ABSTRACT
Preeclampsia (PE) is the most common complication in the pregnancy of women. PE progression was found to be associated with dysregulated circular RNAs (circRNAs), and we aimed to explore the pathological mechanism with circ_0004904 in PE. The circ_0004904, microRNA-19a-3p (miR-19a-3p) and DNA damage inducible alpha (GADD45A) were quantified via reverse transcription-quantitative polymerase chain reaction assay. Trophoblast cell behaviors were examined by cell viability using Cell Counting Kit-8 assay, cell proliferation using EdU assay, cell apoptosis using flow cytometry, cell invasion using transwell assay and migration using wound healing assay. Western blot was used for protein analysis of epithelial mesenchymal transition (EMT) and GADD45A. Dual-luciferase reporter assay and RNA immunoprecipitation assay were used for target interaction analysis. The circ_0004904 upregulation was detected in placenta tissues from PE patients. Trophoblast cell proliferation, invasion, migration and EMT were repressed but cell apoptosis was promoted after overexpression of circ_0004904. Circ_0004904 acted as a miR-19a-3p sponge in trophoblast cells, and all regulatory effects of circ_0004904 on trophoblast cell behaviors were reversed by miR-19a-3p upregulation. The miR-19a-3p directly targeted GADD45A and miR-19a-3p downregulation inhibited trophoblast cell development through elevating the GADD45A level. Moreover, circ_0004904 enhanced the expression of GADD45A via sponging miR-19a-3p. Our results elucidated that circ_0004904 reduced proliferation and cell motility of trophoblast cells via the miR-19a-3p-mediated GADD45A level elevation, indicating the involvement of circ_0004904/miR-19a-3p/GADD45A in PE progression.
Introduction
Hypertensive disorders are common in pregnant females, and preeclampsia (PE) is the most feared complication affecting both mother and fetus [Citation1]. Since PE patients usually develop a severe prothrombotic state and has a higher cardiovascular risk, the early detection and treatment are important for maternal and fetal health [Citation2,Citation3]. The research suggests that early-onset PE is closely associated with poor placentation [Citation4]. The placentation is dependent on the normal proliferation, migration and invasion of trophoblasts [Citation5,Citation6]. Thus, exploring the pathogenic mechanism in trophoblast cell development may improve the diagnosis and treatment of PE.
Non-coding circular RNAs (circRNAs) are closed-loop transcripts with the sponge property of microRNAs (miRNAs), thereby affecting the downstream genes and disease progression [Citation7]. Circ_0111277 was shown to enhance trophoblast cell viability and invasion partly by mediating miR-424-5p/NFAT5 axis [Citation8]. Circ_0026552 targeted the miR-331-3p/TGF-βR1 axis to suppress invasive and migratory abilities in trophoblast cells [Citation8]. CircRNA/miRNA/mRNA regulatory network plays a pivotal role in trophoblast cell development of PE.
Circ_0004904 was identified as a upregulated molecule in placenta tissues of PE patients, and silence of circ_0004904 promoted trophoblast cell function by interacting with miR-570/ATG12 axis [Citation9,Citation10]. Wang et al. reported that miR-19a-3p contributed to trophoblast cell growth and metastasis [Citation11]. DNA damage inducible alpha (GADD45A) was a upregulated gene in PE and it attenuated the normal growth of trophoblast cells [Citation12,Citation13]. It is unclear whether circ_0004904 can affect GADD45A level via combining with miR-19a-3p in PE.
The target relation between circ_0004904 and miR-19a-3p, as well as GADD45A and miR-19a-3p, was assumed in this study. Our research was performed to discover a novel molecular mechanism circ_0004904/miR-19a-3p/GADD45A in the PE progression.
Material and methods
Placenta tissues
The placenta tissues were collected from PE patients (n = 22) and normal pregnant women (n = 22) at Shengjing Hospital of China Medical University, based on the acquisition of written informed consent forms. PE was diagnosed by two experienced pathologists. Before this study was started, those samples were conserved at −80°C. Tissue collection was in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Shengjing Hospital of China Medical University.
