ABSTRACT
The aim is to explore the underlying mechanism of basic leucine zipper ATF-like transcription factor 2 (BATF2) in tongue squamous cell carcinoma (TSCC). The expression of BATF2 in TSCC tissues and corresponding adjacent normal TSCC tissues, human TSCC cell lines (SCC-15 and CAL-27) and human normal tongue epithelial cells NTEC was detected. Then, SCC-15 cells with stable BATF2 knockdown and CAL-27 cells with BATF2 overexpression were established to investigate the functional effect of BATF2 on TSCC. Thereafter, the effect of BATF2 on TSCC angiogenesis and BATF2 m6A methylation was also examined. BATF2 was significantly downregulated in TSCC tissues and cell lines, and BATF2 overexpression could suppress growth, metastasis and angiogenesis of TSCC. Mechanistically, vascular endothelial growth factor A (VEGFA) was identified as a downstream gene of BATF2, and it was confirmed that BATF2 suppressed growth, metastasis and angiogenesis of TSCC via inhibiting VEGFA. In addition, the N6-methyladenosine (m6A) modification of BATF2 mRNA mediated by METTL14 suppressed its expression in TSCC. METTL14/BATF2 axis could serve as a novel promising therapeutic candidate against angiogenesis for TSCC.
Introduction
Oral cavity and oropharynx cancers are the eighth most common human malignant neoplasm worldwide and exhibit poor prognosis [Citation1]. Tongue squamous cell carcinoma (TSCC) is classified as the most common type of oral cancer [Citation2], accounting for 25%–40% of all oral cancer cases worldwide [Citation3] and leading to approximately 2,830 deaths annually [Citation1]. In addition, TSCC is characterized by unlimited growth and high incidence, which may cause disorders of speech, chewing, and swallowing [Citation4,Citation5]. Currently, TSCC can be alleviated by therapeutic strategies mainly including radiotherapy, surgical resection, and chemotherapy [Citation6], whereas the prognosis of TSCC remains poor due to unpredictable long-term survival, distant metastasis and frequent recurrence [Citation7]. Moreover, the scarcity of understanding of the molecular mechanism underlying TSCC tumorigenesis is responsible for limited therapeutic development. Hence, novel biomarkers for predicting TSCC recurrence are urgently required to ameliorate its prognosis.
Enormous evidence has proven that the formation of new blood vessels significantly contributes to tumor growth and metastasis [Citation8], resulting from the capacity of blood vessels to deliver nutrients and oxygen to tumor cells, which is dispensable for tumor survival and metastasis [Citation9]. It is generally believed that the angiogenesis of tumor includes the following four processes: the dissolution of basement membrane under vascular endothelium and activation of endothelial cells, migration of endothelial cells to tumor tissues, proliferation of endothelial cells, and formation of blood vessels [Citation10,Citation11]. It has been demonstrated that vascular endothelial growth factor (VEGF) family proteins are crucial for angiogenesis, especially protein vascular endothelial growth factor A (VEGFA), which is the leading driving force for angiogenesis [Citation12,Citation13]. Recently, Chen S et al. found that AEG-1 promoted angiogenesis and might be a novel treatment target for tongue squamous cell carcinoma [Citation14], and Huang C et al. reported the association of increased ligand cyclophilin A and receptor CD147 with hypoxia, angiogenesis, metastasis and prognosis of TSCC [Citation15]. Therefore, inhibiting the formation of blood vessels provides a new direction for the treatment of TSCC; however, novel biomarkers involved in angiogenesis, which mediate cell proliferation and metastasis of TSCC, remain to be investigated.
Basic leucine zipper ATF-like transcription factor 2 (BATF2), also known as suppressor of AP-1 regulated by interferon (SARI), is a member of the BATF family [Citation16]. Initially, members of the BATF family were identified as inhibitors of AP-1 [Citation16], but recent reports revealed that they were involved in the cancer progression through diverse mechanisms [Citation17–20]. Zhang X et al. suggested that BATF2 prevented glioblastoma multiforme progression by inhibiting recruitment of myeloid-derived suppressor cells [Citation21]. In addition, mA modification-mediated BATF2 acted as a tumor suppressor in gastric cancer through inhibition of ERK signaling [Citation22]. Specifically, decreased expression of BATF2 was found in TSCC and is significantly associated with poor prognosis [Citation23]. However, whether and how BATF2 regulates TSCC progression remain elusive.
m6A (N6-methyladenosine) is the most prevalent post-transcriptional modification in eukaryotic mRNA, accounting for 80% of RNA methylation modifications [Citation24]. Mechanistically, m6A modification has been proven to affect mRNA stability, splicing, and translation [Citation25]. From the perspective of functional effect, previous studies have demonstrated that m6A methylation participates in regulating cancer progression [Citation24,Citation26,Citation27]. For example, IFN-α-2a reduces the stability of pgRNA by increasing its m6A RNA modification level, thereby inhibiting the development of HBV-related HCC [Citation28], and m6A could predict immune phenotypes and therapeutic opportunities in renal clear cell carcinoma [Citation29]. m6A methylases are widely involved in immunity [Citation30], tumor metastasis [Citation31] and stem cell renewal [Citation32]. Among them, methyltransferase-like 14 (METTL14) can promote the self-renewal of hematopoietic stem cells [Citation33], and regulate the level of m6A in glioblastoma [Citation34] and liver cancer [Citation35]. However, the mechanism by which METTL14 mediated the m6A modification of BATF2 that affects TSCC progression has not been reported.
In this study, to investigate the functional effect of BATF2 on TSCC progression and its underlying mechanism, we performed a variety of assays and found that BATF2 could inhibit the proliferation, metastasis and angiogenesis of TSCC cell lines in vitro and in vivo via VEGFA. Besides, METTL14 could mediate m6A methylation modification of BATF2, thereby inhibiting BATF2 expression.
