ABSTRACT
Severe hydronephrosis increases the risk of urinary tract infection and irretrievable renal fibrosis. While TGFβ1-mediated fibrotic changes in proximal tubular epithelial cells and fatty acid oxidation (FAO) deregulation contribute to renal fibrosis and hydronephrosis. Firstly, a few elements were analyzed in this paper, including differentially-expressed long non-coding RNAs (lncRNAs), and miRNAs correlated to CPT1A, RXRA, and NCOA1. This paper investigated TGFβ1 effects on lncRNA FABP5P3, CPT1A, RXRA, and NCOA1 expression and fibrotic changes in HK-2 cells and FABP5P3 overexpression effects on TGFβ1-induced changes. Moreover, this paper predicted and proved that miR-22 binding to lncRNA FABP5P3, 3’UTR of CPT1A, RXRA, and NCOA1 was validated. The dynamic effects of the FABP5P3/miR-22 axis on TGFβ1-induced changes were investigated. A Renal fibrosis model was established in unilateral ureteral obstruction (UUO) mice, and FABP5P3 effects were investigated. Eventually, this paper concluded that TGFβ1 inhibited lncRNA FABP5P3, CPT1A, RXRA, and NCOA1 expression, induced fibrotic changes in HK-2 cells, and induced metabolic reprogramming within HK-2 cells, especially lower FAO. FABP5P3 overexpression partially reversed TGFβ1-induced changes. miR-22 targeted lncRNA FABP5P3, CPT1A, RXRA, and NCOA1. LncRNA FABP5P3 counteracted miR-22 inhibition of CPT1A, NCOA1, and RXRA through competitive binding. TGFβ1 stimulation induced the activation of TGFβ/SMAD and JAG/Notch signaling pathways; Nocth2 knockdown reversed TGFβ1 suppression on lncRNA FABP5P3. FABP5P3 overexpression attenuated renal fibrosis in unilateral ureteral obstruction mice. The LncRNA FABP5P3/miR-22 axis might be a potent target for improving the FAO deregulation and fibrotic changes in proximal TECs under TGFβ1 stimulation.
Introduction
Severe hydronephrosis can increase the risk of urinary tract infection, leading to kidney infection (pyelonephritis). Severe pyelonephritis, if left untreated, can lead to permanent kidney damage (scarring), high blood pressure, and finally, kidney failure [Citation1–3]. Based on previous reports, hydronephrosis is not only related to injury but also related to renal fibrosis [Citation4,Citation5]. Fibrosis is often thought to be a long-term manifestation of tissue damage, and as the disease progresses, kidney function gradually deteriorates to kidney failure [Citation6–8]. What’s worse, it has been reported that even when hydronephrosis is relieved by surgery, the fibrotic lesion cannot be recovered, as the renal fibrosis caused by the obstruction continues to develop [Citation9]. Therefore, it is necessary to treat renal fibrosis even after the hydronephrosis has been treated. However, the lack of knowledge of the mechanism of renal fibrosis has hindered the progress of its treatment.
Renal fibrosis is a major event in the progression of various chronic kidney diseases (CKD) [Citation10]. Elevated expression of genes encoding ECM (extracellular matrix proteins), reduced matrix degradation, cell-matrix interaction dysregulation, infiltration of inflammatory cells, and resident cell transformation are the major characteristic features of renal fibrosis [Citation11,Citation12]. In fibrosis, the loss of proximal TECs (tubular epithelial cells) is attributed to cell death. De-differentiation of the remaining cells, a critical characteristic of CKD, could inhibit characteristic epithelial marker expression while enhancing mesenchymal marker expression. The increase in tubular epithelial Notch signaling pathway may cause tubular epithelial cell de-differentiation [Citation13]. Besides, transforming growth factor-beta (TGFβ1) is recognized as a vital mediator in renal fibrosis, which can induce fibrillary collagen secretion and enhance cell death and de-differentiation [Citation14]. Thus, these two signaling pathways have been regarded as pro-fibrotic signaling pathways in renal fibrosis progression, which might play a role in severe hydronephrosis.
Changes in cell metabolism, such as alterations in fuel source preferences (glucose, fatty acids, or ketones), have become a critical cell differentiation mechanism, particularly under the circumstances of stem cells and carcinogenesis [Citation15]. Proximal TECs have large baseline energy consumption and abundant mitochondrial supply. Compared to glucose oxidation, FAO (fatty acid oxidation) produces more ATP, making it the prime energy source for hypermetabolic cells. Long-chain fatty acid transporters promote the uptake of long-chain fatty acids [Citation16], and fatty acid metabolism needs transport to mitochondria, mediated by CPT1 (carnitine palmitoyl-transferase 1), which binds fatty acids to carnitine [Citation17]. Thus, CPT1 is regarded as the rate-limiting enzyme for FAO. PPAR (peroxisome proliferator-activated receptors) and PPARGC1A (PPARγ coactivator-1a) are considered critical for regulating protein levels during the uptake and oxidation of fatty acids. Retinoid X receptor alpha (RXRA)/PPARA heterodimer is indispensable for the transcriptional activity of PPARA. In the meantime, the nuclear receptor coactivator 1 (NCOA1) regulates lipid metabolism as a transcriptional coactivator [Citation18–22]. In general, the uptake, oxidation, and synthesis of fatty acids achieve a tight balance to prevent intracellular lipid deposition; however, upon injury, FAO is impaired, metabolic reprogramming occurs, essential enzymes’ levels and regulatory factors are reduced, and intracellular lipid accumulation is increased, finally leading to the progression of renal fibrosis and CKD.
MicroRNAs (miRNAs) are a class of small, endogenous RNAs. They play an important regulatory role in gene expression by targeting specific mRNAs for degradation or translation repression, therefore greatly influencing the regulation of multiple biological processes [Citation23]. In addition, miRNAs have been implicated in the regulation of renal cancer, diabetic nephropathy, obstructive nephropathy, acute kidney injury, and some other kidney diseases [Citation24]. As we have mentioned, the expression of key enzymes and regulatory factors are reduced during the metabolic reprogramming in renal fibrosis, suggesting that there may be epigenetic mechanisms regulating these factors at the post-transcriptional level. Another series of crucial non-coding RNAs, namely long non-coding RNAs, are known for serving as ceRNAs (competing endogenous RNAs) for miRNAs to regulate miRNAs with each other dually [Citation25]. Thus, we hypothesize that lncRNA-miRNA interaction might play a role in regulating FAO during renal fibrosis, possibly through regulating the translation of related key factors and signaling pathways.
In the present study, we first downloaded an online microarray profile (GSE20247), analyzed the differentially-expressed genes (DEGs) in proximal TECs in response to TGFβ1 stimulation, and constructed a CPT1A gene co-expression network. Data reported in GSE20247 [Citation26] appeared a positive correlation between CPT1A and RXRA and NCOA1. Moreover, we also investigated differentially-expressed lncRNAs in TGFβ1-induced HK-2 cells and selected lncRNAs positively correlated to CPT1A and examined their expression within HK-2 cells upon TGFβ1 treatment. Next, fibrotic changes and metabolic reprogramming indexes were examined within HK-2 cells under TGFβ1 stimulation; the impacts of lncRNA FABP5P3 upon fibrotic changes and FAO showed to be evaluated within HK-2 cells under TGFβ1 stimulation. With online tools, we predicted miRNAs that could bind to lncRNA FABP5P3 and CPT1A, RXRA, and NCOA1; miR-22 was selected, and the predicted bindings were verified. The dynamic effects of lncRNA FABP5P3 and miR-22 were examined. In addition, the expressions of lncRNA FABP5P3, miR-22, and CPT1A, RXRA, and NCOA1 in clinical renal fibrosis tissues were examined, which validated the dynamic effects further. Finally, the involvement of TGFβ/SMAD and JAG/Notch signaling pathways was examined. In summary, we attempted to provide a solid experimental basis for understanding the mechanism of FAO deregulation in proximal TECs during renal fibrosis by non-coding RNA regulation.