Cell culture and transfection
JEG-3 and HTR-8/SVneo (BioVector NTCC Inc., Beijing, China) are common trophoblast cells used for PE research. For cell culture, cells were maintained in Roswell Park Memorial Institute-1640 (RPMI-1640) containing 1% antibiotics and 10% fetal bovine serum (FBS) at 37° with 5% CO2. All culture reagents were commercially obtained from Gibco (Carlsbad, CA, USA). The pCD5-ciR vector (GENESEED, Guangzhou, China) was cloned with circ_0004904 sequence to construct the overexpression vector pCD5-ciR-circ_0004904 (circ_0004904). The small interfering RNA (siRNA) for circ_0004904 or GADD45A and negative control (si-circ_0004904, si-GADD45A, si-NC), miR-19a-3p mimic and control (miR-19a-3p, miR-NC), miR-19a-3p inhibitor and control (anti-miR-19a-3p, anti-miR-NC) were purchased from Ribobio (Guangzhou, China). 1 × 104 cells/well were planted into the 96-well plates overnight. Lipofectamine™ 3000 transfection reagent (Invitrogen, Carlsbad, CA, USA) was mixed with the above vectors or RNAs, then incubated with cells at 37°C for further use.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assay
All detection kits were bought from Sangon (Shanghai, China). Total RNA extraction by Trizol reagent was performed as per the user’s manuals, followed by reverse transcription by One Step RT-PCR Kit. 2 × TaqMan Fast qPCR Master Mix and MicroRNAs SYBR Green qPCR Kit were utilized for level quantification. Glyceraldehyde-phosphate dehydrogenase (GAPDH) was normalized to circ_0004904 and GADD45A, but U6 acted as an internal reference for miR-19a-3p. The 2−∆∆Ct method was applied to analyze the relative gene levels [Citation14]. The primers used for RT-qPCR were synthesized by Sangon ().
Table 1. Primer sequences used for RT-qPCR.
CircRNA identification assays
Total RNA was treated with 4 U/μg RNase R (GENESEED) at 37°C for 2 h, then circ_0004904 and GAPDH levels were assayed by RT-qPCR. Reverse transcription assay was performed using Random primers or Oligo (dT)18 primers, then expression analysis of circ_0004904 and GAPDH was conducted through RT-qPCR. In addition, gene levels (circ_0004904, U6 and GAPDH) were measured via RT-qPCR after isolation of nuclear and cytoplasmic RNAs by PARIS™ Kit (Invitrogen).
Cell counting kit-8 (CCK-8) assay
JEG-3 and HTR-8/SVneo cells were transfected for 48 h, followed by incubating with 10 μL CCK-8 solution (Beyotime, Shanghai, China) for 2 h and measuring the absorbance at 450 nm via a microplate reader. The percentage of viable cells was calculated to display cell viability.
EdU assay
EdU Detection Kit (Ribobio) was used for proliferation analysis. EdU solution was diluted with cell medium, and cells were incubated with 100 µL EdU medium for 2 h. After cell fixation by 4% paraformaldehyde (Sangon), cells were stained with 100 μL 1 × Apollo staining solution and nuclei were stained with diamidine phenylindole (DAPI; Sangon). Then, EdU and D API merged cells were counted as the EdU-positive cells through a fluorescence microscope (Olympus, Tokyo, Japan).
Flow cytometry
At 72 h after transfection, Annexin V-fluorescein isothiocyanate (FITC) and Propidium Iodide (PI) were applied for cell staining according to the Apoptosis Detection Kit (Beyotime). The stained cells were incubated in the dark for 15 min, and apoptotic cells were examined under the flow cytometer (BD Biosciences, San Diego, CA, USA).
