ABSTRACT
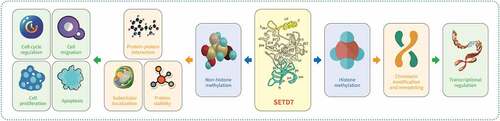
SET domain containing lysine methyltransferase 7 (SETD7) belongs to the protein lysine methyltransferase family and can catalyze the monomethylation of histone H3K4, which plays a vital role in the regulation of cell cycle, cell differentiation, DNA damage response and chromatin remodeling through K/R-S/T-K (K is lysine residue) sites and the recognition of substrates mediated by SET, i-SET, and n-SET domains and electrostatic action. SETD7 also can regulate the transcription of several genes including β-catenin, Cullin l and lin-28 homolog A (LIN28A), etc. In addition, the abnormal expression of SETD7 can promote the proliferation, migration, invasion of tumor cells, predict the poor prognosis of tumor patients, and may be a potential target for tumor therapy. This paper reviews the structure of SETD7, its role in tumor genesis and development, and the current research progress of relevant targeted drugs to explore its regulatory mechanism in tumor genesis and development and the prospect of targeted therapy.
KEYWORDS:
Regulation of post-translational chemical modifications of proteins plays an important role in the development of organisms and the process of disease. Among them, methyltransferase-mediated protein methylation is a crucial and essential way of chemical modification. Lysine methylation is a unique post-translational modification of proteins. It causes small changes in the size and electrostatic state of lysine residues, in turn, it regulates the fate and function of target proteins [Citation1]. Lysine methylation is catalyzed by protein lysine methyltransferases (PKMTs), which play a regulatory role in the formation of single-methylation, dimethylation, triple-methylation of the lysine ɛ group in a S-adenosyl methionine (SAM)-dependent manner [Citation2].
To date, more than 50 PKMTs have been discovered and divided into 8 subfamilies: KMT1~8. Among them, glucagon like peptide (GLP), Su(var)3–9, enhancer-of-zeste and trithorax (SET) domain bifurcated histone lysine methyltransferase 1 (SETDB1), SET domain protein containing 1A (SETD1A), nuclear receptor binding SET domain protein (NSD1), SET and MYND domain containing 2 (SMYD2), SET and MYND domain containing 3 (SMYD3), SET domain containing lysine methyltransferase 8 (SETD8) and SETD7 not only catalyze methylation of H3K9, H3K27, H3K4 and H4K20 histone substrates [Citation3], but also catalyze methylation of non-histone substrates such as p53, retinoblastoma-associated protein (Rb), vascular endothelial growth factor receptor (VEGFR), nuclear factor kappa B (NF-κB) and DNA methyltransferase 1 (DNMT1), which is closely related to the occurrence and development of cancer [Citation4].
SETD7 is similar to the most PKMTs, and it belongs to the family of protein lysine methyltransferases and contains the evolutionarily conserved SET domain, which is an important member of the SAM-dependent protein lysine methyltransferase family that includes nearly 50 different subtypes.
The initial research on SETD7 mainly focused on the areas of protein structure, cell cycle, tissue differentiation and development, etc. With the deepening development of tumor research, it has been found that SETD7 is abnormally expressed during the occurrence and development of tumors, suggesting that SETD7 plays a vital role in the course of tumors. This review will discuss the structure of SETD7, its role in tumor genesis and development, and the current research progress in the area of SETD7 inhibitors.
1 Structure and regulation mechanism of SETD7
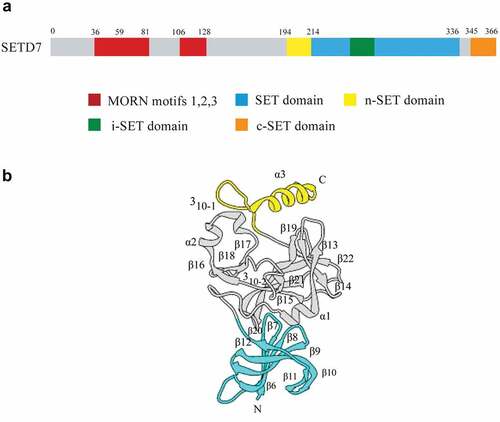
SETD7, also known as KMT7, SET7, SET7/9, SET9,is located on human chromosome 4q31.1, encoding protein contains 366 amino acids with a molecular weight of 40721 Da. SETD7 is composed of several β chains in two domains: the N-terminal domain and C-terminal domain (that is, the SET domain). The N-terminal domain is a non-conservative structure formed by a series of anti-parallel β chains, and the C-terminal domain is a discontinuous spherical structure composed of β chain, on both sides of which are a set of variable regions, namely, the front SET (n-SET), i-set and the back SET (c-SET) [Citation5–7]. Unlike other methyltransferase structures, the binding sites of the substrate and the cofactor SAM are located on opposite sides of the SET domain ().
