ABSTRACT
Drug delivery and triggered release in tumor cells would realize the ultimate goal of precise cancer treatment. An APE1 triggered DNA nanoprism was designed, aiming at the applications of both drug delivery and precise triggered drug release in cancer cell. We demonstrate that the AP-Prism was successfully used as a vehicle based on the intracellular endogenous enzyme APE1 triggered for controlled drug delivery and triggered release. The box like DNA prism was self-assembled by annealing process and Doxorubicin molecules were then inserted into the GC base pairs. The reaction of AP-Prism enzymolysis and stability of DNA prism were investigated. Encouraged by the demonstration of AP-Prism as a drug delivery carrier, the cellular uptake and Dox release were with investigated in a human cervical cancer cell HeLa and human embryonic kidney cell HEK-293 T. Thanks to the overexpression level of APE1 in cancer cells, DNA prism could selectively release the trapped doxorubicin in response to APE1 activity in cancer cells, and provide a new strategy for the development of precision medicine.
1. Introduction
Apurinic/apyrimidinic endonuclease/redox effector factor 1 (APE1) plays key roles in both the short-patch and long-patch pathways of base excision repair (BER) pathway [Citation1,Citation2]. It can also respond to intracellular oxidative stress conditions. Abnormal expression of APE1 has been found in a couple of tumor cells, which demonstrate APE1 as an emerging biomarker for some cancer types [Citation3]. Utilizing the precise cleavage of APE1 in mammalian cells, a number of approaches have been developed for detecting APE1 activity [Citation4,Citation5], and intracellular enzyme regulation [Citation6,Citation7].
Nanotechnology is receiving attention in many fields of chemistry, engineering, biology, and medicine [Citation8,Citation9]. Nanoparticle-based technology has opportunities in therapeutic and diagnostic applications, especially for drug and gene delivery, photothermal therapy, and recognition in cancer [Citation10,Citation11]. Targeted delivery and drug release have become a hot topic in precision medicine. Nanodevices are biocompatible and nontoxic and they have diagnostic and therapeutic properties with wide applications in this field [Citation12,Citation13]. Researches developed lots of ingenious system to achieve drug target delivery and release which are successfully used in cancer therapy applications, such as pH-sensitive [Citation14,Citation15], X-ray-responsive [Citation16] drug delivery systems, et al. These advanced approaches have made important breakthroughs in reducing multidrug-resistance [Citation17], reducing side effects and upregulating the drug accumulation at the target site [Citation18]. Due to the programmability and biocompatibility, DNA nanostructures have potential applications in the fields of biosensor development [Citation19,Citation20], biomedical imaging [Citation21], drug delivery [Citation22], etc. DNA prism has been used to create functional components by a DNA-minimal approach [Citation23–26]. It has been demonstrated that DNA prism exhibited significant biostability and more drug or siRNA delivery to the tumor sites [Citation27].
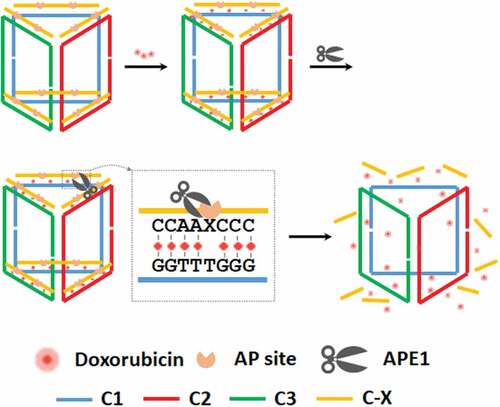
Herein, we develop an APE1 responsive DNA prism as a nanovehicle for both drug delivery and precise triggered drug release in cancer cells (). APE1 recognizes the double strand edges and then cleaves the AP-site, releasing short fragments which activate doxorubicin (Dox) dissociated. DNA nanostructures, such as DNA prism, not only have been shown to gain access to the cytoplasm without a co-carrier agent but also have high robustness due to the rigid structure [Citation25,Citation28]. This DNA prism shows high specificity toward APE1 and the kinetics analysis suggests that the enzymatic cleavage is the rate-limiting step, making the rate of drug release highly dependent on digest progress. APE1 has often been seen to be overexpressed or to exhibit a sub-cellular distribution pattern, in many cancer types that is not observed in normal pre-cancerous tissue [Citation29,Citation30] The cytoplasmic distribution of APE1 provides a key to drug release in living cancer cells. This cleavage process triggers the release of the trapped Dox for accurate diagnosis and precise drug delivery in cancer cells, which suggests a potential application in drug target therapy.