Materials and methods
Human renal tissue samples
The calyceal tissues from renal parenchymal atrophy were obtained from patients with chronic kidney disease with unilateral severe obstructive hydronephrosis and renal fibrosis who had received the nephrectomy in the Third Xiangya Hospital of Central South University. The normal calyceal tissues were obtained from regions distant from tumor foci from fifteen patients diagnosed with clear cell renal cell carcinomas. All procedures were approved by the Ethics Committee of the Third Xiangya Hospital of Central South University (approval number: 2019-S291). All operations were performed with patient consent.
Cell line, cell treatment, and cell transfection
HK-2 (ATCC® CRL-2190™) cell line was obtained from ATCC and cultured in keratinocyte serum-free medium +5 ng/ml epidermal growth factor and 50 mg/ml bovine pituitary extract and antibiotics (100 U/ml of penicillin G, 100 μg/ml of streptomycin, and 0.25 μg/ml of amphotericin B) at 37°C in a 5% CO2 humidified atmosphere.
At 80% confluence, cells were treated with 50 ng/ml TGFβ1 for 48 h. The in vitro renal fibrosis model in HK-2 cells was identified by light microscope and IF staining for Fn protein.
The knockdown of lncRNA FABP5P3 or Notch2 was achieved by transfection of si-FABP5P3 or si-Notch2 (Genepharma, Shanghai, China). The overexpression of FABP5P3 was achieved by the FABP5P3-overexpressing vector (Genepharma) using Lipofectamine 3000 (Invitrogen).
RNA extraction and real-time PCR
Total RNA was extracted using TRIzol reagent (Invitrogen, Waltham, MA, USA), and cDNA was synthesized using a high-capacity cDNA reverse transcriptase kit (Thermo Fisher Scientific, Waltham, MA, USA) as previously described [Citation27]. SYBR Green quantitative PCR reagent (Beijing Transgen Biotech, Beijing, China) was used to detect relative RNA expression. The 2−ΔΔCt method [Citation28] was used to analyze gene expression.
Immunoblotting
The protein levels of CPT1A, NCOA1, RXRA, Col1A1, Bcl-2, CASP3, SMAD3, p-SMAD3, JAG1, Notch2, ICN2 (intracellular fragment of Notch2), and Fn were determined by Immunoblotting taking β-actin (plasma) or histone (nucleus) as endogenous control. All the antibodies were obtained from Abcam (Cambridge, MA, USA) unless otherwise stated. Proteins extracted from cells were separated by SDS-PAGE, transferred to a polyvinylidene difluoride membrane (Millipore, Burlington, MA, USA), and then incubated with the primary antibody. They were then incubated with anti-rabbit or anti-mouse IgG conjugated to horseradish peroxidase (Abcam, Cambridge, MA, USA). The primary antibodies used are as follows: anti-CPT1A (ab128568), anti-NCOA1 (SRC1, ab5407), anti-RXRA (ab125001), anti-Col1A1 (ab34710), anti-Bcl2 (ab32124), anti-cleaved CASP3 (ab2302), anti-SMAD3 (ab40854), anti-p-SMAD3 (ab52903), anti-JAG1 (ab7771), anti-Notch2 (#5732, CST, MA, USA), anti-Fibronectin (Fn; ab2413), anti-β-actin (ab8226), anti-histone (ab213257).
Oxygen consumption rate (OCR) determination
To determine the OCR changes, an XFe24 Analyzer (Seahorse-Bioscience, Santa Clara, CA, USA) was employed. Briefly, cells were seeded in an XF24 V7 cell culture microplate at 1.0 × 104 cells per well under the various conditions designated in the experiments. The rate change of dissolved O2 in the medium was measured as OCR (pmol/min). The baseline OCR was measured at first, followed by the addition of palmitate (180 μM). The final state was determined after the addition of oligomycin (ATP synthase inhibitor, 1 μM) to check the minimal OCR without oxidative phosphorylation.
ATP measurement
ATP concentration in the cells was measured using the ATP colorimetric assay kit (K354; Biovision, Milpitas, CA, USA). Briefly, cells were lysed in assay buffer, followed by the addition of deproteinizing buffer. Samples were mixed with ATP probe, converter, and developed and incubated at 25℃ for 30 min. Absorbance (OD570 nm) was measured and calculated based on the ATP standard curve.
Fn protein determined by Immunofluorescence (IF) analysis
Fn protein content and distribution were analyzed by performing IF staining using anti-Fn (ab2413, Abcam). The FITC-labeled Secondary antibody was also obtained from Abcam. The nucleus was stained with DAPI. The green fluorescence represents Fn protein, and the blue fluorescence represents the nucleus.
Unilateral ureteral obstruction (UUO) mice models and intrarenal lentivirus delivery
Male C57BL/6J mice (8–10 weeks old, 20–25 g) were purchased from SLAC Laboratory Animal Co., Ltd. (Changsha, China). UUO mice were established as described below. After anesthetized with sodium pentobarbital, the left ureter of one group was exposed and ligated. Meanwhile, the control group was operated in the same procedure, except for the ligation. On day 14 of experiment, the mice were euthanized for the renal tissue harvest. As for the lentivirus delivery, mice were administered with a recombinant lentivirus harboring FABP5P3 overexpression (Genechem, Shanghai, China). Two days before UUO surgery, 10 μl of FABP5P3 overexpression or vector lentivirus (1 × 105 IU/μl) was injected into the renal cortex of experimental mice. All animal experiments were conducted in accordance with the Guidance for Care and Use of Laboratory Animals of Central South University and approved by the Ethics Committee of the Third Xiangya Hospital of Central South University.
H&E and Masson trichrome staining
To pathologically confirm the diagnosis of renal fibrosis, the collected human and mice renal tissues were fixed in 10% formalin, embedded in paraffin, sectioned consecutively at 5 µm, and stained by hematoxylin and eosin by two independent clinical pathologists in a double-blinded manner. For Masson Trichrome Staining, mice renal tissues were dehydrated in a series of increasing concentrations of ethanol, embedded in paraffin after having been fixed in 4% buffered paraformaldehyde, and cut into 5 µm slices. The slices were rehydrated with 100, 95, 70, and 50% alcohol and stained with Masson trichrome reagent for 15 min, followed by counterstaining with hematoxylin.