Materials and methods
Tissue collection
TSCC tissues (n = 15) and corresponding adjacent normal tissues (n = 10) were obtained from the First People’s Hospital of Chenzhou (Affiliated Chenzhou Hospital, Southern Medical University). After that, tissue samples were immediately frozen in liquid nitrogen and stored at −80°C for further research. The research was performed in accordance with the guiding principles of the World Medical Association Declaration of Helsinki and was approved by the First People’s Hospital of Chenzhou (Affiliated Chenzhou Hospital, Southern Medical University) ethics committee. Signed informed consent was obtained from the patients.
Cell culture and transfection
Human TSCC cell lines SCC-15, CAL-27, human normal tongue epithelial cell line NTEC1 and human umbilical vein endothelial cells (HUVECs) were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). SCC-15 and CAL-27 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Thermo Fisher Scientific, USA) with 10% fetal bovine serum (FBS), 100 U/ml streptomycin and 100 U/ml penicillin. HUVECs were cultured routinely with endothelial cell growth Kit-VEGF including rh VEGF, rh EGF, rh FGF basic, rh IGF-1, L-glutamine, heparin sulfate, hydrocortisone hemisuccinate, 2% FBS and ascorbic acid.
The overexpression vectors of BATF2 (pcDNA-BATF2) and METTL14 (pcDNA-METTL14) as well as shRNAs against BATF2 and METTL14 were purchased and designed by GeneChem Corporation (Shanghai, China) (Supplementary Table S1). SCC-15 and CAL-27 cells were transfected with indicated plasmids according to the instructions of lipofectamine 2000 (Invitrogen, NY, CA, USA) and were incubated for 48 h for the subsequent experiments.
Cell counting kit-8 (CCK-8)
CCK-8 was used to assess the cell viability. Briefly, the SCC-15 and CAL-27 cells were seeded in a 96-well plate supplemented with 10% CCK-8 (Sigma, USA). Cell counting kit reagent (Beyotime Biotechnology Co., Ltd., Shanghai, China) was employed to examine the optical density (OD) value at 450 nm after incubation for 24, 48, 72 and 96 h.
Colony formation assay
At a density of 5 × 102 cells per well, SCC-15 and CAL-27 cells were incubated in 6-well plates for 2 weeks at 37°C with 5% CO2. Then, the number of colonies was counted and images were recorded after the formed colonies were fixed with 4% paraformaldehyde and stained with 1% crystal violet.
Methyl thiazolyl tetrazolium (MTT) assay
The proliferation of HUVECs was estimated with a MTT cell viability assay kit (Roche, Basel, Switzerland). Transfected HUVECs were plated into 96‐well plates and cultured at 37°C. At different times (24, 48, 72 and 96 h) post-incubation, each well was supplemented with 10 μL of MTT (5 mg/mL), followed by 4 h of incubation and then added with 100 μL of dimethyl sulfoxide (DMSO). Finally, absorbance was tested using a microplate reader at 570 nm.
Transwell assay
Transwell Chamber (Corning Glass Works, Corning, NY, USA) was used to evaluate the cell invasion ability. In brief, HUVECs and TSCC cells with a density of 4 × 104 mL −1 were added into the upper chambers coated with Matrigel (BD, Franklin Lakes, USA). Then, cells were mixed with DMEM containing 10% FBS in 24-well plates, followed by incubation for 24 h at 37°C with 5% CO2. After the removal of media, cells in the bottom chamber were fixed with 4% paraformaldehyde, stained with 0.1% crystal violet and counted in inverted fields under a microscope (Zeiss, Germany).
Wound scratch assay
SCC-15 and CAL-27 cells were cultured in a 12-well plate at 37°C with 5% CO2 for 24 h. Then, cells were incubated for another 48 h after being scratched using a sterile pipette tip. Next, images were captured using a microscope, and the migration percentage was evaluated using ImageJ software.
ELISA
ELISA was performed to evaluate the concentrations of VEGFA. Briefly, a VEGFA ELISA kit (Thermo Fisher Scientific, USA) was used according to the manufacturer’s guidelines and a microplate reader (BioTek Instruments, Inc., USA) was adopted to measure the absorbance at 450 nm.
Tube formation
SCC-15 or CAL-27 cells were incubated for 24 h after different treatments, and then the supernatants (also called conditioned medium) of culture media were collected for analysis of angiogenesis. HUVECs were treated with these supernatants for 24 h. The tube formation assay was performed using an In Vitro Angiogenesis Assay Kit (AmyJet Scientific Inc, Wuhan, China) in accordance with the manufacturer's instructions. The endothelial tube formation was examined using light microscopy (Leica, Allendale, NJ, USA).
Methylated RNA immunoprecipitation (MeRIP)
Total RNAs were first extracted from METTL4-overexpressed SCC-15 and CAL-27 cells. For BATF2 m6A immunoprecipitation, chemically fragmented RNA was incubated with m6A antibody in accordance with the manufacturer’s protocol of the Magna MeRIP m6A Kit (Merck Millipore, USA), and quantitative reverse-transcription polymerase chain reaction (qRT-PCR) was performed to analyze the immunoprecipitated RNA extracts.