2. Materials and methods
2.1. Materials
Human apurinic/apyrimidinic endonuclease I (APE1), DNase I, Exonuclease I, Exonuclease III, T7 Exonuclease and λ Exonuclease were obtained from NEB (Beverly, MA). Doxorubicin hydrochloride was from Sigma-Aldrich (St. Louis, MO). All of the oligonucleotides used in this work were synthesized by Sangon Co. (Shanghai, China) and their sequences are listed in Table S1. All modified oligonucleotides were purified by HPLC, while unmodified oligonucleotides were purified by PAGE. DNase/RNase free deionized water from Tiangen Biotech Co. (Beijing, China) was used in all experiments.
2.2. Design, preparation and characterization of the DNA prism by gel
The software Tiamat was used for DNA structure design and sequence generation. To prepare the DNA prism, the oligonucleotides (final concentration of each strand 1 μM) were mixed in 1× TAE-Mg2+ buffer (40 mM Tris, 12.5 mM MgCl2, and 2 mM EDTA, pH 8.0). The solution was denatured on a thermocycler as the following procedure: 95°C for 5 min, 65°C for 30 min, 50°C for 30 min, 37°C for 30 min, 22°C for 30 min and 4°C for 55 min. For the assembly of dSpacer-labeled probe, equimolar probe was added to the DNA prism solution and denatured following procedure: 50°C for 30 min, 37°C for 30 min, 22°C for 30 min and 4°C for 55 min. The DNA prism was characterized by 8% native PAGE which was operated at 4°C for 4 h at a constant voltage of 120 V. The gel was subsequently stained with SYBR Gold.
2.3. Characterization of the APE1 activity by fluorescence dequenching assay
All reactions in homogeneous solution were carried out in 0.2 mL sealed PCR tube. Once APE1 was added to reach final concentrations of 0.658 nM. Then fluorescence was recorded immediately in the FAM channel (ex: 480 nm, em: 510 nm) of a real-time PCR cycler (Rotor-Gene Q, QIAGEN, Germany) at 37°C, using a gain of 8, with a time interval of 5 s.
2.4. DOX loading to DNA prism
0.1 mL Dox (1 mM) was added to the DNA prism solution with continuous shaking under dark condition at room temperature for 24 h. To remove excessive Dox, DNA prism-Dox was washed with 1× TAE-Mg2+ buffer using an Amicon-2 kD cutoff filter.
2.5. Drug delivery to living cells
HeLa and HEK-293 T cell lines were cultured in 1640 medium supplemented with 1% Penn/Strep and 10% fetal bovine serum and incubated at 37°C in a humidified atmosphere of 5% CO2/95% air. The cells were transferred to a laser confocal culture dish for fluorescence imaging in an appropriate density. The cells were incubated with DNA prism-Dox (labeled with Cy5) and other reagents in a low-fluorescence culture media (FluoroBrite DMEM, Thermo Fisher) for 4 h. Fluorescence imaging was carried out on an inverted fluorescence microscope equipped with a mercury light source (Nikon). The filter with 470 nm/ 585 nm, 640 nm/660 nm and 360 nm/ 447 nm were used to detecting the emission of Dox, Cy5 and Hoechst33342 respectively. The images were acquired using a 40 × objective and recorded by the EMCCD (50 ms, gain 3).
2.6. Western blot analysis of APE1 in different cells
Cells were collected by trypsinization and centrifuged to remove supernatant. Cell pellet was resuspended in lysis buffer with 1× protease inhibitor, 1 mM NaF and 1 mM Na3VO4 for 30 min, at 4°C. After centrifugation at 13000 rpm for 15 min, at 4°C, the supernatant was collected as total cell lysate. The protein concentrations of cells were determined with BSA standard curve method from absorbance. For western blot analysis, 30 μg of proteins were resolved on 10% SDS-PAGE, transferred onto nitrocellulose membranes, and probed with anti-APE1 antibodies (1: 5000) (ab194, abcam). The membranes were incubated with goat anti-mouse IgG labeled with horseradish peroxidase (HRP) (1: 5000) (ab6789, abcam). Bands on blots were visualized using an enhanced chemiluminescence (ECL) detection system (5200, Tanon, China).