Immunohistochemical staining
Paraffin-embedded renal tissue sections were deparaffinized. After treatment with 3% H2O2 in methanol, the sections were hydrated with gradient alcohol and PBS, incubated in 10 mM citrate buffer, and finally heated at 100°C for 20 min in PBS. Slides were incubated with the anti-CPT1A (ab128568), anti-NCOA1 (SRC1, ab5407), and anti-RXRA (ab125001), respectively, for 20 min at room temperature, followed by incubation with horseradish peroxidase/Fab polymer conjugate for 30 min. Then, slides were thoroughly washed with PBS, and the sites of peroxidase activity were visualized using 3, 3-diamino-benzidine tetrahydrochloride. Afterward, the sections were counterstained with hematoxylin. Stainings were observed with an Olympus microscope.
Statistical analysis
Data were processed using GraphPad software and are presented as the mean standard deviation of results from at least three independent experiments. The student’s t-test was used for statistical comparison between means where applicable. Differences among more than two groups in the above assays were estimated using one-way ANOVA. *P < 0.05; **P < 0.01.
Results
Differentially-expressed genes in response to TGFβ1 stimulation in HK-2 cells and clinical renal fibrosis tissues
To figure out the critical genes of FAO which could be affected by TGFβ1 in proximal tubular cells, we downloaded and analyzed online microarray profiles (GSE20247) reporting DEGs in HK-2 cells under TGFβ1 stimulation [Citation29]. Of all the DEGs, 144 genes showed to be dramatically increased (fold change >4, p < 0.001), and 168 genes were decreased (fold change <0.25, p < 0.001); CPT1A, a critical rate-limiting enzyme of FAO, was included in the remarkably decreased genes (fold change = 0.42, p < 0.05). FAO requires fatty acids to translocate into mitochondria, a rate-limited step regulated by CPT1 during the long-chain fatty acid oxidation process [Citation30]. To find out how the FAO pathway is impaired during the kidney injury process, we established a co-expression network of CPT1A with FAO-related genes using Pearson’s correlation analysis based on the raw data of GSE20247. As shown in , in TGFβ1-induced proximal tubular cells, CPT1A was significantly positively correlated with RXRA and NCOA1, respectively (r > 0.80, p < 0.05). LCFA mediates the transcriptional activation of CPT via a heterodimeric complex composed of PPARα and the RXR [Citation31,Citation32]; in the meantime, NCOA1 acts as a coactivator for PPARα-mediated transcriptional activation [Citation33]. Thus, we speculate that TGFβ1 may cause the deregulation of the lipid metabolism transcriptional activation system.
Figure 1. Differentially-Expressed genes in response to TGFβ1 stimulation in HK-2 cells and in clinical renal fibrosis tissues. (a) Interaction networks of CPT1A-related proteins in TGFβ1-induced proximal tubular cells. (b) Co-expression networks of lncRnas (green bars) and CPT1A, NCOA1, and RXRA in TGFβ1-induced proximal tubular cells. (c) HK-2 cells were stimulated with 0, 2, 5, 10, and 50 ng/ml TGFβ1 for 48 h and examined for the expression of lncRnas (FABP5P3, DGCR5, MEIS3P1, EIF3IP1). (d) H&E staining examined the histopathological features of human renal fibrosis and normal renal tissues. (e) the expression of FABP5P3 in human renal tissues was examined by real-time PCR (n = 15). (f) HK-2 cells were stimulated with 0, 2, 5, 10, and 50 ng/ml TGFβ1 for 48 h and examined for the expression of CPT1A, NCOA1, and RXRA. (g) Immunohistochemical staining detected the protein abundance of CPT1A, NCOA1, and RXRA. **p < 0.01.
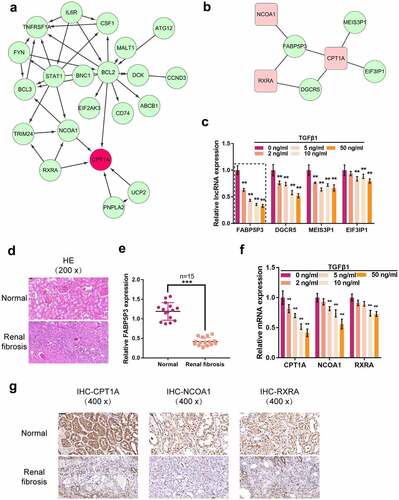
As mentioned previously, non-coding RNAs, including lncRNAs and miRNAs, exert an important effect on lipid metabolism [Citation34]. To further investigate the mechanism of TGFβ1-induced FAO deregulation, we analyzed differentially-expressed lncRNAs in TGFβ1-induced HK-2 cells based on GSE20247. Of the 47 remarkably decreased lncRNAs (fold change <0.67, p < 0.05), FABP5P3, DGCR5, MEIS3P1, and EIF3IP1 had a positive correlation with CPT1A (r > 0.80, p < 0.05) (). To confirm the candidate factors further, we treated HK-2 cells with 0, 2, 5, 10, and 50 ng/ml TGFβ1 for 48 h and examined for the expression of these four lncRNAs (FABP5P3, DGCR5, MEIS3P1, EIF3IP1); as shown in , four lncRNAs could be significantly downregulated by TGFβ1 stimulation in a dose-dependent manner, and FABP5P3 was more downregulated. Next, the renal fibrosis and normal renal tissues were subjected to analyses of clinical characteristics by H&E staining.
The images showed that pathological tissues exhibited disorganized renal structures and were filled with fibrotic areas (). Real-time PCR was used to detect lncRNA FABP5P3 in tissues. The results showed that FABP5P3 expression was down-regulated in renal fibrotic tissues, which was consistent with the expression of HK2 cells (). Moreover, CPT1A, RXRA, and NCOA1 could be significantly downregulated by 10 and 50 ng/ml TGFβ1 stimulation (). Consistently, immunohistochemistry results showed the abundance of CPT1A, RXRA, and NCOA1 protein were also downregulated in renal fibrosis tissues (). Thus, we hypothesize that FABP5P3 might be related to CPT1A, RXRA, and NCOA1 and play roles in TGFβ1-induced FAO deregulation and renal fibrosis.
TGFβ1 induces fibrotic changes and metabolic reprogramming in proximal tubular cells
Before investigating the role and mechanism of FABP5P3 in FAO deregulation, we first stimulated the proximal tubular cell line, HK-2 cell line, with 50 ng/ml TGFβ1 for 48 h and then monitored the alterations within HK-2 cells. As shown on the upper panels in , the HK-2 cells within the control group possessed a paving stone-like appearance and had adherent growth ability; in the TGFβ1 stimulation group, the intercellular space increased, and the appearance of the cells changed into a fusiform and fibroblast-like shape. Moreover, IF staining showed that TGFβ1 stimulation increased the fluorescence intensity representing the Fn protein (). These data indicate that TGFβ1 stimulation successfully induced fibrotic changes in HK-2 cells.
Figure 2. TGFβ1 induces fibrotic changes and metabolic reprogramming in proximal tubular cells (a) HK-2 cells were stimulated with 50 ng/ml TGFβ1 for 48 h and examined for morphologic changes under a light microscope and Fn protein localization and level by if staining. (b) the protein levels of CPT1A, NCOA1, and RXRA were examined by Immunoblotting. (c) the oxygen consumption rate (OCR) of HK-2 cells was determined with or without TGFβ1 stimulation. When indicated palmitate (180 μM) and oligomycin (1 μM) were added. (d) ATP levels of HK-2 cells were determined with or without TGFβ1 stimulation. **P <0.01.