Western blotting
The total protein in tissues and cells was extracted using lysis buffer (Beyotime, China) and quantified by a BCA protein assay kit (Beyotime, China). Then, the protein samples were separated using SDS-PAGE gels (Jinsirui, China) and transferred onto PVDF membranes (Merck Millipore, USA), which were cultured with 10% skim milk in PBS for 1 h, then mixed with the primary antibodies at 4°C overnight and washed with TBST (three times). These membranes were cultured with horseradish peroxidase (HRP)-labeled secondary antibody (mouse anti-rabbit IgG, #5127, 1:2000, Cell Signaling Technology, Inc., Boston, MA, USA). The Odyssey Infrared Imaging System (LI-COR Biosciences, USA) was used to observe blots, and the Image J software (National Institutes of Health, USA) was used to measure the protein expression level. The rabbit anti-human primary antibody for GAPDH (ab9485, 1:2500, Abcam, UK) was used to normalize the loading. The rabbit anti-human primary antibody BATF2 (PA5-37138, 1:500) was obtained from Thermo Fisher Scientific (USA), and VE-cadherin (ab33168, 1 µg/ml), E-cadherin (ab40772, 1:10,000), N-cadherin (ab76011, 1:5000), vimentin (ab92547, 1:1000), slug (ab27568, 1:1000), snail (ab216347, 1:1000), MMP2 (ab92536, 1:2000), TIMP-1 (ab211926, 1:1000), pRb (ab184796,1:1000), Cyclin D (ab16663, 1:200), VEGFA (ab214424, 1:1000) and METTL14 (ab252562, 1:1000) were purchased from Abcam (UK).
qRT-PCR
TRIzol reagent (Takara, Japan) was used to extract the total RNA from tissues and cells, and then RNA was reverse transcribed into cDNA by PrimeScript RT Reagent Kit (Takara, Japan). The mRNA levels were measured by Real-time PCR (RT-PCR) using the SYBR Premix Ex Taq Reagent Kit (Takara, Japan) on the StepOne RT-PCR System (Life Technologies, USA) in accordance with manufacturer’s instructions and were normalized to the GAPDH level. The formula 2−ΔΔCt method was used to calculate the relative expression of the mRNA: ΔΔCt = (Ct target gene − Ct reference gene) experimental group − (Ct target gene − Ct reference gene) control group. The primer sequences are listed in Supplementary Table S2.
Animal experiments
Animal assays were approved by the Ethics Committee of the First People’s Hospital of Chenzhou (Affiliated Chenzhou Hospital, Southern Medical University). BALB/c nude mice (aged 4–6 weeks) were fed under specific pathogen-free conditions at 26–28°C under 40–60% humidity. For tumorigenicity assay in vivo, CAL-27 cells transfected with BATF2 overexpression plasmids or empty plasmids or untreated CAL-27 cells were subcutaneously inoculated into the right armpit of mice. Following 4 weeks, mice were sacrificed by cervical dislocation under isoflurane anesthesia and neoplasms were isolated and weighted. The volume of neoplasms was monitored once a week. For the in vivo metastasis model, the tail vein of mice was intravenously injected with BATF2-overexpressed or control CAL-27 cells or untreated CAL-27 cells. After euthanasia of mice, lung tissues were collected and then histologically analyzed with hematoxylin and eosin (H&E) assay to count the number of lung metastatic nodules.
Immunohistochemistry (IHC) assay
After fixation in 4% paraformaldehyde, tumor specimens were embedded in paraffin, sliced into 5 μm‐thick sections, followed by deparaffinization, rehydration and antigen retrieval, and blocked with 3% BSA-PBS at room temperature. Then, IHC staining was conducted by using rabbit anti-human primary antibodies against BATF2 (ab204510, 1:500, Abcam), Ki67 (ab15580, 1 µg/ml, Abcam), CD31 (ab28364, 1:50, Abcam), and VEGFA (ab52917, 1:100, Abcam). Thereafter, sections were incubated with HRP-conjugated secondary antibody goat anti-rabbit IgG (ab6721, 1:1000, Abcam) for 1 h, treated with diaminobenzidine (Sigma, USA), counterstained with hematoxylin and observed under a light microscope.
Statistical analysis
Statistical analysis was performed using SPSS software (Version 20.0, Chicago, USA), GraphPad Prism Software (Version 7, USA). The differences between two or multiple groups were analyzed using Student’s t-test and one-way analysis of variance (ANOVA). Spearman correlation analysis was implemented to assess the relationship between BATF2 and METTL14. All experiments were repeated in triplicate. The data were represented as mean ± SD in our study. P < 0.05 indicated significant differences between groups.
Results
BATF2 is lowly expressed in TSCC
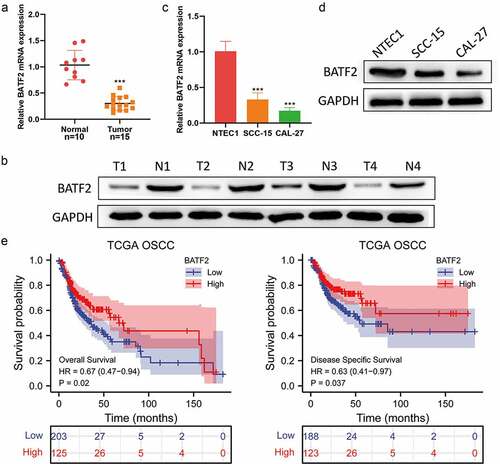
We first examined the expression level of BATF2 in TSCC tissues and corresponding adjacent normal tissues using western blotting and qRT-PCR. The expression level of BATF2 in TSCC tissues was significantly lower than that in adjacent normal tissues (). For further confirmation, the expression level of BATF2 was detected in TSCC cells (SCC-15 and CAL-27) and human normal tongue epithelial cells NTEC, and it was found that BATF2 was lowly expressed in SCC-15 and CAL-27 cells, compared with NTEC cells (). In the OSCC data set of TCGA, we found that low expression of BATF2 was correlated with poor survival (). Collectively, these data suggested that BATF2 was lowly expressed in TSCC tissues and cell lines.
Figure 1. Low BATF2 expression in TSCC tissues and cells.