Figure 1. Sequence and three-dimensional structure of SETD7. a) Sequence diagram of SETD7. The N-terminal of SETD7 is responsible for its stability, and it contains three MORN motifs, which are responsible for protein binding to plasma membrane phospholipids. The C-terminal is mainly responsible for its catalytic function, which contains a highly conserved SET domain. It is flanked by the n-SET domain and c-SET domain, and contains an insert region i-SET domain. b) Three-dimensional structure of SETD7. The N-terminal domain is colored cyan, the SET domain is gray and the C-terminal segment is yellow.
SETD7 is characterized by the catalysis of lysine methylation through an ordered double-substrate sequence mechanism, and then the substrate binds to form a ternary cofactor-enzyme-substrate complex. The initial step is SAM binding, and through a narrow hydrophobic channel, the substrate lysine binds to the cofactor at the core of the enzyme. This structure protects lysine from the effects of solvents and is essential for catalysis and methyl transfer from SAM to the lysine ε-group of the substrate [Citation8,Citation9].
Research on the structure of SETD7 shows that compared with the first methyl transfer of histone H3K4 (≈17–18 kcal/mol), the free energy barrier of the second methyl transfer is 5 kcal/mol higher. Therefore, it is currently believed that SETD7 mainly plays the role of monomethyltransferase. Recent studies have found that SETD7 used its N-terminal occupation and recognition connection (MORN) repeat sequence to dock with its substrate, and then juxtapose its lys methylation sequence, and the highly efficient and specific methylation was carried out by catalyzing the SET domain. This docking site-mediated methylation mechanism rationalizes the binding and methylation of previously known substrates, and be able to predict new SETD7 substrates [Citation10]。
Structure and function analysis suggests that SETD7 recognizes the substrate through the K/R-S/T-K (K is a lysine residue) site, and is mediated by SET, i-SET and n-SET domains and electrostatic interactions. Furthermore, it is involved in post-transcriptional modification of discontinuous sites through different and independent mechanisms, affecting the stability or biological activity of their substrates, and participating in a variety of cell biological processes, such as cell cycle, cell differentiation, DNA damage response and chromatin remodeling [Citation11,Citation12] ().
2 Targets of SETD7
2.1 Histone
It was initially discovered that SETD7 plays a transcriptional regulatory role as a methyltransferase of histone H3K4 [Citation11,Citation13]. SETD7-mediated H3K4 methylation plays a role in transcriptional activation by competing with histone deacetylases and blocking H3K9 methylation through Suv39h1 [Citation14]. SETD7 activates the expression of splicing regulatory glutamic acid and lysine rich protein 1 interacting protein 1 (SREK1IP1), coiled-coil domain containing 28B (CCDC28B) and progastricsin (PGC) through H3K4me1 transcription. Data from SETD7 and its target genes showed that the knockdown of SETD7 and SREK1IP1 in gastric cancer cells significantly promoted cell proliferation, migration and invasion [Citation15].
2.2 Non-histone substrates
An increasing body of evidence suggests that the modification of lysine methylation of non-histone substrates plays an important role in the genesis and development of tumors. In recent years, anti-tumor drugs targeting lysine methyltransferase have entered clinical research. For example, enhancer of zeste homolog 2 (EZH2) inhibitor EPZ-6438 for the treatment of INI1-negative tumor synovial sarcoma [Citation16], GSK126 for the treatment of relapsed or refractory diffuse B-cell lymphoma and disruptor of telomeric silencing 1-like (DOT1 L) inhibitor EPZ-5676 is used for the treatment of acute leukemia [Citation17,Citation18]. While the in-depth study of the molecular mechanism of the action of lysine methyltransferase plays a key role in elucidating the mechanism of action of such drugs, and will also provide new information and evidence for the search for new anti-tumor therapeutic drug targets.
