) was applied to analyze CRC cell proliferation. (c-d) The CRC cell invasion was assessed using a transwell assay (Scale bar: 100 μm). (e-f) A wound-healing experiment was carried out to detect CRC cell migration (Scale bar: 200 μm). **P < 0.01, ***P < 0.001 vs. si-NC. NC: Negative control. All experiments were performed at least in triplicate.
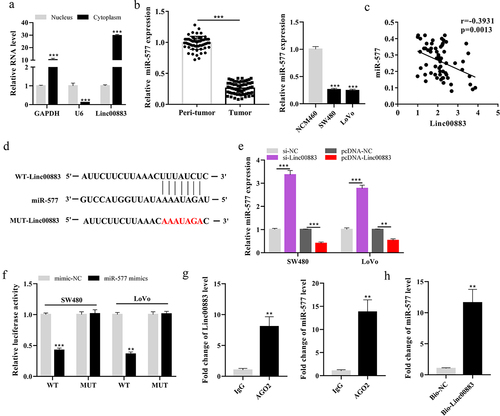
Linc00883 negatively regulates miR-577 expression
As exhibited in ), Linc00883 expression was elevated in the cytoplasm of cells, implying that Linc00883 was primarily distributed in the cytoplasm (p < 0.001). Furthermore, we found that Linc00883 and miR-577 had binding sites through the online bioinformatics (microRNA.org) and then measured the miR-577 expression in CRC tissue samples and cell lines. The results confirmed that miR-577 was decreased in CRC tissues (n = 64) and cell lines compared with the matched normal tissues adjacent to cancer and colon epithelial cell line NCM460 (p < 0.001; )). ) showed that Linc00883 was significantly negatively correlated with miR-577 (p = 0.0013). The online bioinformatics (microRNA.org) predicted the binding sites between Linc00883 and miR-577 ()). Next, si-Linc00883, pcDNA-Linc00883, and their corresponding controls were transfected into SW480 and LoVo cells. As shown in ), Linc00883 negatively regulated the expression of miR-577 (p < 0.001). Furthermore, the dual-luciferase reporter gene results corroborated that miR-577 negatively regulated the Linc00883 luciferase activity (p < 0.001; )). Therefore, we speculated that miR-577 might be negatively regulated by Linc00883, which would hinder RISC-mediated downstream mRNA silencing. To address this possibility, we further conducted RIP and RNA pull-down experiments. As shown in the RIP analysis, Linc00883 and miR-577 were enriched more in Ago2 precipitated particles than in the IgG control (p < 0.01; )). Subsequent RNA pull-down tests further confirmed this result (p < 0.01; )). Biotin-labeled Linc00883 (Bio-LinC00883) has a large number of miR-577 enrichment in its pull-down complex. Collectively, Linc00883 can combine with miR-577, and these data suggested the molecular sponge role of Linc00883 for miR-577 via the ceRNA mechanism.
Figure 3. Effect of Linc00883 on miR-577 expression. (a) The Linc00883 expression in the nucleus or cytoplasm of cells was measured using qRT-PCR. (b) qRT-PCR was applied to detect miR-577 expression in CRC tissue samples (n = 64) and cell lines. (c) The correlation analysis between Linc00883 and miR-577. (d) The online bioinformatics (microRNA.org) was performed to predict the binding sites between Linc00883 and miR-577. The si-Linc00883, pcDNA-Linc00883, and their corresponding controls were transfected into SW480 and LoVo cells. (e) Analysis of the miR-577 expression by qRT-PCR. (f) A dual-luciferase reporter gene assay was conducted to assess the influence of miR-577 on Linc00883 luciferase activity. (g-h) RIP and RNA pull-down assays were performed. **P < 0.01 vs. lgG or Bio-NC; ***P < 0.001 vs. nucleus, Peri-tumor, NCM460, si-NC, pcDNA-NC or mimics-NC. All experiments were performed at least in triplicate.