Transwell assay
The transwell chamber (Corning Inc., Corning, NY, USA) was coated with matrigel (BD Biosciences), then the upper chamber was seeded with 1 × 105 cells in serum-free medium and the lower chamber was added with 600 µL cell medium. After incubation for 24 h, the invaded cells were stained with 0.1% crystal violet (Sangon) and cells were counted through the inverted microscope (Olympus). Cell pictures were saved under 100 × magnification.
Wound healing assay
The monolayer JEG-3 and HTR-8/SVneo cells were created with two straight scratches through a sterile 200 μL pipette tip, then the 12-well plates were washed with phosphate buffer solution (PBS; Gibco). Subsequently, cells were cultured with serum-free medium for 24 h. The migration distance was measured at 0 h and 24 h.
Western blot
The protein extraction from tissues and cells was implemented via radioimmunoprecipitation assay (RIPA) buffer (Beyotime), followed by examining the protein concentrations using BCA Protein Assay Kit (Beyotime). The protein blot analysis was carried out following the previous description [Citation15]. The primary antibodies targeting E-cadherin (#3195), N-cadherin (#13116), GADD45A (#4632) or GAPDH (#8884) were diluted at 1:1000 and incubated at 4°C overnight. Anti-rabbit IgG, HRP-linked Antibody (#7074) was diluted at 1:3000 and incubated at 25°C for 1 h. These antibodies were from Cell Signaling Technology (CST, Boston, MA, USA). After blots were exhibited by Electrochemiluminescence (ECL) regent (Beyotime), the expression was analyzed via Image J software (NIH, Bethesda, MD, USA).
Dual-luciferase reporter assay
The luciferase plasmids were constructed by cloning the molecular sequences into the basic pmirGLO plasmid (Promega, Madison, WI, USA). The wild-type (WT) plasmids WT-circ_0004904 and WT-GADD45A 3”UTR contained the binding sites of miR-19a-3p, while the mutant (MUT) plasmids MUT-circ_0004904 and MUT-GADD45A 3”UTR contained the mutated sites aiming at miR-19a-3p. JEG-3 and HTR-8/SVneo cells were co-transfected with miR-NC or miR-19a-3p and WT or MUT plasmid, then cells were collected at 48 h post-transfection. By performing Dual-Luciferase Reporter Detection Kit (Promega), the luciferase activity of each group was detected.
RNA immunoprecipitation (RIP) assay
The target interaction by Imprint® RNA Immunoprecipitation Kit (Sigma, St. Louis, MO, USA) was conducted through cell incubation with Anti-Argonaute-2 (anti-Ago2)-coated magnetic beads. Anti-immunoglobulin G (anti-IgG) was employed as the negative control group. After total RNA acquisition, RT-qPCR was performed for level examination of circ_0004904, miR-19a-3p and GADD45A.
Statistical analysis
The linear analysis between targets in PE samples was carried out through Pearson’s correlation coefficient. Data from three independently repeated experiments were indicated as the mean ± standard deviation. After data analysis by SPSS 22.0 (SPSS Inc., Chicago, IL, USA), statistical difference was assessed via Student’s t-test and analysis of variance (ANOVA) followed by Tukey’s test. It was significant when P < 0.05.
Results
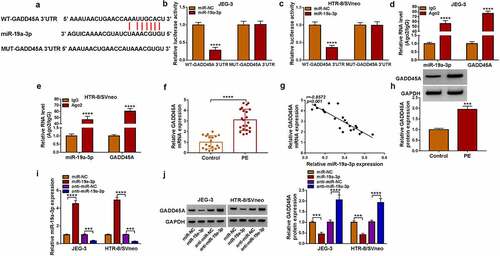
Circ_0004904 was upregulated in placenta tissues of PE patients
RT-qPCR showed that circ_0004904 was highly expressed in placenta samples from PE patients relative to normal pregnancy females (). In addition, clinicopathological characteristics showed that high expression of circ_0004904 was related to proteinuria, systolic blood pressure and diastolic blood pressure of PE patients (). The stability of circ_0004904 was analyzed via RNase R assay. As exhibited in () circ_0004904 was more stable than linear GAPDH after RNase R treatment for total RNA from JEG-3 and HTR-8/SVneo cells. In addition, circular structure was affirmed through reverse transcription using random primers and oligo (dT)18 primers. The results indicated that circ_0004904 was lower in oligo (dT)18 primers group than that in random primers group while GAPDH was no significant change, suggesting that circ_0004904 was circular formation without 3’-polyA tail (). Nuclear and cytoplasmic RNAs were isolated from JEG-3 and HTR-8/SVneo cells, and the expression levels of GAPDH and U6 validated that the isolation was successful. Through the comparison with GAPDH and U6, circ_0004904 was identified to be localized in the cytoplasm (). The aberrant upregulation of circ_0004904 was detected in PE patients.