SETD7 can regulate the function of non-histone substrates through methylation, which includes common non-histone substrates, such as p53, forkhead box O3 (FOXO3), Rb, centromere protein C 1 (CENPC1), methyl-CpG binding protein 2 (MeCP2) and Cullin1, etc. For example, SETD7 methylates β-catenin at K180, which reduces the stability of β-catenin, prevents the transcription of its downstream target genes and indirectly reduces cell proliferation. While the deletion of SETD7 significantly enhanced the expression of Wnt/β-catenin target genes such as c-myc and Cyclin D1, and promoted the growth of tumor cells [Citation19]. SETD7 methylates Cullin1 at K73 [Citation20]. The increase in Cullin1 expression is significantly related to the poor prognosis of breast cancer patients. Cullin1 may be an important marker and therapeutic target for breast cancer [Citation21]. In colon cancer, the overexpression of Cullin1 may occur in the early stage of carcinogenesis, mainly affecting p53(+) colorectal cells, blocking them in the G1 phase of the cell cycle and promoting tumorigenesis [Citation22]. In embryonic stem cells, LIN28A is monomethylated by SETD7 lysine methyltransferase at K135. Moreover, the methylation of LIN28A mediated by SETD7 at K135 increases the stability of LIN28A [Citation23]. Studies have shown that LIN28A promotes the proliferation and metastasis of colon cancer by inhibiting the biogenesis of let-7 family microribonucleic acid [Citation24,Citation25]. SETD7 methylates Smad7 at K70 [Citation26]. Recent studies have shown that Smad7 plays a dual function in the development of colorectal cancer, which may depend on cell subpopulations and the analyzed biological environment [Citation27]. SETD7 can methylate two highly conserved lysine (K) residues, K173 and K411. The methylation of yin and yang 1 protein (YY1) mediated by SETD7/9 has been shown to be closely related to gene transcription and cell proliferation regulated by YY1 [Citation28]. Therefore, it will be valuable to study whether YY1 methylation is abundant and therefore a biomarker of cancer. The role of YY1 in promoting or inhibiting tumor growth is still controversial, and its regulatory effect, at least in experimental systems suggests that it may depend on tumor cell types.
3 SETD7 and tumors
In recent years, studies have found that SETD7 is associated with the occurrence and development of breast cancer, liver cancer, stomach cancer, colorectal cancer and prostate cancer [Citation29–35].
3.1 Promote tumor effect
Through the analysis of the Gene Expression Omnibus (GEO) GSE9893 and GSE12276 database, it is found that the expression of SETD7 in breast tumor tissues is significantly higher, and it has a negative impact on the overall survival rate and local recurrence time of patients. Through Transwe11 analysis, the study also proved that SETD7 is involved in regulating the proliferation, invasion and metastasis of breast cancer cells by activating runt-related transcription factor 2 (RUNX2) [Citation29]. Other studies have shown that SETD7 guides the epigenetic plasticity of breast cancer cells and serves as an early biomarker for predicting metastasis [Citation33].
After immunohistochemical analysis of 68 hepatocellular carcinoma tissue samples, it was found that the expression of SETD7 and E2F transcription factor 1 (E2F1) was up-regulated, and compared with the levels of SETD7 and E2F1 in matched healthy liver samples, and the expression levels of SETD7 and E2F1 significantly correlated with pathological stage and tumor size [Citation30].
It has recently been discovered that insulin gene enhancer binding protein 1 (ISL1) can predict the poor prognosis of patients with gastric cancer. The complex of SETD7 and ISL1 can combine with the promoter region of zinc finger e-box binding homeobox 1 (ZEB1) to promote its expression, thereby promoting the metastasis of gastric cancer [Citation31]. The above results suggest that SETD7 may be involved in the occurrence and development of tumors as an oncogene.
Menuo’s studies of mice found that SETD7 modifies the Yes-associated protein (YAP) through methylation and then affects the accumulation of β-catenin in the nucleus. The results suggest that the SETD7-dependent regulatory mechanism plays a key role in the process of intestinal epithelial cell regeneration and tumorigenesis in mice [Citation32].