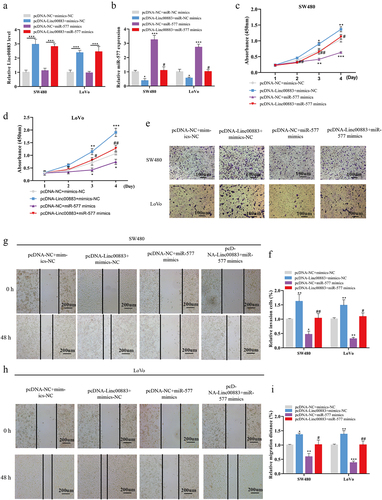
Linc00883 regulates CRC cell proliferation, invasion, and migration via miR-577
Subsequently, we transfected pcDNA-Linc00883 or/and miR-577 mimics into SW480 and LoVo cells to investigate whether Linc00883 influenced CRC cell proliferation, invasion, and migration through miR-577. As shown in ) (a–4, the transfection of pcDNA-Linc00883 elevated the Linc00883 expression, and the transfection of miR-577 mimics had no remarkable effect on the Linc00883 expression (p < 0.001). Meanwhile, the transfection of pcDNA-Linc00883 restrained the miR-577 expression, and the transfection of the miR-577 mimics canceled the effect of pcDNA-Linc00883 (p < 0.05). From the results of the CCK-8, the Linc00883 overexpression facilitated the proliferation of CRC cells, while this facilitation was reversed after the transfection of miR-577 mimics (p < 0.05; )(c–4). The transwell assay also indicated that the Linc00883 overexpression promoted the invasion of CRC cells, while this promotion was reversed after the transfection of the miR-577 mimics (p < 0.05; ) (e–4. Moreover, the wound-healing experiment indicated that the Linc00883 overexpression facilitated CRC cell migration, while this trend was reversed after the transfection of the miR-577 mimics (p < 0.05; )(g–4). In summary, the Linc00883 overexpression facilitated CRC cell proliferation, invasion, and migration by decreasing miR-577.
Figure 4. Linc00883 influences CRC cell proliferation, invasion, and migration via miR-577. The pcDNA-Linc00883 or/and miR-577 mimics and their corresponding controls were transfected into SW480 and LoVo cells. (a-b) The expressions of Linc00883 and miR-577 were measured using qRT-PCR. (c-d) CCK-8 was applied to assess CRC cell proliferation. (e-f) The CRC cell invasion was measured using transwell assay (Scale bar: 100 μm). (g-i) A wound-healing experiment was performed to assess CRC cell migration (Scale bar: 200 μm). *P < 0.05, **P < 0.01, ***P < 0.001 vs. pcDNA-NC + mimics-NC. #P < 0.05, ##P < 0.01 vs. pcDNA-NC + miR-577 mimics. NC: Negative control. All experiments were performed at least in triplicate.
Linc00883 regulates CRC cell proliferation, invasion, and migration through the miR-577/FKBP14 axis
Next, we further probed into the downstream target genes of miR-577. Online bioinformatics software (starBase) predicted the potential binding sites between miR-577 and FKBP14 ()), indicating that FKBP14 might be one of the downstream target genes of miR-577. From the TCGA database, we found that FKBP14 was more elevated in colon cancer (n = 286) than that in the normal group (n = 41); the elevated FKBP14 (n = 70) was more relevant to poor prognosis than the low-/medium-expressed FKBP14 (n = 209; p = 0.012; )(b–5). The western blot and qRT-PCR analysis also corroborated that the levels of FKBP14 were elevated in CRC tissues (p < 0.001; )(d–5). Additionally, ) showed that FKBP14 was significantly negatively correlated with miR-577 in CRC tissues (p = 0.0079). Subsequently, we transfected si-Linc00883 or/and the miR-577 inhibitor into SW480 cells. The results of the qRT-PCR clarified that the transfection of si-Linc00883 decreased the Linc00883 expression, while the co-transfection of the miR-577 inhibitor had no remarkable effect on the Linc00883 expression. Moreover, the transfection of si-Linc00883 increased the expression of miR-577, while the co-transfection of the miR-577 inhibitor canceled the effect of si-Linc00883 (p < 0.05; ) (g–5. As exhibited in ), the interference with Linc00883 decreased the mRNA and protein levels of FKBP14, while these effects were reversed after the transfection of the miR-577 inhibitor (p < 0.01). Moreover, we transfected pcDNA-Linc00883 or/and the miR-577 mimics into SW480 cells. As shown in ), the Linc00883 overexpression elevated the mRNA and protein levels of FKBP14, while these effects were reversed after the transfection of the miR-577 mimics (p < 0.05). From the results of the dual-luciferase reporter gene assay, miR-577 negatively regulated the luciferase activity of FKBP14, while the Linc00883 overexpression reversed the regulation of miR-577 on FKBP14 luciferase activity (p < 0.01; )). Next, we transfected si-FKBP14 into SW480 and LoVo cells to investigate the effect of FKBP14 on CRC cell proliferation, invasion, and migration. Our experimental results indicated that the interference with FKBP14 restrained the proliferation (p < 0.001; )), invasion (p < 0.001; )), and migration (p < 0.01; )) of CRC cells. Overall, Linc00883 regulated CRC cell proliferation, invasion, and migration through the miR-577/FKBP14 axis.