BATF2 inhibits cell proliferation, migration, invasion and EMT in TSCC
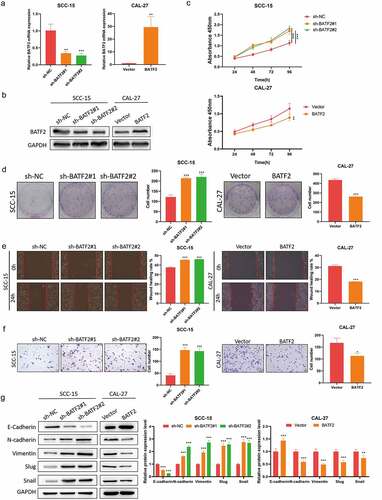
To evaluate the functional effect of BATF2 on TSCC, we established SCC-15 cells with stable BATF2 knockdown and CAL-27 cells with BATF2 overexpression (). Downregulation of BATF2 significantly promoted TSCC cell proliferation, while overexpression of BATF2 resulted in the inhibition of TSCC cell proliferation (). Similarly, wound scratch and transwell assays () demonstrated that BATF2 knockdown facilitated the migration and invasion of SCC-15 cells; conversely, BATF2 overexpression in CAL-27 cells elicited the opposite effects. Moreover, it was disclosed that silencing of BATF2 weakened E-cadherin expression and elevated the levels of N-cadherin, vimentin, slug and snail, while upregulation of BATF2 triggered the opposite results (). Taken together, these results indicated that BATF2 remarkably suppressed the proliferation, migration and invasion of TSCC cells.
Figure 2. Effect of BATF2 on TSCC cell proliferation, migration and invasion.
BATF2 suppresses TSCC angiogenesis
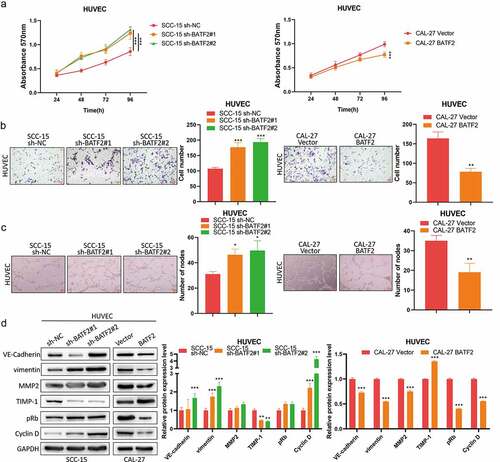
To further explore the effect of BATF2 on TSCC angiogenesis, HUVECs were treated with supernatants from BATF2-silenced SCC-15 cells and BATF2-overexpressed CAL-27 cells, respectively. MTT and transwell assays revealed that downregulation of BATF2 in SCC-15 cells remarkably promoted HUVEC proliferation and invasion, while overexpression of BATF2 in CAL-27 cells played a suppressive role in the proliferation and invasion of HUVECs (). Moreover, it was also found that knockdown of BATF2 in TSCC cells increased the number of nodes in HUVECs, whereas upregulated BATF2 in TSCC cells exerted an inhibitory effect on the tube formation of HUVECs. Specifically, western blotting assay () showed that BATF2 knockdown in TSCC cells caused the upregulation of VE-cadherin, vimentin, MMP2, pRb and cyclin D and enhanced expression of TIMP-1 in HUVECs. In contrast, upregulated BATF2 in CAL-27 cells induced reverse outcomes (). These results suggested that BATF2 suppressed the angiogenesis of TSCC.
Figure 3. Effect of BATF2 on TSCC angiogenesis.
BATF2 suppresses TSCC angiogenesis via downregulating VEGFA
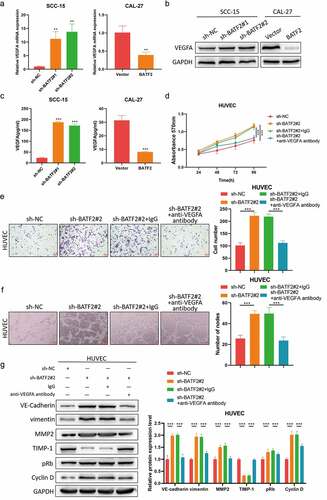
Considering VEGFA is the primary driving force for angiogenesis, to further investigate whether BATF2 mediated TSCC angiogenesis via VEGFA, SCC-15 cells were transfected with BATF2 shRNA plasmids, and CAL-27 cells were transfected with BATF2 overexpression plasmids. qRT-PCR western blotting and ELISA assays () indicated that VEGFA was highly expressed in SCC-15 cells with low BATF2 expression, but was downregulated in BATF2-overexpressed in CAL-27 cells, implying that BATF2 inhibited the expression of VEGFA in TSCC cells. Next, HUVECs were treated with supernatants derived from BATF2-silenced SCC-15 cells or underwent incubation with anti-VEGFA antibody. MTT and transwell assays () indicated that downregulation of BATF2 promoted HUVEC proliferation and invasion, which could be blocked by anti-VEGFA antibody. Furthermore, BATF2 knockdown facilitated the formation of tube nodes on HUVECs, while anti-VEGFA antibody restored the angiogenic capacity of HUVECs (). In addition, BATF2 knockdown resulted in increased expression levels of VE-cadherin, vimentin, MMP2, pRb and cyclin D, and reduced TIMP-1 expression, which could also be blocked by anti-VEGFA antibody (). Taken together, these data demonstrated that BATF2 inhibited TSCC angiogenesis via downregulating VEGFA.
Figure 4. VEGFA mediates the role of BATF2 in TSCC angiogenesis.