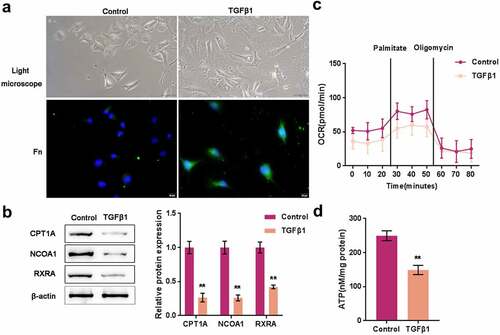
Consistent with the online microarray data and real-time PCR results, CPT1A, NCOA1, and RXRA proteins were significantly reduced by TGFβ1 stimulation (). Thus, next, we investigated the effects of TGFβ1 stimulation on FAO process. The oxygen consumption rate (OCR) of HK-2 cells was measured with or without TGFβ1 stimulation. We presented palmitate (180 μM) and oligomycin (1 μM) when indicated. When palmitate was presented in HK-2 cells, OCR showed to be significantly increased, indicating that HK-2 cells had a higher palmitate metabolic efficiency. The ATP synthase inhibitor oligomycin could significantly reduce OCR elevation. The oxygen consumption levels of cells treated with TGFβ1 were lower; in these cells, the inducible effects of palmitate on the increase in OCR were reduced, suggesting a decreased fatty acid metabolic activity (). Consistently, ATP levels of HK-2 cells showed to be significantly decreased by TGFβ1 stimulation (). These data indicated that TGFβ1 stimulation elicited a metabolic reprogramming within HK-2 cells, especially lower FAO.
TGFβ1 promotes renal fibrosis and inhibits fatty acid oxidation (FAO) via inhibiting FABP5P3 in vitro
After establishing the TGFβ1-induced metabolic reprogramming model in HK-2 cells, we generated knockdown or overexpression of FABP5P3 within HK-2 cells to investigate the specific effects of FABP5P3 within FAO and renal fibrosis. Real-time PCR was used to detect the transfection efficiency of si-FABP5P3 #1/2 and FABP5P3 overexpression vectors (). Under TGFβ1 stimulation, FABP5P3 knockdown significantly enhanced the fluorescence intensity representing Fn protein; on the contrary, FABP5P3 overexpression attenuated the fluorescence intensity (). Consistently, the protein level of Col1A1 was significantly increased by FABP5P3 knockdown while decreased by FABP5P3 overexpression (), suggesting that the inducible effects of TGFβ1 on HK-2 cell fibrotic changes could be attenuated by FABP5P3 overexpression.
Figure 3. TGFβ1 promotes renal fibrosis and inhibits fatty acid oxidation (FAO) via inhibiting FABP5P3 in vitro (a) the knockdown or overexpression of FABP5P3 was generated in HK-2 cells by transfection of si-FABP5P3 #1, #2 or FABP5P3-overexpressing vector, as confirmed by real-time PCR. Next, HK-2 cells were stimulated with TGFβ1 and transfected with si-FABP5P3 or FABP5P3-overexpressing vector and examined for (b) the localization and protein level of Fn were determined by if staining; (c) the protein levels of Col1A1, CPT1A, NCOA1, and RXRA by Immunoblotting; (d) ATP levels by an ATP colorimetric assay kit; (e) OCR by an XFe24 Analyzer; (f) the protein levels of Bcl-2 and CASP3 by Immunoblotting. *P <0.05, **P <0.01.
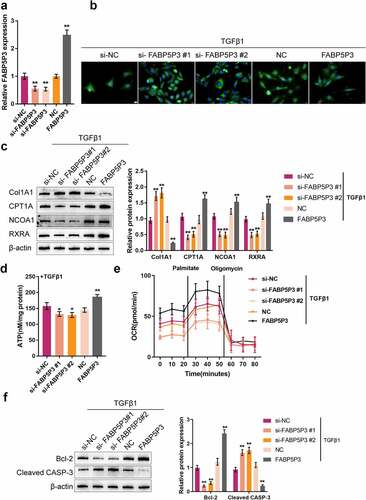
In the meantime, the protein levels of FAO key factors were monitored; FABP5P3 knockdown significantly decreased, while FABP5P3 overexpression increased the protein levels of CPT1A, NOCA1, and RXRA (). Consistently, FABP5P3 knockdown was significantly reduced, whereas FABP5P3 overexpression increased the ATP level in HK-2 cells (). Regarding the OCR levels under TGFβ1 stimulation, cells transfected with si-FABP5P3 had an even lower baseline of oxygen consumption levels, and FABP5P3 knockdown also reduced the inducible effects of palmitate on the increase in OCR, which indicated that FABP5P3 knockdown reduced fatty acid metabolic activity (). On the contrary, FABP5P3 vector-transfected HK-2 cells had a higher baseline of oxygen consumption levels, and FABP5P3 overexpression increased fatty acid metabolic activity (). These data suggested that FABP5P3 overexpression could partially reverse TGFβ1-caused suppression on FAO.
Then, the protein levels of apoptosis-related factors, Bcl-2 and CASP3, were also monitored in HK-2 cells under TGFβ1 stimulation. FABP5P3 knockdown significantly decreased Bcl-2 protein and increased CASP3 protein; conversely, FABP5P3 overexpression exerted opposite effects on these two proteins (), which demonstrated that the overexpression of FABP5P3 might partially reverse the inducible effects of TGFβ1 on HK-2 cell apoptosis.
FABP5P3 promotes the expression of critical genes of FAO within HK-2 cells through miR-22 under TGFβ1 stimulation
Due to the crucial effects of miRNAs on FAO and their mode of function, we speculate that miRNA might be involved in the FABP5P3 functioning process. Here, with the use of the online tool TargetScan, we screened for miRNAs that could bind to CPT1A, NCOA1, and RXRA, and 147 miRNAs were found; among these miRNAs, 4 were previously reported to promote fibrosis, and 2 were further predicted by lncTar to target FABP5P3 (FigureS1A), namely miR-22 and miR-27a. Ago2-RIP assays also indicated that FABP5P3 and miR-22 could bind to ago2 protein (FigureS1B).
To validate the specific role of these two miRNAs, we treated HK-2 cells with TGFβ1 for 48 h and examined their expression levels; both miR-27a and miR-22 were significantly induced by TGFβ1, and miR-22 was more upregulated (). In response to FABP5P3 overexpression or FABP5P3 knockdown, miR-22 expression was significantly inhibited or promoted, whereas miR-27a expression was only moderately changed ().
Figure 4. FABP5P3 promotes the expression of critical genes of FAO in HK-2 cells via TGFβ1-induced miR-22 (a) HK-2 cells were stimulated with TGFβ1 for 48 h and examined for the expression levels of miR-27a and miR-22. (b) HK-2 cells were stimulated with TGFβ1 for 48 h, transfected with si-FABP5P3 or FABP5P3-overexpressing vector, and examined for the expression levels of miR-27a and miR-22. (c) the expression levels of miR-27a and miR-22 in renal fibrotic and normal tissues were examined(n = 15). (d) miR-22 inhibition or overexpression was generated in HK-2 cells by transfection of miR-22 inhibitor or miR-22 mimics, as confirmed by real-time PCR. (e) HK-2 cells were stimulated with TGFβ1 for 48 h, co-transfected with si-FABP5P3 and miR-22 inhibitor, and examined for the protein levels of CPT1A, NCOA1, and RXRA. (f) Wild-type and mutant-type luciferase reporter vectors containing predicted miR-22 binding site or mutated miR-22 binding site were constructed. (g-j) These vectors were co-transfected to 293T cells with miR-22 mimics or miR-22 inhibitor; the luciferase activity was determined. *P <0.05, **P <0.01.