3. Results and discussion
As illustrated in and Fig. S1 in SI, the boxlike DNA prism was constructed with three clip strands. The two 10-base ends of the first clip hybridized to the back of the next clip through their complementary domain. Likewise, the second clip bound to the third clip and the third clip bound to the first clip as the same way which formed a closed DNA triangular-prism. The three vertical edges of the prism are of double-stranded structure, which guarantees nanoassembly with a rigid 3D structure. The strands with AP sites hybridized to the six edges of the top and bottom faces of DNA prism which were rich of GC base pairs (AP-Prism). Doxorubicin (Dox) molecules were then inserted into the GC base pairs. Specifically, the double strand edges contained AP sites which were recognized and cleaved by APE1, resulting in a single strand nick as well as short fragments The structure of DNA prism changed back to the rigid 3D structure with three hybridized clips. Dox molecules then were released because APE1 destroys the double stranded structure. Native gel analysis confirmed formation of the expected assembled structures (Fig. S2). Successive additions of the three clip strands and AP site labeled edge generated bands of reduced mobility, indicating that successful hybridization of each clip (lane 2–5) produced the 3D triangular-prism nanostructure in high yield.
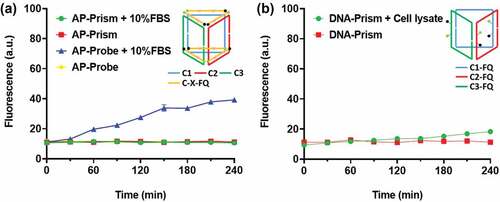
To examine the enzyme activity, we labeled the DNA prism with a fluorophore and quencher as the reporter ()). The fluorescence of intact AP-Prism was quenched. The APE1 cleaves the AP sites yielding short fragments release and fluorescence restoration. As the concentration of APE1 increases, the reaction speed increased ()). By considering these results, we speculated that the catalytic hydrolysis is the rate-limiting step of the process. The kinetics of APE1 catalysis directly reflected the velocity of drug release. To test the specificity of the AP-Prism toward APE1, we examined the possible nonspecific interactions of the AP-Prism with some nucleases that can digest dsDNA or ssDNA. Weak fluorescence increase was observed when the AP-Prism was incubated with these nucleases ()). The degradation of AP-Prism by these enzymes is below 15% after 4 h ()). In addition, the stability of AP-Prism in fetal bovine serum ()) and DNA-Prism in cell lysate ()) were investigated. Almost no detectable fluorescence increase was detected of Prism present into these solutions. The resistance of nuclease activity is mainly attributed to the biostability of AP-Prism achieved by the steric hindrance effect of the backbone [Citation31,Citation32]. Accordingly, the rational design of the AP-Prism allows for high specificity to APE1 with potential to response the intracellular AP endonuclease activity.
Figure 2. Exploring the speed and selective of DNA prism by ensemble fluorescence measurement. (a) Scheme of the fluorogenic AP-Prism. (b) The fluorescence response generated by the fluorogenic AP-Prism in the presence of APE1 at different concentrations. (c) Fluorescence response of the AP-Prism digested by APE1 and non-specific enzymes. (d) Percentage degradation of the AP-Prism by the nonspecific enzymes for 4 h.
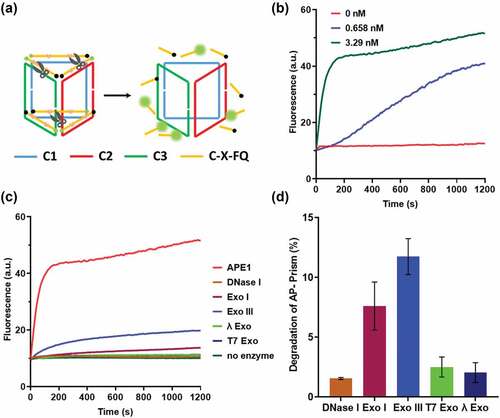
Figure 3. The stability assay of the DNA Prism over time in (a) 10% fetal bovine serum (FBS) and (b) HeLa cell lysate. Error bars represent standard deviations from three measurements.