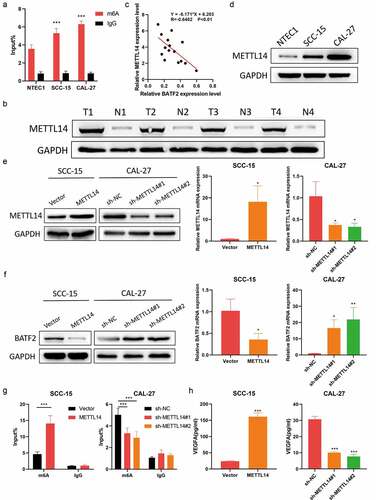
METTL14-Mediated m6a modification of BATF2 mRNA in TSCC
The mechanisms of aberrant BATF2 expression in TSCC remain unclear. Given the low mRNA level of BATF2 in TSCC, we investigated whether m6A modification regulated the mRNA stability of BATF2. MeRIP further confirmed BATF2 m6A methylation in NTEC1, SCC-15 and CAL-27 cells (). Subsequently, western blotting showed that METTL14 expression was higher in TSCC samples than that in normal tissues (). Moreover, correlation analysis revealed the negative association between METTL14 level and BATF2 expression (). Similarly, it was found that METTL14 was upregulated in SCC-15 and CAL-27 cells compared to NTEC1 cells (). After METTL14 was overexpressed in SCC-15 cells and METTL14 was silenced in CAL-27 cells (), it was observed that METTL14 overexpression caused reduced BATF2 expression at mRNA and protein levels, while METTL14 knockdown raised the mRNA and protein expression of BATF2 (). To prove that METTL4 targeted BATF2 mRNA for m6A modification, we carried out MeRIP assay. The results indicated that METTL14 overexpression strongly increased the enrichment of BATF2 in compounds precipitated by m6A antibody, while it was inhibited by depletion of METTL14 (). In addition, upregulated METTL14 caused an obvious rise of the VEGFA level, but it was decreased by METTL14 silencing (). These results demonstrated that m6A maintained the mRNA stability of BATF2 by METTL14 in TSCC.
Figure 5. METTL14 inhibits BATF2 expression in TSCC via m6a modification.
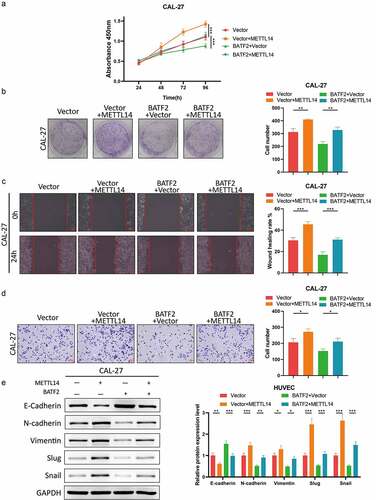
METTL14 induces the malignancy of TSCC via m6a modification of BATF2 mRNA
Based on the above findings, we explored the role of the METTL14/BATF2 axis in the malignant behaviors of TSCC cells. CCK-8 assay indicated that the viability of TSCC cells was increased by overexpression of METTL14 and then recovered by BATF2 upregulation (). Colony formation assay further validated that BATF2 mediated the promoting function of METTL14 in TSCC cell proliferation (). Additionally, wound healing and transwell assays demonstrated that forced expression of METTL14 facilitated cell migration and invasion in TSCC and upregulation of BATF2 led to the recovery of TSCC cell migration and invasion (). Consistently, western blotting revealed that upregulating the METTL14 reduced E-cadherin level and increasing the expression of N-cadherin, vimentin, slug and snail, and BATF2 abolished the role of METTL14 upregulation in the EMT process (). Collectively, METTL14 acted as a stimulative factor in TSCC progression by regulating BATF2.
Figure 6. METTL14 acts as an oncogene in TSCC through promoting m6a modification of BATF2 mRNA.
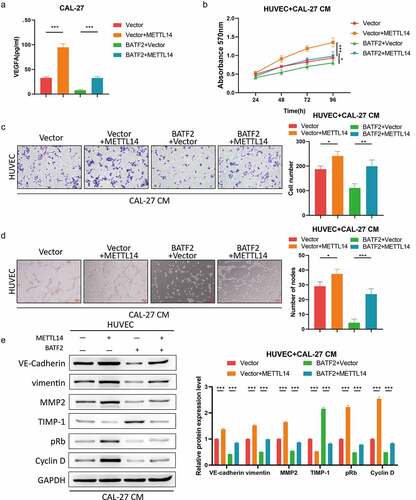
METTL14 contributes to TSCC angiogenesis through regulating BATF2
Subsequently, rescue assays were implemented to validate the potency of METTL14/BATF2 axis in TSCC angiogenesis. The ELISA assay showed that the expression of VEGFA was elevated by enhanced expression of METTL14 and the restoration of the VEGFA level occurred owing to upregulation of BATF2 (). As expected, MTT and transwell assays confirmed that co-culture of HUVECs and the culture medium of METTL14-overexpressed TSCC cells led to the promotion of HUVEC proliferation and invasion, while the proliferative and invasive capacity of HUVECs was relieved when BATF2 was also upregulated in TSCC cells (). In concert with these findings, tube formation experiment illustrated that HUVECs treated with the culture medium of TSCC cells transfected with pcDNA-METTL14 vector exhibited the heightened angiogenic ability, and overexpression of BATF2 in TSCC cells resulted in the inhibited tube formation of co-cultured HUVECs (). Moreover, upregulating METTL14 in TSCC cells increased the expression of VE-cadherin, vimentin, MMP2, pRb and cyclin D and decreased the TIMP-1 level in co-cultured HUVECs, and the role of METTL14 overexpression in these proteins was abrogated by forced expression of BATF2 in TSCC cells (). In a word, METTL14 facilitated angiogenesis in TSCC via suppressing BATF2 in an m6A-dependent manner.
Figure 7. METTL14 induced TSCC angiogenesis via modulation of BATF2.