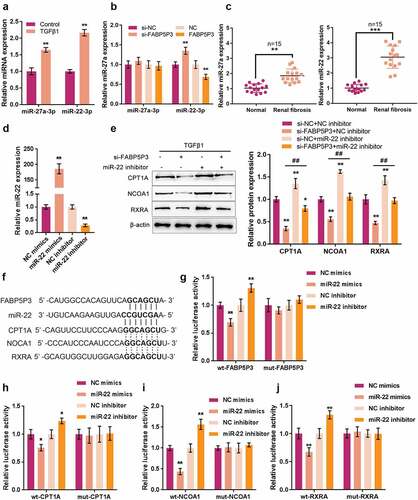
In human renal fibrosis tissues, the expression of miR-22 and miR-27a were both up-regulated, compared to normal renal tissues. The miR-22 expression showed a greater fold change (). Afterward, to further confirm that FABP5P3 might compete with miR-22 targets for miR-22 binding, we transfected miR-22 inhibitor/mimics to generate miR-22 inhibition or overexpression in HK-2 cells and performed real-time PCR to verify the transfection efficiency (). Next, HK-2 cells were treated with TGFβ1 for 48 h, co-transfected with si-FABP5P3 and miR-22 inhibitors, and evaluated for CPT1A, NCOA1, and RXRA proteins. As shown in , FABP5P3 knockdown was significantly suppressed, whereas the inhibition of miR-22 enhanced CPT1A, NCOA1, and RXRA proteins; the inhibition of miR-22 might significantly attenuate the effects of FABP5P3 knockdown. In summary, FABP5P3 may compete with the 3’UTR of CPT1A, NCOA1, and RXRA to counteract miR-22-mediated suppression on CPT1A, NCOA1, and RXRA.
To further confirm the bindings between miR-22 and related factors, luciferase reporter assays were performed. We constructed two different types of luciferase reporter vectors, wild-type and mutant-type, namely FABP5P3, CPT1A, NCOA1, and RXRA, which contain putative or mutated miR-22 bindings (). We co-transfected these vectors with miR-22 mimics/inhibitor into 293T cells and examined the luciferase activity. showed that the luciferase activity of all wild-type vectors was remarkably reduced via the overexpression of miR-22 and increased via the inhibition of miR-22. Mutating the putative miR-22 binding could abolish the alterations within the luciferase activity. These findings indicate that FABP5P3 counteracts the inhibitory effects of miR-22 upon CPT1A, NCOA1, and RXRA via competitive binding to miR-22.
FABP5P3/mir-22 axis regulates the FAO in HK-2 cells to affect fibrotic changes
After confirming the binding of miR-22 to FABP5P3, CPT1A, NCOA1, and RXRA, we further investigated the dynamic effects of FABP5P3 and miR-22 upon HK-2 cell FAO. HK-2 cells were treated with TGFβ1 for 48 h and co-transfected with si-FABp5P3 and miR-22 inhibitors, and then the related indexes were determined. As shown in , the fluorescence intensity representing Fn was significantly enhanced via FABP5P3 knockdown, while it was inhibited via the inhibition of miR-22. The inhibition of miR-22 could significantly attenuate the effects of FABP5P3 knockdown. The protein levels of Col1A1 were dramatically enhanced via FABP5P3 knockdown while reduced via miR-22 inhibition. Similarly, the effects of FABP5P3 knockdown were partially reversed by miR-22 inhibition ().
Figure 5. Fabp5p3/mir-22 axis regulates the FAO in HK-2 cells to affect fibrotic changes HK-2 cells were stimulated with TGFβ1 for 48 h, co-transfected with si-FABP5P3 and miR-22 inhibitor, and examined for (a) the localization and protein level of Fn by if staining; (b) the protein level of Col1A1 by Immunoblotting; (c) ATP levels by an ATP colorimetric assay kit; (d) OCR by an XFe24 Analyzer; (e) the protein levels of Bcl-2 and CASP3 by Immunoblotting. *P <0.05, **P <0.01, compared to control group; ##P <0.01, compared to si-FABP5P3 + NC inhibitor group.
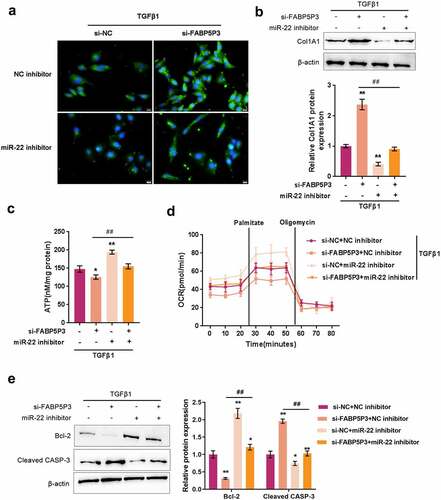
Regarding the FAO process, FABP5P3 knockdown was significantly reduced, while miR-22 inhibition increased the ATP level in HK-2 cells upon TGFβ1 stimulation (). As earlier revealed, cells transfected with si-FABP5P3 had a lower baseline of oxygen consumption levels, and FABP5P3 knockdown also reduced palmitate-induced elevation in OCR (). On the contrary, miR-22 inhibitor-transfected HK-2 cells had a higher baseline of oxygen consumption levels, and miR-22 inhibition increased the activity of fatty acid metabolism (). The inhibition of miR-22 could significantly attenuate the effects of FABP5P3 knockdown (). In summary, lncRNA FABP5P3 modulates FAO under TGFβ1 stimulation in HK-2 cells through miR-22.
To monitor the apoptosis of HK-2 cells, we also examined Bcl-2 and CASP3 proteins by Immunoblotting. As shown in , FABP5P3 knockdown significantly reduced Bcl-2 protein while increased CASP3 protein. The inhibitory effect of miR-22 was opposite. The effects of FABP5P3 knockdown were partially reversed by miR-22 inhibition ().
TGFβ1 induces the activation of JAG1/Notch2 signaling to inhibit FABP5P3 expression
We first examined the changes in JAG1/Notch2 signaling in response to TGFβ1 stimulation to further investigate the underlying mechanism. After 48 h of TGFβ1 stimulation, the protein levels of JAG1 and the ratio of p-SMAD3/SMAD3 were significantly increased within HK-2 cells (). Moreover, ICN2 protein was dramatically increased in the nucleus by TGFβ1 stimulation (). These data indicate that TGFβ1 stimulation induces the activation of JAG1/Notch2 signaling within HK-2 cells.
Figure 6. TGFβ1 induces the activation of JAG1/Notch2 signaling to inhibit FABP5P3 expression HK-2 cells were stimulated with TGFβ1 for 48 h and examined for (a) the protein levels of SMAD3, p-SMAD3, and JAG1 by Immunoblotting; (b) cytoplasm and nucleus protein levels of ICN2 by Immunoblotting. Next, HK-2 cells were stimulated with TGFβ1 for 48 h, transfected with si-Notch2, and examined for (c) the transfection efficiency by Immunoblotting; (d) the expression of FABP5P3 by real-time PCR. **P <0.01.