In the study of prostate cancer, it was found that modification of retinoid-related orphan receptor (ROR) α2 by SETD7 methylation increased the oncogenic potential of RORα2. The methylated RORα2 binds to the coactivator complex of pontin and tip60, enhances the function of target genes in the nucleus of transcriptional regulation, thereby promoting the progression and proliferation of prostate tumors [Citation34].
Hong’s group found that there was a negative correlation between the expression of SETD7 and right open reading frame kinase 1 (RIOK1) in colorectal cancer and gastric cancer tissues. SETD7 can affect the stability of RIOK1 through methylation modification and then participate in the progression of colorectal cancer and gastric cancer [Citation35].
3.2 Anti-tumor effect
Some scholars have reported that SETD7 can up-regulate the expression level of histone deacetylase 6 (HDAC6)-mediated acetylated -α-tubulin, inhibit the activity of the extracellular signal-regulated kinase (ERK) signaling pathway, and play a role in inhibiting the proliferation and migration of colon cancer cells [Citation36]. Similarly, it has been reported that SETD7 acts as a tumor inhibitor in lung cancer, and the low expression of SETD7 promotes the progression of lung cancer through the Janus kinase 2 (JAK2) signal transducer and activator of transcription 3 (STAT3) signaling pathway [Citation37].
4 SETD7 and drugs
The abnormal expression of SETD7 has been linked to a variety of diseases, including cancer, so SETD7 is considered as a target for the development of new epigenetic drugs. Few selective small molecule inhibitors have been reported to target SETD7, among them, (R)-PFI-2 is the most effective. (R)-PFI-2 is a histone competitive inhibitor, which has shown inhibitory ability in vitro and cell tests [Citation38]. X-ray complex structure analysis showed that (R)-PFI-2 can occupy the catalytic pathway of histone lysine binding, and (R)-PFI-2 binds to SETD7 only in the presence of SAM, the residues H252, D256, L267, Y335, G336, and H339 are responsible for the binding of SETD7 to (R)-PFI-2 [Citation39,Citation40]. The IC50 value of (R)-PFI-2 measured by mass spectrometry is 2 Nm [Citation40]. The study of (R)-PFI-2 provides new ideas for the design of novel inhibitors, such as (R)-PFI-2 analogs.
Studies have found that anti-allergic drug cyproheptadine can be used in the treatment of breast cancer. About 70% of breast cancers express estrogen receptor alpha (ERα), and the methylation of Erα by SETD7 contributes to the stabilization of ERα and activates its transcriptional activity, which is involved in the carcinogenesis of breast cancer [Citation41]. Using fluorescent substrate-based HMT assay, in RIKEN natural products depository (NPDepo), high-throughput screening identified the clinically approved anti-allergic drug cyproheptadine as a SETD7 inhibitor. The IC value by c analysis is 1 μm [Citation42]. Cyproheptadine may enter the catalytic center of SETD7 from the substrate-binding side and interact with the catalytic residue through a unique binding mechanism [Citation41]. This suggests that the regulation of SETD7 activity may be a new strategy for breast cancer treatment. However, pharmacological evidence on the adequacy of SETD7 as a therapeutic target remains limited.
In the latest study, biophysical and biochemical evidence show that compounds 2–79 can bind to SETD7 and inhibit it in vitro. Among the synthetic derivatives of compound 2–79, DC21 is considered to be an effective SETD7 inhibitor, half maximal inhibitory concentration (IC50) value is 15.93 μM. DC21 can be used as a lead compound to develop more potential SETD7 inhibitors and as a chemical probe for the study of the biological function of SETD7, which provides a new idea for the development of anti-tumor drugs [Citation43].
Summary and outlook
In summary, SETD7 plays the role of chemical modification and regulates multiple biological functions by regulating transcriptional activity. Meanwhile, SETD7 may promote or inhibit the occurrence and development of tumors to some extent through methylation of histone and non-histone targets, which may be related to tumor types and different research methods, and its mechanism of action needs to be further clarified in the future. Therefore, SETD7 is a potential target for tumor therapy. Although there are several SETD7 inhibitors, they have not been used in tumor therapy. Recently discovered compounds has relatively strong inhibition effect on SETD7, which may promote the development of novel regimens. Therefore, in-depth screening and study of its inhibitors are promising in the future.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Additional information
Funding
References
- Luo M. Chemical and biochemical perspectives of protein lysine methylation. Chem Rev. 2018;118(14):6656–6705.