Figure 5. Linc00883 regulates CRC cell proliferation, invasion, and migration via the miR-577/FKBP14 axis. (a) Bioinformatics online software (StarBase) was applied to forecast the potential binding sites between miR-577 and FKBP14. TCGA database was applied to analyze the expression in colon cancer (B, Normal: n = 41 and Primary tumor: n = 286) or the effect of FKBP14 on the prognosis of colon cancer (C, high expression FKBP14: n = 70; low/medium expression FKBP14: n = 209). (d) The protein level of FKBP14 in CRC tissue samples was detected by western blot. (e) The mRNA level of FKBP14 in CRC tissue samples was detected by qRT-PCR. (f) The correlation analysis between FKBP14 and miR-577. The si-Linc00883 or/and miR-577 inhibitor were transfected into SW480 cells. (g-h) The expressions of Linc00883 and miR-577 were detected by qRT-PCR. (i) Detection of FKBP14 mRNA and protein levels using qRT-PCR and western blot. The pcDNA-Linc00883 or/and miR-577 mimics was transfected into SW480 cells. (j) Detection of FKBP14 mRNA and protein levels by qRT-PCR and western blot. (k) Detection of the luciferase activity of FKBP14 using dual-luciferase reporter gene assay. The si-FKBP14 was transfected into SW480 and LoVo cells. (l) The CRC cell proliferation was assessed by CCK-8. (m) Transwell assay was applied to analyze CRC cell invasion (Scale bar: 100 μm). (n) The CRC cell migration was assessed using a wound-healing experiment (Scale bar: 200 μm). *P < 0.05, **P < 0.01, ***P < 0.001 . All experiments were performed at least in triplicate.
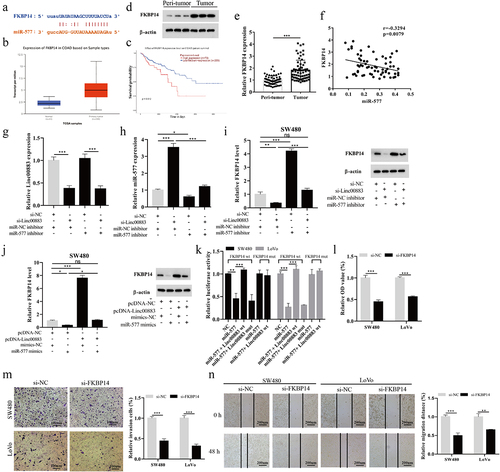
Linc00883 facilitates CRC tumor growth in vivo
To clarify whether Linc00883 also exerted as an oncogene in vivo, we constructed a xenograft nude mouse model of CRC by subcutaneously injecting CRC cells SW480 transfected with lenti-Linc00883. As shown in ) (a–6, the Linc00883 overexpression increased the volume and weight of the CRC tumors (p < 0.01). Meanwhile, the Linc00883 overexpression increased the expressions of Linc00883 and FKBP14, decreased miR-577 (p < 0.001; )(d–6), and elevated the protein level of FKBP14 ()). Moreover, the Linc00883 overexpression elevated the protein levels of α-SMA, vimentin, and fibronectin and decreased E-cadherin, supporting the conclusion that the Linc00883 overexpression facilitated the epithelial–mesenchymal transition (EMT) of CRC ()). In conclusion, our experimental results corroborated that Linc00883 facilitated the growth of CRC tumors in vivo.