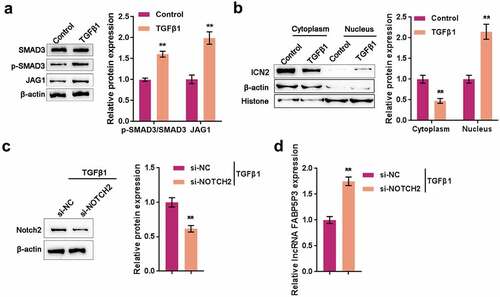
Next, we transfected HK-2 cells with si-Notch2 and stimulated them with TGFβ1 for 48 h to detect the expression of FABP5P3. The transfection efficiency under TGFβ1 stimulation was first approved by Immunoblotting (). As shown in , TGFβ1-induced suppression on lncRNA FABP5P3 expression was significantly reversed by Notch2 knockdown, indicating that TGFβ1 inhibits lncRNA FABP5P3 expression through JAG1/Notch2 signaling.
TGFβ1-Mediated Nocth2 signaling activation affects FAO and fibrotic changes in HK-2 cells through FABP5P3
After confirming that TGFβ1 induces the activation of JAG1/Notch2 signaling to inhibit FABP5P3 expression, we investigated the involvement of TGFβ1-induced JAG1/Notch2 signaling activation in the FAO process and fibrotic changes in HK-2 cells. Under TGFβ1 stimulation, Notch2 knockdown remarkably suppressed Fn and Col1A1 proteins whereas enhanced CPT1A, NCOA1, and RXRA proteins, while FABP5P3 knockdown exerted opposite effects on these five proteins. The effects of Notch2 knockdown could be significantly reversed by FABP5P3 knockdown (). These data suggest that TGFβ1-induced JAG1/Notch2 signaling activation participates in the fibrotic changes in HK-2 cells.
Figure 7. TGFβ1-Mediated Nocth2 signaling activation affects FAO and fibrotic changes in HK-2 cells through FABP5P3 HK-2 cells were stimulated with TGFβ1 for 48 h, co-transfected with si-FABP5P3 and si-Notch2, and examined for (a) the protein levels of Fn and Col1A1 by Immunoblotting; (b) the protein levels of CPT1A, NCOA1, and RXRA by Immunoblotting; (c) ATP levels by an ATP colorimetric assay kit; (d) OCR by an XFe24 Analyzer; (E) the protein levels of Bcl-2 and CASP3 by Immunoblotting. *P <0.05, **P <0.01, compared to control group; ##P <0.01, compared to si-Notch2 group.
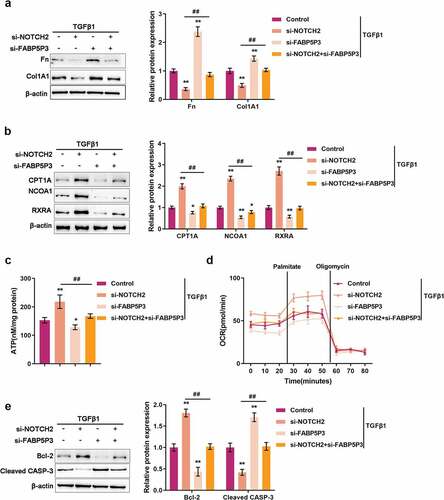
As for the FAO process, Notch2 knockdown was significantly increased, while FABP5P3 knockdown reduced the ATP level in HK-2 cells upon TGFβ1 stimulation (). In sharp contrast to FABP5P3 knockdown, Notch2 knockdown led to a higher baseline of oxygen consumption levels and increased the activity of fatty acid metabolism (). The effects of Notch2 knockdown were partially reversed by FABP5P3 knockdown (). These data suggest that TGFβ1-induced JAG1/Notch2 signal activation is involved in the process of lncRNA FABP5P3 modulating FAO in HK-2 cells.
Regarding the changes in apoptosis-related factors, Notch2 knockdown significantly increased Bcl-2 protein while decreasing CASP3 protein, whereas FABP5P3 knockdown exerted opposite effects on these two proteins. The effects of Notch2 knockdown were partially reversed by FABP5P3 knockdown ().
FABP5P3 alleviates the renal fibrosis of UUO mice model
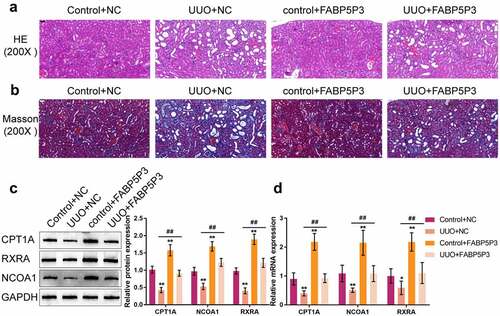
To identify the effect of lncRNA FABP5P3 on renal fibrosis in vivo further, lentiviruses carrying FABP5P3 overexpression or empty lentivirus were injected into mice renal parenchyma before UUO surgery. HE and Masson staining showed that the obstructive kidney tissues displayed typical fibrosis characteristics, such as proximal tubule dilation, atrophy, and extracellular matrix (ECM) accumulation. Overexpression of FABP5P3 in UUO group mice alleviated the typical fibrosis characteristics of UUO mice (). Western blotting and real-time qPCR results showed that UUO surgery significantly decreased while FABP5P3 overexpression increased the expression of CPT1A, NCOA1, and RXRA. The effects of UUO surgery on CPT1A, NCOA1, and RXRA expression were partially reversed by FABP5P3 overexpression ().
Figure 8. FABP5P3 alleviates the renal fibrosis of Unilateral Ureteral Obstruction (UUO) mice model Mice were treated with an intrarenal parenchymal injection of either FABP5P3-overexpressing or empty lentivirus 2 days before UUO. Two weeks after UUO. (a) the histopathological features of renal tissues were determined by H&E staining; (b) Masson’s trichrome staining was used for fibrosis analysis; (c) the protein levels of CPT1A, NCOA1, and RXRA in tissues were determined by Immunoblotting; (d) the mRNA levels of CPT1A, NCOA1, and RXRA in tissues were determined by real-time PCR. n = 9, *P <0.05, **P <0.01, compared to Control+NC group; ##P <0.01, compared UUO+NC with FABP5P3+NC group.
Discussion
In recent years, lipid metabolism has been demonstrated to play an important role in multiple kidney diseases [Citation35,Citation36]. Lipid deposition within non-adipose tissues has been recently implicated in causing organ damage [Citation37]. Clinical studies have suggested an underlying association between renal lipid deposition and the development of CKD [Citation38]. Moreover, extensive animal data have demonstrated the correlation between renal lipid accumulation and renal insufficiency, including metabolic and non-metabolic kidney disease models [Citation39,Citation40]. Our results indicated that upon TGFβ1 stimulation, the levels of both Fn and Col1A1 were significantly increased, and the metabolic reprogramming, specifically lower FAO, occurred in HK-2 cells, suggesting the appearance of deregulation of lipid metabolism is related to the progression of fibrotic changes.