Figure 1. Circ_0004904 was upregulated in placenta tissues of PE patients. (a) Circ_0004904 level detection was conducted using RT-qPCR in placenta tissues from PE patients and normal controls. (b-c) Circ_0004904 and GAPDH stabilities were assessed by RT-qPCR after RNase R treatment in total RNA from JEG-3 and HTR-8/svneo cells. (d-e) the levels of circ_0004904 and GAPDH were examined by RT-qPCR after Random primers or Oligo (dT)18 primers were used in reverse transcription. (f-g) Circ_0004904, U6 and GAPDH were quantified by RT-qPCR after cytoplasmic and nuclear RNAs were isolated from JEG-3 and HTR-8/svneo cells. Experiments were repeated for three times with three parallels. ****P <0.0001.
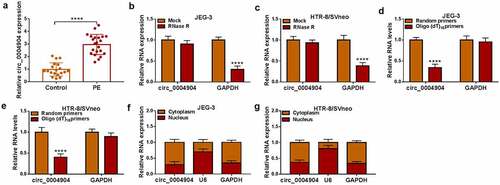
Table 2. The correlation between circ_0004904 expression and clinicopathological characteristics of PE patients.
Circ_0004904 inhibited proliferation, invasion, migration and EMT but enhanced apoptosis of trophoblast cells
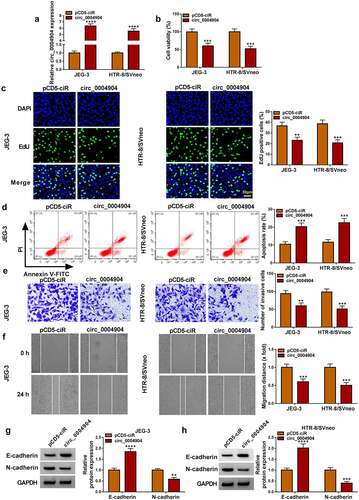
The circ_0004904 was overexpressed by transfection of circ_0004904, and the efficiency was significant compared with pCD5-ciR control group (). As the results of circ_0004904 overexpression, cell viability () and proliferation ability () were reduced but apoptosis rate was increased () in JEG-3 and HTR-8/SVneo cells. The data from transwell assay and wound healing assay revealed that cell invasion () and migration () were suppressed in circ_0004904 group contrasted to pCD5-ciR group. The protein detection exhibited that E-cadherin was upregulated and N-cadherin was downregulated after circ_0004904 was overexpressed, showing that circ_0004904 blocked the process of epithelial mesenchymal transition (EMT) ().Circ_0004904 knockdown promoted cell proliferation, invasion and migration while inhibited cell apoptosis in JEG-3 and HTR-8/SVneo cells (Supplementary Fig. S1). The above evidence demonstrated that circ_0004904 impeded the biological behaviors of trophoblast cells.
Figure 2. Circ_0004904 inhibited proliferation, invasion, migration and EMT but enhanced apoptosis of trophoblast cells. the pCD5-ciR and circ_0004904 were transfected into JEG-3 and HTR-8/svneo cells, respectively. (a) Circ_0004904 expression was determined through RT-qPCR. (b-c) Cell viability (b) and proliferation (c) were evaluated through CCK-8 assay and EdU assay. (D) the apoptosis rate was examined through flow cytometry. (e-f) Cell invasion (e) and migration (f) were assessed through transwell assay and wound healing assay. (g-h) E-cadherin and N-cadherin protein levels were detected through western blot. Experiments were repeated for three times with three parallels. **P <0.01, ***P <0.001, ****P <0.0001.