Figure 6. Effect of Linc00883 on CRC tumor growth in vivo. A xenograft nude mouse model of CRC by subcutaneously injecting CRC cells SW480 transfected with lenti-Linc00883 was established. n = 6. (a-b) Detection of the volume of CRC tumors. (c) Detection of the weight of CRC tumors. (d-f) Detection of Linc00883, miR-577, and FKBP14 expressions by qRT-PCR. (g) Detection of the FKBP14 protein level using western blot. (h) Detection of the protein levels of epithelial-mesenchymal transition (EMT)-related proteins E-cadherin, vimentin, fibronectin, and α-SMA by western blot. **P < 0.01, ***P < 0.001 vs. lenti-NC.
Discussion
Due to the lack of effective clinical treatment and biomarkers with diagnostic value, CRC is one of the most malignant tumors recognized worldwide [Citation32]. Thus, there is an urgent need to investigate the molecular mechanism of CRC to excavate novel effective targets. In this research, we found that Linc00883 expression was elevated in CRC tissue samples and cells; the patients with high Linc00883 expression had a low survival rate and were prone to distant metastasis. Our further investigation illustrated that Linc00883 facilitated CRC cell proliferation, invasion, and migration via the miR-577/FKBP14 axis.
Previous studies demonstrated that lncRNAs play crucial regulatory functions in the evolution of tumors [Citation33,Citation34], and lncRNAs negatively regulate the expression of miRNAs via the sponge function of miRNAs [Citation35]. As has been reported, LOC646329 restrains the progression of CRC by exerting the sponge function of miR-29b [Citation36]. Moreover, Yu et al. reported that Linc00461 exerts oncogenic functions in CRC by sponging miR-323b-3p [Citation37]. In our research, we also clarified that Linc00883 sponged miR-577 by binding to miR-577, thus playing crucial regulatory functions in CRC cell proliferation, invasion, and migration, which was consistent with the results of the aforementioned studies.
LncRNAs also play an important role in the regulation of EMT in cancer cells [Citation38]. Accumulating evidence shows that EMT facilitates CRC cell invasion and metastasis [Citation39]. As reported, lncRNA VIM-AS1 is elevated in CRC tissues and cells and promotes tumor metastasis through the Vim-EMT pathway [Citation40]. In addition, lncRNA CASC21 has also been reported to promote CRC cell proliferation, migration, EMT, and to further aggravate CRC [Citation41]. In this study, we also corroborated that Linc00883 facilitated the EMT of CRC. Previous studies also indicated that EMT exerts crucial regulatory functions in tumor immunosuppression and immune escape [Citation42,Citation43] and that low infiltration of lymphocytosis in tumors is associated with the high expression of EMT-related genes [Citation44]. These results confirmed that elevated Linc00883 might participate in the regulation of tumor immunity, which is worthy of further investigation in this study.
Increasing evidence from recent studies indicates that miRNAs exert important functions in human diseases [Citation45,Citation46]. For instance, miR-130a promotes malignant tumor growth by targeting PTEN [Citation47]. Shen et al. found that miR-145 is down-regulated in CRC cells and miR-145 overexpression restrains CRC cell invasion and migration via targeting TWIST1 [Citation48]. Jiang et al. also proved that miR-577 is decreased in CRC tissue samples and cells, which might act as a tumor suppressor in CRC by targeting HSP27 to restrain the HSP27 expression [Citation21]. Likewise, our results indicated that miR-577 was down-regulated in CRC tissues and cells, and overexpressing miR-577 inhibited cell invasion and migration in CRC cells.
FKBP14 belongs to the FKBP family, which has a cancer-promoting function and abnormal expression in various malignant tumors [Citation23,Citation49]. FKBP14 can promote the progress of gastric cancer, ovarian cancer, and osteosarcoma [Citation23,Citation50,Citation51]. Furthermore, FKBP14 also facilitates CRC cell proliferation and migration [Citation24]. In this study, we also corroborated that FKBP14 was up-regulated in CRC tissues, and interference with FKBP14 inhibited cell proliferation, invasion, and migration in CRC cells. Furthermore, FKBP14 was negatively regulated by miR-577. Our in-depth research confirmed that miR-577 restrained CRC cell proliferation, invasion, and migration via decreasing FKBP14.