- Hamamoto R, Saloura V, Nakamura Y. Critical roles of non-histone protein lysine methylation in human tumorigenesis. Nat Rev Cancer. 2015;15(2):110–124.
- Husmann D, Gozani O. Histone lysine methyltransferases in biology and disease. Nat Struct Mol Biol. 2019;26(10):880–889.
- Carlson SM, Gozani O. Nonhistone lysine methylation in the regulation of cancer pathways. Cold Spring Harb Perspect Med. 2016;6(11):a026435.
- Batista IAA, Helguero LA. Biological processes and signal transduction pathways regulated by the protein methyltransferase SETD7 and their significance in cancer. Signal Transduct Target Ther. 2018;3(1):19.
- Wilson JR, Jing C, Walker PA, et al. Crystal structure and functional analysis of the histone methyltransferase SET7/9. Cell. 2002;111(1):105–115.
- Qian C, Zhou MM. SET domain protein lysine methyltransferases: structure, specificity and catalysis. Cell Mol Life Sci. 2006;63(23):2755–2763.
- Xiao B, Jing C, Wilson JR, et al. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003;421(6923):652–656.
- Ibanez G, McBean JL, Astudillo YM, et al. An enzyme-coupled ultrasensitive luminescence assay for protein methyltransferases. Anal Biochem. 2010;401(2):203–210.
- Liu H, Li Z, Yang Q, et al. Substrate docking–mediated specific and efficient lysine methylation by the SET domain–containing histone methyltransferase SETD7. J Biol Chem. 2019;294(36):13355–13365.
- Keating ST, El-Osta A. Transcriptional regulation by the Set7 lysine methyltransferase. Epigenetics. 2013;8(4):361–372.
- Cornett EM, Ferry L, Defossez PA, et al. Lysine methylation regulators moonlighting outside the epigenome. Mol Cell. 2019;75(6):1092–1101.
- Wang H, Cao R, Xia L, et al. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell. 2001;8(6):1207–1217.
- Nishioka K, Chuikov S, Sarma K, et al. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 2002;16(4):479–489.
- Akiyama Y, Koda Y, Byeon SJ, et al. Reduced expression of SET7/9, a histone mono-methyltransferase, is associated with gastric cancer progression. Oncotarget. 2016;7(4):3966–3983.
- Knutson SK, Warholic NM, Wigle TJ, et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc Natl Acad Sci U S A. 2013;110(19):7922–7927.
- McCabe MT, Ott HM, Ganji G, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492(7427):108–112.
- Daigle SR, Olhava EJ, Therkelsen CA, et al. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood. 2013;122(6):1017–1025.
- Shen C, Wang D, Liu X, et al. SET7/9 regulates cancer cell proliferation by influencing β-catenin stability. Faseb J. 2015;29(10):4313–4323.
- Dhayalan A, Kudithipudi S, Rathert P, et al. Specificity analysis-based identification of new methylation targets of the SET7/9 protein lysine methyltransferase. Chem Biol. 2011;18(1):111–120.
- Zhou YH, Xia J, Xu WH, et al. Cullin-1 promotes cell proliferation in human breast cancer and is related to diabetes. Int J Biol Markers. 2016;31(4):e375–e381.
- Michail O, Moris D, Theocharis S, et al. Cullin-1 and -2 protein expression in colorectal cancer: correlation with clinicopathological variables. Vivo. 2018;32(2):391–396 .
- Kim SK, Lee H, Han K, et al. SET7/9 methylation of the pluripotency factor LIN28A is a nucleolar localization mechanism that blocks let-7 biogenesis in human ESCs. Cell Stem Cell. 2014;15(6):735–749.
- Balzeau J, Menezes MR, Cao S, et al. The LIN28/let-7 pathway in cancer. Front Genet. 2017;8:31.