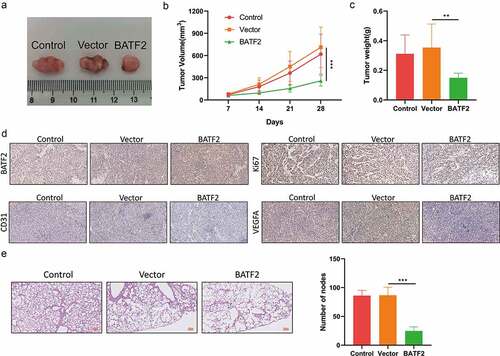
BATF2 restrains cell growth and metastasis of TSCC in vivo
Finally, we carried out animal experiments to certify the effect of BATF2 on tumor growth and metastasis in vivo. It was disclosed that overexpression of BATF2 diminished the size of neoplasms (). Likewise, our observations showed that the weight of neoplasms from vector group and control group were lower than that from BATF2 group (). Results of IHC assay illustrated that enhanced expression of BATF2 increased the BATF2 level and reduced the expression of Ki67, CD31 and VEGFA in tumor tissues (). Moreover, the number of pulmonary metastatic nodules in mice injected with BATF2-upregulated CAL-27 cells was fewer than that in mice from the vector group and control group (). On the whole, BATF2 inhibited in vivo tumor growth and metastasis of TSCC.
Figure 8. Effect of BATF2 on TSCC cell growth and metastasis in vivo.
Discussion
In this study, our results illustrated that BATF2 was lowly expressed in TSCC tissues and cell lines, and BATF2 overexpression suppressed cell proliferation, migration, invasion and EMT in TSCC, while BATF2 silencing exerted opposite effects. Moreover, BATF2 inhibited angiogenesis of TSCC via decreasing VEGFA, and the m6A modification of BATF2 mRNA was mediated by METTL14 in TSCC. More than that METTL14 acted as an oncogene in TSCC development via reducing the mRNA stability of BATF2. Furthermore, we validated the tumor suppressor role of BATF2 in TSCC cell growth and metastasis in vivo.
BATF2 gene is a newly discovered tumor-suppressor gene. Extensive studies have shown that BATF2 plays an important role in regulation of the cell cycle [Citation36], inflammatory response [Citation37], and epithelial-to-mesenchymal transition (EMT) [Citation38] in vitro. Consistent with previous reports, we found that BATF2 functionally suppressed cell proliferation, migration invasion and EMT of TSCC, thereby identifying BATF2 as a cancer cell inhibitor in TSCC. In vivo, BATF2 has been proven to exert inhibited effect on cancer cell growth and metastasis [Citation22,Citation39–41]. According to Zebing Liu et al., BATF2 is a negative regulator of hepatocyte growth factor (HGF)/mesenchymal-epithelial transition factor (MET) axis, and BATF2 overexpression dramatically blunted tumor xenograft growth [Citation42]. Except for cell and animal experiments, BATF2 has prognostic value clinically. According to Tianci Han et al., low expression of BATF2 was closely related to poor tumor differentiation and lymph node metastasis and predicted poor survival [Citation43]. BATF2 expression was also reported in TSCC. In our previous study, low expression of BATF2 was associated with poor tumor differentiation, and predicted reduced survival [Citation23]. In this study, the role of BATF2 in TSCC was clarified.
As referred above, VEGFA is an important regulator and involved in tumor angiogenesis [Citation44], and accumulating reports have demonstrated that VEGFA significantly contributes to tumor angiogenesis, which induces poor prognosis of different cancers, such as pancreatic ductal adenocarcinoma (PDAC) [Citation44], glioblastoma multiforme [Citation45], head and neck cancer (HNC) [Citation46] and osteosarcoma (OS) [Citation47]. Thus, to suppress tumor angiogenesis, it is crucial to downregulate the expression level of VEGFA. Herein, we found that BATF2 inhibited the expression level of VEGFA in TSCC cells, which could partially explain why BATF2 was negatively associated with TSCC angiogenesis. Besides, our results manifested that BATF2 inhibited tumor growth and metastasis of TSCC in vivo.
As the most prevalent modification in over 100 types of chemical modifications, m6A modification is involved in the regulation of mRNA and non-coding RNA [Citation48]. Generally, m6A modification mainly occurs on adenine of the RRACH sequence, and its biological function is primarily determined by m6A methylases, which includes writers (METTL3, METTL14 and WTAP), erasers (FTO, ALKBH5) and readers (YTHDFs) [Citation49]. Among them, METTL14 has been proven to be participating in cancer progression via mediating methylation levels of cancer-related proteins [Citation50–52]. For instance, METTL14 inhibited hepatocellular carcinoma metastasis through regulating eGFR/PI3K/AKT signaling pathway in an m6A-dependent manner [Citation53] and METTL14-mediated HNF3γ reduction rendered hepatocellular carcinoma dedifferentiation [Citation35]. However, whether METTL14 regulates m6A methylases modification in TSCC remains elusive. Herein, we found that METTL14 mediated the m6A modification of BATF2 mRNA and suppressed its expression in TSCC cells. More importantly, our findings confirmed that METTL14 promoted the malignancy and angiogenesis of TSCC via m6A modification of BATF2. Besides, animal assays validated the inhibitory effect of BATF2 on TSCC cell growth and metastasis in vivo.
In conclusion, our study demonstrated that BATF2 was downregulated in TSCC and suppressed TSCC cell growth, metastasis and angiogenesis via inhibiting VEGFA. Moreover, it was found that BATF2 mRNA was mediated by METTL14 in an m6A methylases modification-dependent manner. These findings provided the rationale for considering the METTL14/BATF2 axis as a novel therapeutic strategy against angiogenesis of TSCC.
Supplemental Material
Download MS Word (12.9 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data used to support the findings of this study are available from the corresponding author upon request.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384101.2022.2109897.
Additional information
Funding
References
- Siegel R, Miller K, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30.
- Zhang J, Wang L, Xie X. RFC4 promotes the progression and growth of Oral Tongue squamous cell carcinoma in vivo and vitro. J Clin Lab Anal. 2021;35(5):e23761.