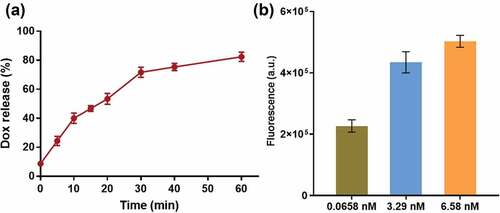
Anticancer drug doxorubicin hydrochloride (Dox) is commonly used to treat several kinds of cancer. It is also known to be fluorescent which has often been used to characterize concentrations, and has opened the possibility of using the molecule as a theranostic agent [Citation33,Citation34]. Further, we investigated the performance of our AP-Prism-Dox in response to APE1. The drug load and release behavior of the AP-Prism-Dox by measuring the fluorescence intensity of Dox in solution was studied. After remove excessive Dox with a cutoff filter, the fluorescence intensity in the solution was obviously decreased which suggesting successful drug load (Fig. S3A). The encapsulation efficiency of Dox was about 0.65 nM according to the calibration curve (Fig. S3B). In the present of APE1, the significant fluorescence increasing in the solution was detected after 1 h. The Dox release curve was shown in ). It is noteworthy that the fluorescence intensity in ) was taken at 10 min after introduced several series concentration of APE1 so that the AP-Prism-Dox with less APE1 exhibits relatively dim fluorescence. As the concentration of APE1 increases, the fluorescence of Dox in the solution increased ()). We speculated that the speed of drug release depends on catalytic hydrolysis, on the other hand the fluorescence products are also able to reflect the AP endonuclease activity.
Figure 4. (a) Dox release curve. The concentration of APE1 is 0.0658 nM. (b) Release behavior of AP-Prism-Dox after and 10 min in the presence of APE1 at different concentrations. Error bars represent standard deviations from three measurements.
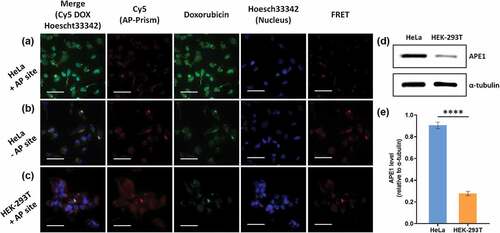
DNA nanostructure can be rapidly internalized in living cells through a caveolin-dependent pathway [Citation35]. Encouraged by the above demonstration of AP-Prism as a drug delivery carrier, we investigated the cellular uptake by HeLa cells. The DNA Prism was labeled with Cy5, incubated with HeLa cells and imaged by fluorescence microscopy. The cytoplasm of the HeLa cell exhibited bright fluorescence within 4 h (Fig. S4) suggested that the DNA Prism began to be internalized by cells and gradually accumulated in cells during the incubation. APE1 plays a central role in base excision repair (BER) pathway throughout the cell cycle [Citation36]. It has long been believed to be located in the nucleus, and cytoplasmic expressions were found in several types of cancer [Citation37]. Intracellular AP endonuclease was exploited as a potential biomarker of cancer cells because there is a significant difference of APE1 leave between cancer and normal cells [Citation4]. The kinetic analysis and drug release in vitro imply that the whole progress highly relies on the amount of APE1 which suggests a potential application in drug target therapy. Further, we detected Dox target release with a human cervical cancer cell HeLa and human embryonic kidney cell HEK-293 T. AP-Prism-Dox with Cy5 labeled prism backbone was incubated for 4 h. The blue fluorescence signal indicated the nucleus of cell, the red fluorescence signal denoted DNA Prism, and the green fluorescence signal showed Dox. As shown in , the red fluorescence was both observed in the cytoplasm of different cells suggested that the Prism was internalized by cells. The green signal could be observed both in cytoplasm and nucleus in HeLa, which suggested Dox could be delivered into cells by AP-Prism and subsequently released into nucleus ()). In contrast, no significant fluorescence of Dox in nucleus and weak fluorescence in cytoplasm was found in nucleus when using AP site free Prism-Dox ()) which indicated that the drug release caused by high accuracy of APE1 cleavage. We also compared the behavior of AP-Prism-Dox in cancer cells and normal cells. Weak green signal in HEK-293 T cell was observed, suggesting that drug can be trapped effectively by using this AP-Prism and released in cancer cells with high selectivity ()). In order to characterization the co-localization of carrier and drug, Dox served as a donor and Cy5 was an acceptor. FRET signal intensity implies the distance of DNA Prism and drug. Bright fluorescence of FRET can be observed when using AP site free Prism-Dox ()) or in HEK-293 T cell ()), while the fluorescence was dim in HeLa cell ()). To confirm the different APE1 levels, western blot analysis was carried out for the cytoplasmic fractions of the cells. APE1 expression of HeLa cells was significantly higher than that of HEK-293 T cells ( 5 (e)). Overall, we demonstrate that the AP-Prism was successfully used as a vehicle for controlled drug delivery and triggered release.