Circ_0004904 sponged miR-19a-3p in trophoblast cells
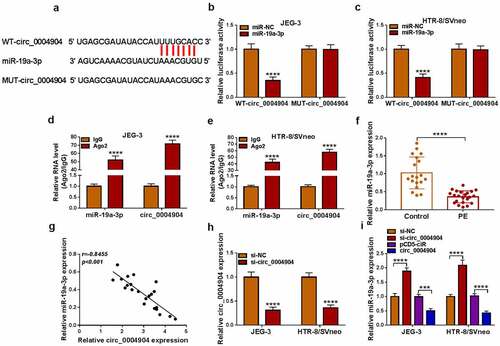
The starbase software predicted the binding sites between circ_0004904 and miR-19a-3p, as shown in . The luciferase activity was inhibited by co-transfection with WT-circ_0004904 and miR-19a-3p but not MUT-circ_0004904 and miR-19a-3p, implying that circ_0004904 interacted with miR-19a-3p in JEG-3 and HTR-8/SVneo cells (). Also, miR-19a-3p and circ_0004904 levels were much higher in Ago2 group than these in IgG group (). The miR-19a-3p level was reduced in PE group compared to the control group (), and circ_0004904 was negatively related to miR-19a-3p (r=-0.8455, P < 0.001) in PE placenta tissues (). The expression of circ_0004904 was effectively knocked down by si-circ_0004904, relative to si-NC group (). Knockdown of circ_0004904 induced the upregulation of miR-19a-3p, while overexpression of circ_0004904 downregulated the level of miR-19a-3p in JEG-3 and HTR-8/SVneo cells (). All in all, circ_0004904 exhibited the sponge effect on miR-19a-3p.
Figure 3. Circ_0004904 sponged miR-19a-3p in trophoblast cells. (a) the binding sites between circ_0004904 and miR-19a-3p in starbase. (b-e) the binding analysis between miR-19a-3p and circ_0004904 was implemented via dual-luciferase reporter assay (b-c) and RIP assay (d-e). (f) the miR-19a-3p expression detection was conducted via RT-qPCR in PE and normal samples. (g) the relationship between circ_0004904 and miR-19a-3p was analyzed via Pearson’s correlation coefficient. (h) the knockdown efficiency of si-GADD45A was measured via RT-qPCR. (i) the miR-19a-3p quantification was performed by RT-qPCR in si-NC, si-circ_0004904, pCD5-ciR, circ_0004904 groups. Experiments were repeated for three times with three parallels. ***P <0.001, ****P <0.0001.
Circ_0004904 suppressed trophoblast cell behaviors through inhibiting miR-19a-3p
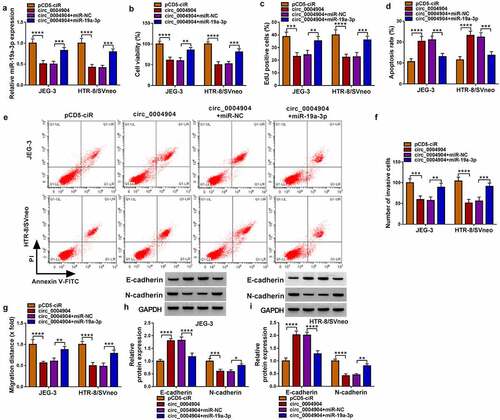
The further experiments were conducted to analyze whether miR-19a-3p sponge function was responsible for circ_0004904 function. The miR-19a-3p level was obviously elevated in circ_0004904+miR-19a-3p group relative to circ_0004904+miR-NC group (). The transfection of miR-19a-3p has abolished the inhibition of viability () and proliferation (), as well as the promotion of apoptosis () caused by circ_0004904 in JEG-3 and HTR-8/SVneo cells. The circ_0004904-mediated inhibitory effects on invasion () and migration () were partly lightened by miR-19a-3p overexpression. Also, co-transfection of circ_0004904 and miR-19a-3p abrogated the protein level changes of E-cadherin and N-cadherin in alone circ_0004904 transfection group (). Thus, circ_0004904 played a pathogenic role in PE via targeting miR-19a-3p.