- Irimie A, Ciocan C, Gulei D, et al. Current insights into oral cancer epigenetics. Int J Mol Sci. 2018;19(3):670. DOI:10.3390/ijms19030670
- Zeng S, Yang J, Zhao J, et al. Silencing Dicer expression enhances cellular proliferative and invasive capacities in human tongue squamous cell carcinoma. Oncol Rep. 2014;31(2):867–873. DOI:10.3892/or.2013.2903
- Yu Z, Zhong L, Ji T, et al. MicroRNas contribute to the chemoresistance of cisplatin in tongue squamous cell carcinoma lines. Oral Oncol. 2010;46(4):317–322. DOI:10.1016/j.oraloncology.2010.02.002
- Ren Z, Wu H, Zhang S, et al. A new surgical strategy for treatment of tongue squamous cell carcinoma based on anatomic study with preliminary clinical evaluation. J Craniomaxillofac Surg. 2015;43(8):1577–1582. DOI:10.1016/j.jcms.2015.07.034
- Blatt S, Krüger M, Ziebart T, et al. Biomarkers in diagnosis and therapy of oral squamous cell carcinoma: a review of the literature. J Craniomaxillofac Surg. 2017;45(5):722–730. DOI:10.1016/j.jcms.2017.01.033
- Saaristo A, Karpanen T, Alitalo K. Mechanisms of angiogenesis and their use in the inhibition of tumor growth and metastasis. Oncogene. 2000;19(53):6122–6129.
- Chen B, Jin H, Wu K. Potential role of vascular targeted therapy to combat against tumor. Expert Opin Drug Deliv. 2009;6(7):719–726.
- Watson EC, Grant ZL, Coultas L. Endothelial cell apoptosis in angiogenesis and vessel regression. Cell Mol Life Sci. 2017;74(24):4387–4403.
- Ingber D. Cancer as a disease of epithelial–mesenchymal interactions and extracellular matrix regulation. Differentiation. 2002;70(9–10):547–560.
- Ferrara N. The role of VEGF in the regulation of physiological and pathological angiogenesis. EXS. 2005;(94):209–231 doi:10.1007/3-7643-7311-3_15.
- Abhinand CS, Raju R, Soumya SJ, et al. VEGF-A/VEGFR2 signaling network in endothelial cells relevant to angiogenesis. J Cell Commun Signal. 2016;10(4):347–354. DOI:10.1007/s12079-016-0352-8
- Chen S, Chen L, Niu Y, et al. AEG-1 promotes angiogenesis and may be a novel treatment target for tongue squamous cell carcinoma. Oral Dis. 2020;26(5):876–884. DOI:10.1111/odi.13300
- Huang C, Sun Z, Sun Y, et al. Association of increased ligand cyclophilin a and receptor CD147 with hypoxia, angiogenesis, metastasis and prognosis of tongue squamous cell carcinoma. Histopathology. 2012;60(5):793–803. DOI:10.1111/j.1365-2559.2011.04130.x
- Su Z, Lee S, Emdad L, et al. Cloning and characterization of SARI (suppressor of AP-1, regulated by IFN). Proc Natl Acad Sci U S A. 2008;105(52):20906–20911. DOI:10.1073/pnas.0807975106
- Li P, Weng Z, Li P, et al. BATF3 promotes malignant phenotype of colorectal cancer through the S1PR1/p-STAT3/miR-155-3p/WDR82 axis. Cancer Gene Ther. 2021;28(5):400–412. DOI:10.1038/s41417-020-00223-2
- Lin Y, Cheng L, Liu Y, et al. Intestinal epithelium-derived BATF3 promotes colitis-associated colon cancer through facilitating CXCL5-mediated neutrophils recruitment. Mucosal Immunol. 2021;14(1):187–198. DOI:10.1038/s41385-020-0297-3
- Yang W, Zhao S, Wu B, et al. BATF2 inhibits chemotherapy resistance by suppressing AP-1 in vincristine-resistant gastric cancer cells. Cancer Chemother Pharmacol. 2019;84(6):1279–1288. DOI:10.1007/s00280-019-03958-4
- Kanemaru H, Yamane F, Fukushima K, et al. Antitumor effect of Batf2 through IL-12 p40 up-regulation in tumor-associated macrophages. Proc Natl Acad Sci U S A. 2017;114(35):E7331–e7340. DOI:10.1073/pnas.1708598114
- Zhang X, Liu Y, Dai L, et al. BATF2 prevents glioblastoma multiforme progression by inhibiting recruitment of myeloid-derived suppressor cells. Oncogene. 2021;40(8):1516–1530. DOI:10.1038/s41388-020-01627-y
- Xie JW, Huang XB, Chen QY, et al. m6A modification-mediated BATF2 acts as a tumor suppressor in gastric cancer through inhibition of ERK signaling. Mol Cancer. 2020;19(1):114. DOI:10.1186/s12943-020-01223-4
- Wen H, Chen Y, Hu Z, et al. Decreased expression of BATF2 is significantly associated with poor prognosis in oral tongue squamous cell carcinoma. Oncol Rep. 2014;31(1):169–174. DOI:10.3892/or.2013.2863
- Lan Q, Liu PY, Haase J. The critical role of RNA m6A methylation in cancer. Cancer Res. 2019;79(7):1285–1292.
- Yang Y, Hsu P, Chen Y, et al. Dynamic transcriptomic mA decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28(6):616–624. DOI:10.1038/s41422-018-0040-8
- Sun T, Wu R, Ming L. The role of m6A RNA methylation in cancer. Biomed Pharmacother. 2019;112:108613.
- Chen M, Nie ZY, Wen XH, et al. m6A RNA methylation regulators can contribute to malignant progression and impact the prognosis of bladder cancer. Biosci Rep. 2019;39(12). DOI:10.1042/BSR20192892
- Ding W, Wang M, Yu J, et al. HBV-pgRNA increases the stemness and promotes the development of HBV-related HCC through reciprocal regulation with IGF2BP3. Hepatology. 2021;74(3):1480–1495.
- Li H, Hu J, Yu A, et al. RNA modification of N6-methyladenosine predicts immune phenotypes and therapeutic opportunities in kidney renal clear cell carcinoma. Front Oncol. 2021;11:642159.