Overall, our experimental results confirmed the promoting effect of Linc00883 on CRC, which might be achieved by regulating the miR-577/FKBP14 axis. We also confirmed the binding relationships between Linc00883 and miR-577, as well as between miR-577 and FKBP14. Our research might provide novel potential targets for CRC treatment. Previous studies indicated that lncRNAs can be applied to identify different clinically relevant CRC molecular subtypes [Citation52,Citation53], and we found the high expression of Linc00883 to be of great significance, These results suggest that we can further investigate which consensus molecular subtypes (CMS) can be classified in other tumors. Meanwhile, with the development of RNA-seq technology and the association between lncRNAs and other pathways [Citation54–56], we would use RNA-seq technology in the future to investigate the relationship between LINC00883 and other targets/pathways, including angiogenesis, mitochondria, and ion channels to further enrich the content of this study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Yeo H, Betel D, Abelson JS, et al. Early-onset colorectal cancer is distinct from traditional colorectal cancer. Clin Colorectal Cancer. 2017;16(4):293–299.e296.
- Qianqian F, Wang S, Zhu C, et al. Di-2-pyridylketone 4, 4-dimethyl-3-thiosemicarbazone effectively induces human colorectal carcinoma cell apoptosis via mTOR pathway. Aging Pathobiol Ther. 2021;3(3):56–62.
- Pan JH, Zhou H, Cooper L, et al. LAYN is a prognostic biomarker and correlated with immune infiltrates in gastric and colon cancers. Front Immunol. 2019;10:6.
- Gupta GP, Massagué J. Cancer metastasis: building a framework. Cell. 2006;127(4):679–695.
- Liu T, Wang H, Yu H, et al. The long non-coding RNA HOTTIP is highly expressed in colorectal cancer and enhances cell proliferation and invasion. Mol Ther Nucleic Acids. 2020;19:612–618.
- Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017;18(1):206.
- Li J, Li Z, Zheng W, et al. LncRNA-ATB: an indispensable cancer-related long noncoding RNA. Cell Prolif. 2017;50(6):e12381.
- Huang Q, Yan J, Agami R. Long non-coding RNAs in metastasis. Cancer Metastasis Rev. 2018;37(1):75–81.
- Donato L, Scimone C, and Alibrandi S, et al. Transcriptome analyses of lncRNAs in A2E-stressed retinal epithelial cells unveil advanced links between metabolic impairments related to oxidative stress and retinitis pigmentosa. Antioxidants (Basel). 2020;9(4):318.
- Wang Q, Feng Y, Peng W, et al. Long noncoding RNA Linc02023 regulates PTEN stability and suppresses tumorigenesis of colorectal cancer in a PTEN-dependent pathway. Cancer Lett. 2019;451:68–78.
- Calanca N, Paschoal AP, É P M, et al. The long non-coding RNA ANRASSF1 in the regulation of alternative protein-coding transcripts RASSF1A and RASSF1C in human breast cancer cells: implications to epigenetic therapy. Epigenetics. 2019;14(8):741–750.
- Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272–283.
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358.
- Zhu H, He G, Wang Y, et al. Long intergenic noncoding RNA 00707 promotes colorectal cancer cell proliferation and metastasis by sponging miR-206. Onco Targets Ther. 2019;12:4331–4340.
- Zhang W, Duan W, Mo Z, et al. Upregulation of SNHG14 suppresses cell proliferation and metastasis of colorectal cancer by targeting miR-92b-3p. Journal of Cellular Biochemistry. 2020;121(2):1998–2008.
- Perry MM, Tsitsiou E, Austin PJ, et al. Role of non-coding RNAs in maintaining primary airway smooth muscle cells. Respir Res. 2014;15(1):58.
- Utnes P, Løkke C, Flægstad T, et al. Clinically relevant biomarker discovery in high-risk recurrent neuroblastoma. Cancer Inform. 2019;18:1176935119832910.
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233.