Recent studies have shown the effects of FAO signaling pathway impairment on renal interstitial fibrosis development [Citation39]; proximal TECs rely heavily on FAO as their energy source, and the decrease in FAO could be related to intracellular lipid deposition. In addition, it has been demonstrated that the damage to FAO could be due to a decline in the PPARα-PGC1α axis in the process of renal fibrosis. Critically, it has been reported by another group that the protein levels of FAO rate-limiting enzyme, CPT1A, were significantly reduced within the renal tubular epithelium [Citation41]. Herein, CPT1A, NCOA1, and RXRA expression and protein levels were all significantly reduced in HK-2 cells upon TGFβ1 stimulation and clinical renal fibrosis tissues, further indicating that the FAO was deregulated. Since we observed significant downregulation in both the gene and protein levels of CPT1A, NCOA1, and RXRA, we speculated that there might be epigenetic mechanisms regulating the expression of CPT1A, NCOA1, and RXRA post-transcriptionally.
LncRNAs are an understudied subclass of non-coding RNA with multifaceted functions in epigenetic regulation, transcriptional activation or repression, remodeling of chromatin architecture, modulation of mRNAs at a post-transcriptional level, and protein activity regulation [Citation42]. To date, it has been reported that lncRNAs exerted an effect on lipid metabolism through their effects on the SREBP transcription factors [Citation43,Citation44], apolipoproteins [Citation45], triglyceride metabolism [Citation46], and macrophage cholesterol uptake and efflux [Citation47]. Based on online microarray profiles, several lncRNAs were differentially expressed in HK-2 cells upon TGFβ1 stimulation, among which lncRNA FABP5P3 was significantly downregulated by TGFβ1 stimulation and was positively correlated with CPT1A, NCOA1, and RXRA.
Since TGFβ1 stimulation causes fibrotic changes and FAO deregulation within HK-2 cells, lncRNA FABP5P3 could critically affect the process. As expected, the overexpression of FABP5P3 within HK-2 cells significantly reversed the inducible effects of TGFβ1 on fibrotic changes, partially reversed TGFβ1-caused suppression on FAO, and partially reversed the inducible effects of TGFβ1 on HK-2 cells apoptosis. Moreover, in vivo studies suggested that overexpression of lncRNA FABP5P3 significantly alleviated the UUO mice renal fibrosis and increased CPT1A, NCOA1, and RXRA protein levels. In summary, lncRNA FABP5P3 could improve fibrotic alterations and TGFβ1-induced deregulation of FAO.
As reported, lncRNAs, which modulate gene transcription and contribute to the regulation at a post-transcriptional level, can not only act as molecular signals or guides for transcription factors but also modulate epigenetic modifiers [Citation48]. As we have mentioned, one of the most widely-recognized mechanisms of lncRNA function is to serve as ceRNAs (competing endogenous RNAs) of miRNAs, which mutually regulate miRNAs and then regulate the expression of miRNA target genes [Citation25]. Herein, miR-22 was predicted by online tools to target lncRNA FABP5P3, as well as three key factors in FAO, CPT1A, NCOA1, and RXRA. As previously reported, miR-22 was related to cancer metabolic reprogramming [Citation49,Citation50]; however, its specific role in FAO deregulation in HK-2 cells remains unclear. As predicted by the online tools, lncRNA FABP5P3 relieved miR-22-induced inhibition on CPT1A, NCOA1, and RXRA via acting as a ceRNA. Moreover, miR-22 inhibition exerted similar effects to lncRNA FABP5P3 overexpression on FAO and fibrotic changes in HK-2 cells; that is, it reversed TGFβ1-caused fibrotic changes, partially reversed TGFβ1-caused suppression on FAO, and partially reversed the inducible effects of TGFβ1 on HK-2 cell apoptosis. More importantly, the inhibition of miR-22 might significantly attenuate the effects of lncRNA FABP5P3 knockdown, indicating that lncRNA FABP5P3 affects FAO and TGFβ1-caused HK-2 cell fibrotic alterations through miR-22.
To further investigate the related pathways, we verified the TGFβ/SMAD activation and the intracellular fragment of Notch2 (ICN2) protein levels in the cytoplasm and nucleus. Under TGFβ1 stimulation, an increased ratio of p-SMAD3/SMAD3 and JAG1 protein levels, and the increased nucleus ICN2 levels suggested that TGFβ1 stimulation induced the activation of TGFβ/SMAD and JAG/Notch signaling pathways. More importantly, the suppressive effects of TGFβ1 on lncRNA FABP5P3 expression was significantly reversed by Notch2 knockdown. These results suggested that the TGFβ/SMAD and JAG/Notch pathways were involved in the lncRNA FABP5P3/miR-22 axis modulating FAO in HK-2 cells under TGFβ1 stimulation. In conclusion, the lncRNA FABP5P3/miR-22 axis might be a potent target for improving the FAO deregulation and fibrotic changes in the fibrotic process of CKD.
Availability of data and material
Data are available in the publications cited in the manuscript.
Ethics approval and consent to participate
The study was conducted by following per under the Declaration of Helsinki and approved by the Ethics Committee of Third Xiangya Hospital, Central South University (No.2019-S291)
Supplemental Material
Download TIFF Image (837.5 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384101.2022.2122286.
Additional information
Funding
References
- Woodward M, Frank D. Postnatal management of antenatal hydronephrosis. BJU Int. 2002;89:149–156.
- Ismaili K, Avni FE, Wissing KM, et al. Long-Term clinical outcome of infants with mild and moderate fetal pyelectasis: validation of neonatal ultrasound as a screening tool to detect significant nephrouropathies. J Pediatr. 2004;144:759–765.
- Shapiro E. Antenatal hydronephrosis: here today, gone tomorrow-one way or another: nYU Case of the Month, May 2017. Rev Urol. 2017;19:138–141.
- Seseke F, Thelen P, Ringert RH. Characterization of an animal model of spontaneous congenital unilateral obstructive uropathy by cDNA microarray analysis. Eur Urol. 2004;45:374–381.
- Vielhauer V, Anders HJ, Mack M, et al. Obstructive nephropathy in the mouse: progressive fibrosis correlates with tubulointerstitial chemokine expression and accumulation of CC chemokine receptor 2- and 5-positive leukocytes. J Am Soc Nephrol. 2001;12:1173–1187.
- Jha V, Garcia-Garcia G, Iseki K, et al. Chronic kidney disease: global dimension and perspectives. Lancet. 2013;382:260–272.
- Kaissling B, Lehir M, Kriz W. Renal epithelial injury and fibrosis. Biochim Biophys Acta. 2013;1832:931–939.
- Singh SP, Tao S, Fields TA, et al. Glycogen synthase kinase-3 inhibition attenuates fibroblast activation and development of fibrosis following renal ischemia-reperfusion in mice. Dis Model Mech. 2015;8:931–940.
- Ito K, Chen J, El Chaar M, et al. Renal damage progresses despite improvement of renal function after relief of unilateral ureteral obstruction in adult rats. Am J Physiol Renal Physiol. 2004;287:F1283–93.
- Brosius FC 3rd, Alpers CE, Bottinger EP, et al. Mouse models of diabetic nephropathy. J Am Soc Nephrol. 2009;20:2503–2512.