Figure 4. Circ_0004904 suppressed trophoblast cell behaviors through inhibiting miR-19a-3p. JEG-3 and HTR-8/svneo cells were transfected with pCD5-ciR, circ_0004904, circ_0004904+mir-NC or circ_0004904+mir-19a-3p. (a) RT-qPCR was used for miR-19a-3p level analysis. (b-c) CCK-8 assay and EdU assay were used for assessment of cell viability (b) and proliferation (c). (d-e) Flow cytometry was used for detection of cell apoptosis rate. (f-g) Transwell assay and wound healing assay were used for examination of cell invasion (f) and migration (g). (h-i) Western blot was used for protein determination of E-cadherin and N-cadherin. Experiments were repeated for three times with three parallels. *P <0.05, **P <0.01, ***P <0.001, ****P <0.0001.
GADD45A worked as a target of miR-19a-3p
GADD45A 3”UTR was predicted to contain the binding sites of miR-19a-3p in starbase (). The miR-19a-3p upregulation reduced the luciferase activity of WT-GADD45A 3”UTR group instead of MUT-GADD45A 3’UTR group in JEG-3 and HTR-8/SVneo cells (). The concurrent enrichment of miR-19a-3p and GADD45A by Ago2 protein further affirmed the interaction between miR-19a-3p and GADD45A (). GADD45A mRNA level was markedly elevated in PE tissues contrasted to control tissues (). Pearson’s correlation coefficient showed a negative relation (r = −0.8572, P < 0.001) between GADD45A and miR-19a-3p levels in PE samples (). The high level of GADD45A in PE was also validated by western blot (). The transfection efficiencies of miR-19a-3p and anti-miR-19a-3p were great in JEG-3 and HTR-8/SVneo cells, compared with miR-NC and anti-miR-NC groups (). Furthermore, GADD45A protein expression was downregulated or upregulated by miR-19a-3p overexpression or inhibition in JEG-3 and HTR-8/SVneo cells (). GADD45A was directly regulated by miR-19a-3p in a negative way.
Figure 5. GADD45A worked as a target of miR-19a-3p. (a) Starbase exhibited the binding sites between miR-19a-3p and GADD45A. (b-e) Dual-luciferase reporter assay (b-c) and RIP assay (d-e) were applied for analysis of interaction between miR-19a-3p and GADD45A in JEG-3 and HTR-8/svneo cells. (f) RT-qPCR was performed to detect the mRNA level of GADD45A in PE tissues. (g) Pearson’s correlation coefficient was employed to analyze the relation between GADD45A and miR-19a-3p. (h) GADD45A protein examination was performed using western blot in PE samples. (i) the miR-19a-3p and anti-miR-19a-3p transfection efficacies were assessed using RT-qPCR. (j) western blot was used for protein detection of GADD45A after JEG-3 and HTR-8/svneo cells were transfected with miR-NC, miR-19a-3p, anti-miR-NC or anti-miR-19a-3p. Experiments were repeated for three times with three parallels. ***P <0.001, ****P <0.0001.