- Lu M, Xue M, Wang H-T, et al. Non-segmented negative-sense RNA viruses utilize N6-methyladenosine (m6A) as a common strategy to evade host innate immunity. J Virol. 2021;95(9):e01939–20.
- Deng R, Cheng Y, Ye S, et al. mA methyltransferase METTL3 suppresses colorectal cancer proliferation and migration through p38/ERK pathways. Onco Targets Ther. 2019;12:4391–4402.
- Cheng Y, Luo H, Izzo F, et al. mA RNA methylation maintains hematopoietic stem cell identity and symmetric commitment. Cell Rep. 2019;28(7):1703–1716.e1706. DOI:10.1016/j.celrep.2019.07.032
- Weng H, Huang H, Wu H, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA mA modification. Cell Stem Cell. 2018;22(2):191–205.e199. DOI:10.1016/j.stem.2017.11.016
- Cui Q, Shi H, Ye P, et al. mA RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18(11):2622–2634. DOI:10.1016/j.celrep.2017.02.059
- Zhou T, Li S, Xiang D, et al. m6A RNA methylation-mediated HNF3γ reduction renders hepatocellular carcinoma dedifferentiation and sorafenib resistance. Signal Transduct Target Ther. 2020;5:296.
- Kassambara A, Herviou L, Ovejero S, et al. RNA-Sequencing data-driven dissection of human plasma cell differentiation reveals new potential transcription regulators. Leukemia. 2021;35(5):1451–1462. DOI:10.1038/s41375-021-01234-0
- Li C, Liu M, Liu K, et al. BATF2 balances the T cell-mediated immune response of CADM with an anti-MDA5 autoantibody. Biochem Biophys Res Commun. 2021;551:155–160.
- Wang Q, Lu W, Yin T, et al. Calycosin suppresses TGF-β-induced epithelial-to-mesenchymal transition and migration by upregulating BATF2 to target PAI-1 via the Wnt and PI3K/Akt signaling pathways in colorectal cancer cells. J Exp Clin Cancer Res. 2019;38(1):240. DOI:10.1186/s13046-019-1243-7
- Huang W, Zhang C, Cui M, et al. Inhibition of bevacizumab-induced epithelial-mesenchymal transition by BATF2 overexpression involves the suppression of Wnt/β-catenin signaling in glioblastoma cells. Anticancer Res. 2017;37(8):4285–4294. DOI:10.21873/anticanres.11821
- Wang C, Su Y, Zhang L, et al. The function of SARI in modulating epithelial-mesenchymal transition and lung adenocarcinoma metastasis. PLoS One. 2012;7(9):e38046. DOI:10.1371/journal.pone.0038046
- Priyadarshini M, Maji S, Samal SK, et al. SARI inhibits growth and reduces survival of oral squamous cell carcinomas (OSCC) by inducing endoplasmic reticulum stress. Life Sci. 2021;287:120141.
- Liu Z, Wei P, Yang Y, et al. BATF2 deficiency promotes progression in human colorectal cancer via activation of HGF/MET signaling: a potential rationale for combining MET inhibitors with IFNs. Clin Cancer Res. 2015;21(7):1752–1763. DOI:10.1158/1078-0432.CCR-14-1564
- Han T, Wang Z, Yang Y, et al. The tumor-suppressive role of BATF2 in esophageal squamous cell carcinoma. Oncol Rep. 2015;34(3):1353–1360. DOI:10.3892/or.2015.4090
- Lao M, Zhang X, Ma T, et al. Regulator of calcineurin 1 gene isoform 4 in pancreatic ductal adenocarcinoma regulates the progression of tumor cells. Oncogene. 2021;40(17):3136–3151. DOI:10.1038/s41388-021-01763-z
- Zheng S, Tao W. Identification of novel transcriptome signature as a potential prognostic biomarker for anti-angiogenic therapy in glioblastoma multiforme. Cancers (Basel). 2021;13(5):1013.
- Dimitrakopoulos F, Koliou G, Kotoula V, et al. Vascular endothelial growth factor (VEGFAGenetic variation in the) gene at rs13207351 is associated with overall survival of patients with head and neck cancer. Cancers (Basel). 2021;13(5):1163. DOI:10.3390/cancers13051163
- Song Y, An W, Wang H, et al. LRH1 acts as an oncogenic driver in human osteosarcoma and pan-cancer. Front Cell Dev Biol. 2021;9:643522.
- Frye M, Jaffrey SR, Pan T, et al. RNA modifications: what have we learned and where are we headed? Nat Rev Genet. 2016;17(6):365–372. DOI:10.1038/nrg.2016.47
- B-B H, Wang X-y, Gu X-Y, et al. N 6-methyladenosine (m6A) RNA modification in gastrointestinal tract cancers: roles, mechanisms, and applications. Mol Cancer. 2019;18:1–8.
- Kong F, Liu X, Zhou Y, et al. Downregulation of METTL14 increases apoptosis and autophagy induced by cisplatin in pancreatic cancer cells. Int J Biochem Cell Biol. 2020;122:105731.
- Wang M, Liu J, Zhao Y, et al. Upregulation of METTL14 mediates the elevation of PERP mRNA N(6) adenosine methylation promoting the growth and metastasis of pancreatic cancer. Mol Cancer. 2020;19(1):130. DOI:10.1186/s12943-020-01249-8
- Yi D, Wang R, Shi X, et al. METTL14 promotes the migration and invasion of breast cancer cells by modulating N6‑methyladenosine and HSA‑miR‑146a‑5p expression. Oncol Rep. 2020;43(5):1375–1386. DOI:10.3892/or.2020.7515
- Shi Y, Zhuang Y, Zhang J, et al. METTL14 inhibits hepatocellular carcinoma metastasis through regulating EGFR/PI3K/AKT signaling pathway in an m6a-dependent manner. Cancer Manag Res. 2020;12:13173–13184.