Figure 5. Uptake and release of AP-Prism-Dox in living cells. (a) HeLa cells. (b) HeLa cells by using DNA-Prism-Dox without AP site. (c) HEK-293T cells. Scale bar=50 μm. (d) Western blot analysis of cytoplasmic APE1 expression level of HeLa and HEK-293T cell. α-tubulin was used as loading control. (e) Relative quantification of cytoplasmic APE1 expression. Error bars represent standard deviations from three measurements. ****p < 0.0001.
4. Conclusions
In summary, we have successfully developed a new drug nanocarrier based on the intracellular endogenous enzyme APE1 triggered DNA prism. By virtue of the structure and function of DNA nanostructure, the AP-Prism showed high robustness and therapeutic efficacy with a drug loaded. Since APE1 is believed to be a potential biomarker for cancers, targeted drug release could be achieved without any external driving force. Therefore, this AP-Prism holds a promising platform to realize enzyme-mediated targeted chemotherapy.
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Supplemental Material
Download MS Word (1.5 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384101.2022.2108567
Additional information
Funding
References
- Tell G, Quadrifoglio F, Tiribelli C, et al. The many functions of APE1/Ref-1: not only a DNA repair enzyme. Antioxid Redox Signal. 2009;11:601–620.
- Vascotto C, Fantini D, Romanello M, et al. APE1/Ref-1 interacts with NPM1 within nucleoli and plays a role in the rRNA quality control process. Mol Cell Biol. 2009;29:1834–1854.
- Abbotts R, Madhusudan S. Human AP endonuclease 1 (APE1): from mechanistic insights to druggable target in cancer. Cancer Treat Rev. 2010;36:425–435.
- Li L, Li N, Fu S, et al. Base excision repair-inspired DNA motor powered by intracellular apurinic/apyrimidinic endonuclease. Nanoscale. 2019;11:1343–1350.
- Yu Y, Li L, Li G, et al. Intracellular enzyme-powered DNA circuit with a tunable amplifier for miRNA imaging. Chem Commun (Camb). 2021;57:3753–3756.
- Zhou T, Luo R, Li Y, et al. Activity assay and intracellular imaging of APE1 assisted with tetrahedral DNA nanostructure modified-dnazyme and molecular beacon. Sens Actuators B Chem. 2020;317:128203.
- Zhang Y, Deng Y, Wang C, et al. Probing and regulating the activity of cellular enzymes by using DNA tetrahedron nanostructures. Chem Sci. 2019;10:5959–5966.
- Tang S, Davoudi Z, Wang G, et al. Soft materials as biological and artificial membranes. Chem Soc Rev. 2021;50(22):12679–12701.
- Zheng C, Li M, Ding J. Challenges and opportunities of nanomedicines in clinical translation. BIO Integration. 2021;2(2):57–60.
- Hua C, Zhang Y, Liu Y. Enhanced anticancer efficacy of chemotherapy by amphiphilic Y-shaped polypeptide micelles. Front Bioeng Biotechnol. 2021;9. DOI:10.3389/fbioe.2021.817143
- Liao J, Peng H, Wei X, et al. A bio-responsive 6-mercaptopurine/doxorubicin based “Click Chemistry” polymeric prodrug for cancer therapy. Mater Sci Eng C. 2020;108:110461.
- Shi L, Zhang J, Zhao M, et al. Effects of polyethylene glycol on the surface of nanoparticles for targeted drug delivery. Nanoscale. 2021;13:10748–10764.
- Wei L, Chen J, Ding J. Sequentially stimuli-responsive anticancer nanomedicines. Nanomedicine. 2021;16:261–264.
- Liao J, Peng H, Liu C, et al. Dual pH-responsive-charge-reversal micelle platform for enhanced anticancer therapy. Mater Sci Eng C. 2021;118:111527.
- Hu R, Zheng H, Cao J, et al. Synthesis and in vitro characterization of carboxymethyl chitosan-CBA-doxorubicin conjugate nanoparticles as ph-sensitive drug delivery systems. J Biomed Nanotechnol. 2017;13:1097–1105.
- Wang J, Xu W, Zhang N, et al. X-ray-responsive polypeptide nanogel for concurrent chemoradiotherapy. J Control Release. 2021;332:1–9.