- Hodgkins KS, Schnaper HW. Tubulointerstitial injury and the progression of chronic kidney disease. Pediatr Nephrol. 2012;27:901–909.
- Kapitsinou PP, Haase VH. Molecular mechanisms of ischemic preconditioning in the kidney. Am J Physiol Renal Physiol. 2015;309:F821–34.
- Bielesz B, Sirin Y, Si H, et al. Epithelial Notch signaling regulates interstitial fibrosis development in the kidneys of mice and humans. J Clin Invest. 2010;120:4040–4054.
- Bottinger EP, Bitzer M. TGF-Beta signaling in renal disease. J Am Soc Nephrol. 2002;13:2600–2610.
- DeBerardinis RJ, Thompson CB. Cellular metabolism and disease: what do metabolic outliers teach us? Cell. 2012;148:1132–1144.
- Susztak K, Ciccone E, McCue P, et al. Multiple metabolic hits converge on CD36 as novel mediator of tubular epithelial apoptosis in diabetic nephropathy. PLoS Med. 2005;2:e45.
- Schug TT, Li X. Sirtuin 1 in lipid metabolism and obesity. Ann Med. 2011;43:198–211.
- Tran M, Tam D, Bardia A, et al. PGC-1alpha promotes recovery after acute kidney injury during systemic inflammation in mice. J Clin Invest. 2011;121:4003–4014.
- Bouchard-Mercier A, Rudkowska I, Lemieux S, et al. Polymorphisms in genes involved in fatty acid beta-oxidation interact with dietary fat intakes to modulate the plasma TG response to a fish oil supplementation. Nutrients. 2014;6:1145–1163.
- Lim D, Chai HH, Lee SH, et al. Gene expression patterns associated with Peroxisome Proliferator-activated Receptor (PPAR) Signaling in the Longissimus dorsi of Hanwoo (Korean Cattle). Asian-Australas J Anim Sci. 2015;28:1075–1083.
- Hostetler HA, Huang H, Kier AB, et al. Glucose directly links to lipid metabolism through high affinity interaction with peroxisome proliferator-activated receptor alpha. J Biol Chem. 2008;283:2246–2254.
- Rost TH, Haugan Moi LL, Berge K, et al. A pan-PPAR ligand induces hepatic fatty acid oxidation in PPARalpha-/- mice possibly through PGC-1 mediated PPARdelta coactivation. Biochim Biophys Acta. 2009;1791:1076–1083.
- Mendell JT, Olson EN. MicroRnas in stress signaling and human disease. Cell. 2012;148:1172–1187.
- Trionfini P, Benigni A, Remuzzi G. MicroRnas in kidney physiology and disease. Nat Rev Nephrol. 2015;11:23–33.
- Yamamura S, Imai-Sumida M, Tanaka Y, et al. Interaction and cross-talk between non-coding RNAs. Cell Mol Life Sci. 2018;75:467–484.
- Du Y, Liu P, Chen Z, et al. PTEN improve renal fibrosis in vitro and in vivo through inhibiting FAK/AKT signaling pathway. J Cell Biochem. 2019;120:17887–17897.
- Xi Y, Nakajima G, Gavin E, et al. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. Rna. 2007;13:1668–1674.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408.
- Hills CE, Willars GB, Brunskill NJ. Proinsulin C-peptide antagonizes the pro-fibrotic effects of TGF-beta1 via up-regulation of retinoic acid and HGF-related signaling pathways. Mol Endocrinol. 2010;24:822–831.
- Casals N, Zammit V, Herrero L, et al. Carnitine palmitoyltransferase 1C: from cognition to cancer. Prog Lipid Res. 2016;61:134–148.
- Brandt JM, Djouadi F, Kelly DP. Fatty acids activate transcription of the muscle carnitine palmitoyltransferase I gene in cardiac myocytes via the peroxisome proliferator-activated receptor alpha. J Biol Chem. 1998;273:23786–23792.
- Yu GS, Lu YC, Gulick T. Co-Regulation of tissue-specific alternative human carnitine palmitoyltransferase Ibeta gene promoters by fatty acid enzyme substrate. J Biol Chem. 1998;273:32901–32909.
- Westin S, Kurokawa R, Nolte RT, et al. Interactions controlling the assembly of nuclear-receptor heterodimers and co-activators. Nature. 1998;395:199–202.
- Smolle E, Haybaeck J. Non-Coding RNAs and lipid metabolism. Int J Mol Sci. 2014;15:13494–13513.
- Trevisan R, Dodesini AR, Lepore G. Lipids and renal disease. J Am Soc Nephrol. 2006;17:S145–7.
- Bobulescu IA. Renal lipid metabolism and lipotoxicity. Curr Opin Nephrol Hypertens. 2010;19:393–402.
- van Herpen NA, Schrauwen-Hinderling VB. Lipid accumulation in non-adipose tissue and lipotoxicity. Physiol Behav. 2008;94:231–241.
- Herman-Edelstein M, Scherzer P, Tobar A, et al. Altered renal lipid metabolism and renal lipid accumulation in human diabetic nephropathy. J Lipid Res. 2014;55:561–572.
- Kang HM, Ahn SH, Choi P, et al. Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat Med. 2015;21:37–46.
- Kume S, Uzu T, Araki S, et al. Role of altered renal lipid metabolism in the development of renal injury induced by a high-fat diet. J Am Soc Nephrol. 2007;18:2715–2723.
- Chung KW, Lee EK, Lee MK, et al. Impairment of PPARalpha and the fatty acid oxidation pathway aggravates renal fibrosis during aging. J Am Soc Nephrol. 2018;29:1223–1237.
- van Solingen C, Scacalossi KR, Moore KJ. Long non-coding RNAs in lipid metabolism. Curr Opin Lipidol. 2018;29:224–232.
- Yan C, Chen J, Chen N. Long non-coding RNA MALAT1 promotes hepatic steatosis and insulin resistance by increasing nuclear SREBP-1c protein stability. Sci Rep. 2016;6:22640.
- Liu C, Yang Z, Wu J, et al. Long non-coding RNA H19 interacts with polypyrimidine tract-binding protein 1 to reprogram hepatic lipid homeostasis. Hepatology. 2018;67:1768–1783.
- Halley P, Kadakkuzha BM, Faghihi MA, et al. Regulation of the apolipoprotein gene cluster by a long non-coding RNA. Cell Rep. 2014;6:222–230.
- Cui M, Xiao Z, Wang Y, et al. Long non-coding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res. 2015;75:846–857.
- Sallam T, Jones M, Thomas BJ, et al. Transcriptional regulation of macrophage cholesterol efflux and atherogenesis by a long non-coding RNA. Nat Med. 2018;24:304–312.
- Ulitsky I, Bartel DP. lincRnas: genomics, evolution, and mechanisms. Cell. 2013;154:26–46.
- Koufaris C, Valbuena GN, Pomyen Y, et al. Systematic integration of molecular profiles identifies miR-22 as a regulator of lipid and folate metabolism in breast cancer cells. Oncogene. 2016;35:2766–2776.
- Pant K, Yadav AK, Gupta P, et al. Butyrate induces ROS-mediated apoptosis by modulating miR-22/SIRT-1 pathway in hepatic cancer cells. Redox Biol. 2017;12:340–349.