- Donato L, Bramanti P, Scimone C, et al. miRNAexpression profile of retinal pigment epithelial cells under oxidative stress conditions. FEBS Open Bio. 2018;8(2):219–233.
- Sun N, Zhang L, Zhang C, et al. miR-144-3p inhibits cell proliferation of colorectal cancer cells by targeting BCL6 via inhibition of Wnt/beta-catenin signaling. Cell Mol Biol Lett. 2020;25:19.
- Jiang H, Ju H, Zhang L, et al. microRNA-577 suppresses tumor growth and enhances chemosensitivity in colorectal cancer. J Biochem Mol Toxicol. 2017;31(6):e21888.
- Ghoorun RA, and Wu XH. Prognostic significance of FKBP14 in gastric cancer. Onco Targets Ther. Vol. 12. 2019. p. 11567–11577.
- Lu M, Miao Y, Qi L, et al. RNAi-Mediated downregulation of FKBP14 suppresses the growth of human ovarian cancer cells. Oncol Res. 2016;23(6):267–274.
- Yang L, Zhang R, Yang J, et al. FKBP14 promotes the proliferation and migration of colon carcinoma cells through targeting IL-6/STAT3 signaling pathway. Onco Targets Ther. 2019;12:9069–9076.
- Zhou Y , Mao L, Wang Y, et al. Increased LncRNA PVT-1 is associated with tumor proliferation and predicts poor prognosis in cervical cancer. Clin Surg Res Commun. 2017;1(1):10–17.
- Shu G, Su H, Wang Z, et al. LINC00680 enhances hepatocellular carcinoma stemness behavior and chemoresistance by sponging miR-568 to upregulate AKT3. J Exp Clin Cancer Res. 2021;40(1):45.
- Roman MG, Flores LC, Cunningham GM, et al. Thioredoxin down-regulation in the cytosol in thioredoxin 2 transgenic mice did not have beneficial effects to extend lifespan in male C57BL/6 mice. Aging Pathobiol Ther. 2020;2(4):203–209.
- Liang W, Wu J, Qiu X. LINC01116 facilitates colorectal cancer cell proliferation and angiogenesis through targeting EZH2-regulated TPM1. J Transl Med. 2021;19(1):45.
- Luo K, Geng J, Zhang Q, et al. LncRNA CASC9 interacts with CPSF3 to regulate TGF-β signaling in colorectal cancer. J Exp Clin Cancer Res. 2019;38(1):249.
- Liang Y, Song X, Li Y, et al. LncRNA BCRT1 promotes breast cancer progression by targeting miR-1303/PTBP3 axis. Molecular Cancer. 2020;19(1):85.
- Luo L, Zhang J, and Tang H. LncRNA SNORD3A specifically sensitizes breast cancer cells to 5-FU by sponging miR-185-5p to enhance UMPS expression. Cell Death Dis. 2020;11(5):329.
- Sadanandam A, Lyssiotis CA, Homicsko K, et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19(5):619–625.
- Chen J, Yu Y, Li H, et al. Long non-coding RNA PVT1 promotes tumor progression by regulating the miR-143/HK2 axis in gallbladder cancer. Mol Cancer. 2019;18(1):33.
- Ma YL, Wang CY, Guan YJ, et al. Long noncoding RNA ROR promotes proliferation and invasion of colorectal cancer by inhibiting tumor suppressor gene NF2 through interacting with miR-223-3p. Eur Rev Med Pharmacol Sci. 2020;24(5):2401–2411.
- Xu Y, Yao Y, Jiang X, et al. SP1-induced upregulation of lncRNA SPRY4-IT1 exerts oncogenic properties by scaffolding EZH2/LSD1/DNMT1 and sponging miR-101-3p in cholangiocarcinoma. J Exp Clin Cancer Res. 2018;37(1):81.
- Javanmard AR, Dokanehiifard S, Bohlooli M, et al. LOC646329 long non-coding RNA sponges miR-29b-1 and regulates TGFbeta signaling in colorectal cancer. Journal of Cancer Research and Clinical Oncology. 2020;146(5):1205–1215.
- Yu H, Ma J, Chen J, et al. LncRNA LINC00461 promotes colorectal cancer progression via miRNA-323b-3p/NFIB axis. Onco Targets Ther. 2019;12:11119–11129.