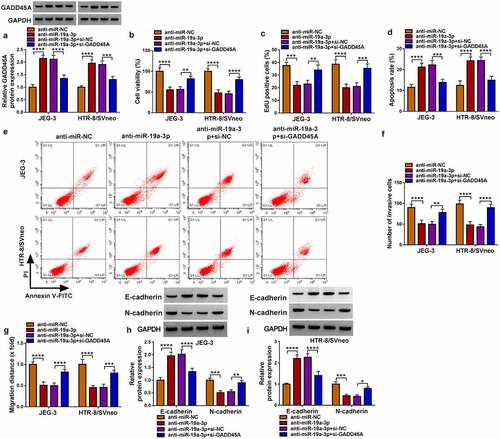
Inhibition of miR-19a-3p suppressed trophoblast cell development via increasing the GADD45A level
GADD45A protein upregulation induced by anti-miR-19a-3p was attenuated by si-GADD45A, showing that the knockdown efficiency of si-GADD45A was significant (). Cell viability () and proliferation () were repressed after transfection of anti-miR-19a-3p, which was then weakened by si-GADD45A. By performing flow cytometry, we found that the introduction of si-GADD45A relieved anti-miR-19a-3p-induced cell apoptosis in JEG-3 and HTR-8/SVneo cells (). The miR-19a-3p inhibitor reduced cell invasion (), migration () and EMT process (). while these impacts were counterbalanced with the downregulation of GADD45A. Altogether, miR-19a-3p repressor inhibited trophoblast cell behaviors by upregulating GADD45A.
Figure 6. Inhibition of miR-19a-3p suppressed trophoblast cell development via increasing the GADD45A level. JEG-3 and HTR-8/svneo cells were performed with transfection of anti-miR-NC, anti-miR-19a-3p, anti-miR-19a-3p+si-NC or anti-miR-19a-3p+gadd45a. (a) Western blot was conducted to examine the protein expression of GADD45A. (b-c) CCK-8 assay and EdU assay were applied to measure cell viability (b) and proliferation (c). (d-e) Flow cytometry was implemented to evaluate cell apoptosis. (f-g) Transwell assay and wound healing assay were performed to determine invasion (f) and migration (g). (h-i) E-cadherin and N-cadherin protein analysis was carried out via western blot. Experiments were repeated for three times with three parallels. *P <0.05, **P <0.01, ***P <0.001, ****P <0.0001.
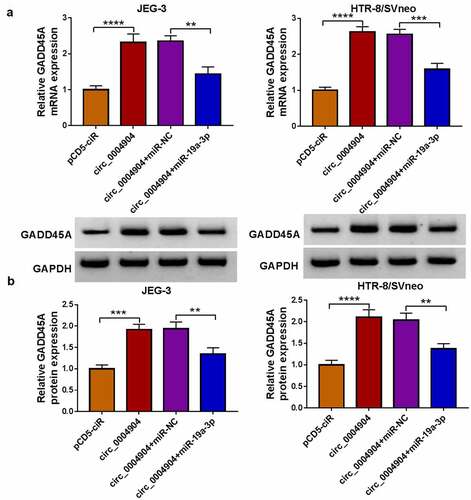
Circ_0004904 regulated GADD45A by targeting miR-19a-3p in trophoblast cell development
The effect of circ_0004904 on GADD45A was assessed via RT-qPCR and western blot. As depicted in (), circ_0004904 overexpression evoked the upregulation of GADD45A mRNA and protein levels but the subsequent miR-19a-3p transfection attenuated these influences in JEG-3 and HTR-8/SVneo cells. The above data manifested that circ_0004904 regulated the level of GADD45A via targeting miR-19a-3p in trophoblast cells. Moreover, GADD45A knockdown reversed the effects of circ_0004904 on cell viability, proliferation, apoptosis, invasion and migration (Supplementary Fig. S2). Thus, circ_0004904 was involved in the development of trophoblast cells via miR-19a-3p/GADD45A axis.
Figure 7. GADD45A was upregulated by circ_0004904/mir-19a-3p axis. (a-b) GADD45A mRNA and protein levels were measured by RT-qPCR and western blot in pCD5-ciR, circ_0004904, circ_0004904+mir-NC or circ_0004904+mir-19a-3p transfection group. Experiments were repeated for three times with three parallels. **P <0.01, ***P <0.001, ****P <0.0001.