- Wang T, He Y, Zhu Y, et al. Comparison of the expression and function of Lin28A and Lin28B in colon cancer. Oncotarget. 2016;7(48):79605–79616.
- Elkouris M, Kontaki H, Stavropoulos A, et al. SET9-Mediated regulation of TGF-β signaling links protein methylation to pulmonary fibrosis. Cell Rep. 2016;15(12):2733–2744.
- Troncone E, Monteleone G. Smad7 and colorectal carcinogenesis: a double-edged sword. Cancers (Basel). 2019;11(5):612.
- Zhang WJ, Wu XN, Shi TT, et al. Regulation of transcription factor Yin Yang 1 by SET7/9-mediated lysine methylation. Sci Rep. 2016;6(1):21718.
- Si W, Zhou J, Zhao Y, et al. SET7/9 promotes multiple malignant processes in breast cancer development via RUNX2 activation and is negatively regulated by TRIM21. Cell Death Dis. 2020;11(2):151.
- Gu Y, Wang X, Liu H, et al. SET7/9 promotes hepatocellular carcinoma progression through regulation of E2F1. Oncol Rep. 2018;40(4):1863–1874.
- Guo T, Wen XZ, Li ZY, et al. ISL1 predicts poor outcomes for patients with gastric cancer and drives tumor progression through binding to the ZEB1 promoter together with SETD7. Cell Death Dis. 2019;10(2):33.
- Oudhoff MJ, Braam MJS, Freeman SA, et al. SETD7 controls intestinal regeneration and tumorigenesis by regulating Wnt/β-Catenin and Hippo/YAP signaling. Dev Cell. 2016;37(1):47–57.
- Montenegro MF, Sanchez-Del-Campo L, Gonzalez-Guerrero R, et al. Tumor suppressor SET9 guides the epigenetic plasticity of breast cancer cells and serves as an early-stage biomarker for predicting metastasis. Oncogene. 2016;35(47):6143–6152.
- Song H, Chu JW, Park SC, et al. Isoform-specific lysine methylation of RORalpha2 by SETD7 is required for association of the TIP60 coactivator complex in Prostate cancer progression. Int J Mol Sci. 2020;21(5):1622.
- Hong X, Huang H, Qiu X, et al. Targeting posttranslational modifications of RIOK1 inhibits the progression of colorectal and gastric cancers. Elife. 2018;7. DOI:10.7554/eLife.29511
- Zhang SL, Du X, Tan LN, et al. SET7 interacts with HDAC6 and suppresses the development of colon cancer through inactivation of HDAC6. Am J Transl Res. 2020;12(2):602–611.
- Cao L, Ren Y, Guo X, et al. Downregulation of SETD7 promotes migration and invasion of lung cancer cells via JAK2/STAT3 pathway. Int J Mol Med. 2020;45(5):1616–1626.
- Lenstra DC, Damen E, Leenders RGG, et al. Structure-activity relationship studies on (R)-PFI-2 analogues as inhibitors of histone lysine methyltransferase SETD7. ChemMedchem. 2018;13(14):1405–1413.
- Niu Y, Shi D, Li L, et al. Revealing inhibition difference between PFI-2 enantiomers against SETD7 by molecular dynamics simulations, binding free energy calculations and unbinding pathway analysis. Sci Rep. 2017;7(1):46547.
- Barsyte-Lovejoy D, Li F, Oudhoff MJ, et al. (R)-PFI-2 is a potent and selective inhibitor of SETD7 methyltransferase activity in cells. Proc Natl Acad Sci U S A. 2014;111(35):12853–12858.
- Takemoto Y, Ito A, Niwa H, et al. Identification of cyproheptadine as an inhibitor of SET domain containing lysine methyltransferase 7/9 (Set7/9) that regulates Estrogen-dependent transcription. J Med Chem. 2016;59(8):3650–3660.
- Gauthier N, Caron M, Pedro L, et al. Development of homogeneous nonradioactive methyltransferase and demethylase assays targeting histone H3 lysine 4. J Biomol Screen. 2012;17(1):49–58.
- Hou Z, Min W, Zhang R, et al. Lead discovery, chemical optimization, and biological evaluation studies of novel histone methyltransferase SET7 small-molecule inhibitors. Bioorg Med Chem Lett. 2020;30(9):127061.