- Xu Q, Deng F, Qin Y, et al. Long non-coding RNA regulation of epithelial-mesenchymal transition in cancer metastasis. Cell Death Dis. 2016;7(6):e2254.
- Brabletz T, Hlubek F, Spaderna S, et al. Invasion and metastasis in colorectal cancer: epithelial-mesenchymal transition, mesenchymal-epithelial transition, stem cells and beta-catenin. Cells Tissues Organs. 2005;179(1–2):56–65.
- Rezanejad Bardaji H, Asadi MH, Yaghoobi MM. Long noncoding RNA VIM-AS1 promotes colorectal cancer progression and metastasis by inducing EMT. Eur J Cell Biol. 2018;97(4):279–288.
- Zhang C, J E, Yu E. LncRNA CASC21 induces HGH1 to mediate colorectal cancer cell proliferation, migration, EMT and stemness. RNA Biol. 2021;18(sup1):369–381.
- Jiang Y, Zhan H. Communication between EMT and PD-L1 signaling: new insights into tumor immune evasion. Cancer Lett. 2020;468:72–81.
- Romeo E, Caserta CA, Rumio C, et al. The vicious cross-talk between tumor cells with an EMT phenotype and cells of the immune system. Cells. 2019;8(5):460.
- Hui D, Chen J, Jiang Y, et al. CD44(+)CD24(-/low) sphere-forming cells of EBV-associated gastric carcinomas show immunosuppressive effects and induce Tregs partially through production of PGE2. Exp Cell Res. 2020;390(2):111968.
- Kong F, Zou H, Liu X, et al. miR-7112-3p targets PERK to regulate the endoplasmic reticulum stress pathway and apoptosis induced by photodynamic therapy in colorectal cancer CX-1 cells. Photodiagnosis Photodyn Ther. 2020;29:101663.
- Liang Y, Zhu D, Hou L, et al. MiR-107 confers chemoresistance to colorectal cancer by targeting calcium-binding protein 39. Br J Cancer. 2020;122(5):705–714.
- Wei H, Cui R, Bahr J, et al. miR-130a deregulates PTEN and stimulates tumor growth. Cancer Res. 2017;77(22):6168–6178.
- Shen X, Jiang H, and Chen Z MicroRNA-145 inhibits cell migration and invasion in colorectal cancer by targeting TWIST , et al. Onco Targets Ther. Vol. 12. 2019. p. 10799–10809 .
- Sun LY, Tao JZ, Yan B, et al. Inhibitory effects of FKBP14 on human cervical cancer cells. Mol Med Rep. 2017;16(4):4265–4272.
- Wang R, Fang H, Fang Q. Downregulation of FKBP14 by RNA interference inhibits the proliferation, adhesion and invasion of gastric cancer cells. Oncol Lett. 2017;13(4):2811–2816.
- Wang K, Qi XJ, Liu HZ, et al. MiR-361 inhibits osteosarcoma cell lines invasion and proliferation by targeting FKBP14. Eur Rev Med Pharmacol Sci. 2018;22(1):79–86.
- Chen H, Xu J, Hong J, et al. Long noncoding RNA profiles identify five distinct molecular subtypes of colorectal cancer with clinical relevance. Mol Oncol. 2014;8(8):1393–1403.
- Eide PW, Eilertsen IA, Sveen A. Long noncoding RNA MIR31HG is a bona fide prognostic marker with colorectal cancer cell-intrinsic properties. International Journal of Cancer. 2019;144(11):2843–2853.
- Donato L, and Scimone C. Possible A2E mutagenic effects on rpe mitochondrial DNA from innovative RNA-Seq bioinformatics pipeline. Antioxidants (Basel). 2020;9(11):1158.
- Scimone C, Alibrandi S, Scalinci SZ, et al. Expression of pro-angiogenic markers is enhanced by blue light in human RPE cells. Antioxidants. 2020. 9(11). 1154
- Donato L, Scimone C, Alibrandi S. New omics-derived perspectives on retinal dystrophies: could ion channels-encoding or related genes act as modifier of pathological phenotype? International Journal of Molecular Sciences. 2020;22(1). DOI:10.3390/ijms22010070