- Ma W, Chen Q, Xu W, et al. Self-targeting visualizable hyaluronate nanogel for synchronized intracellular release of doxorubicin and cisplatin in combating multidrug-resistant breast cancer. Nano Res. 2021;14:846–857.
- Feng X, Xu W, Xu X, et al. Cystine proportion regulates fate of polypeptide nanogel as nanocarrier for chemotherapeutics. Sci China Chem. 2021;64:293–301.
- Jiang Q, Shi Y, Zhang Q, et al. A self-assembled DNA origami-gold nanorod complex for cancer theranostics. Small. 2015;11:5134–5141.
- Fakhoury JJ, McLaughlin CK, Edwardson TW, et al. Development and characterization of gene silencing DNA cages. Biomacromolecules. 2014;15:276–282.
- Tay CY, Yuan L, Leong DT. Nature-inspired DNA nanosensor for real-time in situ detection of mRNA in living cells. ACS Nano. 2015;9:5609–5617.
- Xie N, Liu S, Yang X, et al. DNA tetrahedron nanostructures for biological applications: biosensors and drug delivery. Analyst. 2017;142:3322–3332.
- Meng HM, Liu H, Kuai H, et al. Aptamer-integrated DNA nanostructures for biosensing, bioimaging and cancer therapy. Chem Soc Rev. 2016;45:2583–2602.
- Liu Y, Chen Q, Liu J, et al. Design of a modular DNA triangular-prism sensor enabling ratiometric and multiplexed biomolecule detection on a single microbead. Anal Chem. 2017;89:3590–3596.
- Zheng X, Peng R, Jiang X, et al. Fluorescence resonance energy transfer-based DNA nanoprism with a split aptamer for adenosine triphosphate sensing in living cells. Anal Chem. 2017;89:10941–10947.
- Zhou L, Liu Y, Shi H, et al. Flexible assembly of an enzyme cascade on a DNA triangle prism nanostructure for the controlled biomimetic generation of nitric oxide. Chembiochem. 2018;19:2099–2106.
- Liu Q, Wang D, Xu Z, et al. Targeted delivery of Rab26 siRNA with precisely tailored DNA prism for lung cancer therapy. Chembiochem. 2019;20:1139–1144.
- Wu D, Li BL, Zhao Q, et al. Assembling defined DNA nanostructure with nitrogen-enriched carbon dots for theranostic cancer applications. Small. 2020;16:e1906975.
- Wang D, Xiang DB, Yang XQ, et al. APE1 overexpression is associated with cisplatin resistance in non-small cell lung cancer and targeted inhibition of APE1 enhances the activity of cisplatin in A549 cells. Lung Cancer. 2009;66:298–304.
- Al-Attar A, Gossage L, Fareed KR, et al. Human apurinic/apyrimidinic endonuclease (APE1) is a prognostic factor in ovarian, gastro-oesophageal and pancreatico-biliary cancers. Br J Cancer. 2010;102:704–709.
- Keum JW, Bermudez H. Enhanced resistance of DNA nanostructures to enzymatic digestion. Chem Comm. 2009;7036–7038. DOI:10.1039/b917661f
- Li N, Wang M, Gao X, et al. A DNA tetrahedron nanoprobe with controlled distance of dyes for multiple detection in living cells and in vivo. Anal Chem. 2017;89:6670–6677.
- Karukstis KK, Thompson EH, Whiles JA, et al. Deciphering the fluorescence signature of daunomycin and doxorubicin. Biophys Chem. 1998;73:249–263.
- Mohan P, Rapoport N. Doxorubicin as a molecular nanotheranostic agent: effect of doxorubicin encapsulation in micelles or nanoemulsions on the ultrasound-mediated intracellular delivery and nuclear trafficking. Mol Pharm. 2010;7:1959–1973.
- Liang L, Li J, Li Q, et al. Single-particle tracking and modulation of cell entry pathways of a tetrahedral DNA nanostructure in live cells. Angew Chem. 2014;53:7745–7750.
- Fortini P, Pascucci B, Parlanti E, et al. The base excision repair: mechanisms and its relevance for cancer susceptibility. Biochimie. 2003;85:1053–1071.
- Poletto M, Di Loreto C, Marasco D, et al. Acetylation on critical lysine residues of Apurinic/apyrimidinic endonuclease 1 (APE1) in triple negative breast cancers. Biochem Biophys Res Commun. 2012;424:34–39.