Discussion
The abnormal trophoblast development is the critical factor in the etiology of PE [Citation16]. The current study demonstrated that circ_0004904 prevented the normal development of trophoblast cells by binding to miR-19a-3p to upregulate the GADD45A level, contributing to the understanding of pathogenic mechanism in PE.
The inadequate invasion and growth of trophoblast cells lead to the occurrence and progression of PE [Citation17,Citation18]. The differentially expressed circRNAs are often implicated in the pathogenesis of PE [Citation19]. Zhang et al. showed that circCRAMP1 L promoted cell proliferation and invasion in trophoblasts, and circCRAMP1 L downregulation was related to an increased risk of PE [Citation20]. CircHIPK3 facilitated the trophoblast behaviors in PE [Citation21]. Shen et al. stated that high expression of circTRNC18 reduced trophoblast cell migration and EMT process in PE [Citation22]. The significant upregulated level of circ_0004904 was detected in our placenta specimens collected from PE patients. In vitro results manifested that the overexpression of circ_0004904 repressed proliferation, invasion and migration abilities, as well as EMT of trophoblastic cells. These findings could provide evidence for circ_0004904 as a pathogenic molecule to inhibit the trophoblast development in PE.
Then, we discovered the target interaction of circ_0004904 with miR-19a-3p. Interestingly, circ_0004904 resulted in the direct inhibition of miR-19a-3p level in trophoblast cells. Moreover, all effects of circ_0004904 on the biological behaviors of trophoblasts were partly counteracted by miR-19a-3p. CircRNA/miRNA analysis was widely discovered in human diseases. The upregulation of circRNA -000,911 level promoted apoptosis and reduced migration of breast cancer cells via sponging miR-449a [Citation23]. Circ-CDYL acted as a sponge for miR-1180 to accelerate cell growth of multiple myeloma [Citation24]. Herein, circ_0004904 regulated trophoblast cell behaviors in PE via sequestering miR-19a-3p.
For miR-19a-3p, GADD45A was validated as a downstream target and GADD45A knockdown eliminated the inhibitory effects of miR-19a-3p inhibitor on trophoblast cell development. The small miRNAs are also vital regulators in PE. Su et al. suggested that miR-200 family suppressed invasion and EMT in trophoblasts through targeting ZEB1 [Citation25]. Yang et al. indicated that miR-215-5p attenuated the metastatic capacity of trophoblast cells via inhibiting the CDC6 level [Citation26]. Also, miR-19a-3p promoted the trophoblast behaviors by mediating the GADD45A level.
Commonly, circRNAs can be involved in the crucial pathological processes of PE via the competing endogenous RNA (ceRNA) networks [Citation19]. Circ_0063517 promoted angiogenesis in the pathogenic process of PE by serving as a ceRNA to target miR-31-5p/ETBR axis [Citation27]. Circ_0081343 suppressed trophoblast cell apoptosis and enhanced cell migration via completely binding to miR-210-5p to regulate DLX3 [Citation28]. In addition, circ_0085296 restrained the migration and invasion potentials through the miR-144/E-cadherin axis in trophoblast cells of PE [Citation29]. Overexpression of circ_0004904 was shown to induce the upregulation of GADD45A in trophoblast cells, which was achieved via the miR-19a-3p sponging effect. Hence, circ_0004904 was implicated in the PE progression partly by interacting with miR-19a-3p/GADD45A axis. However, this study has still some limitations. In addition to expression detection in PE patients, all functional experiments were performed in trophoblast cells. Circ_0004904/miR-19a-3p/GADD45A axis remains to be explored in freshly isolated trophoblast cells from PE patients or in vivo assays, which may provide more supports for circ_0004904 functional mechanism in PE progression. It will be performed in future study if possible.
Conclusion
In summary, circ_0004904 upregulated GADD45A by targeting miR-19a-3p to inhibit the development of trophoblast cells. The progression of PE was associated with circ_0004904/miR-19a-3p/GADD45A molecular network, at least in part. The circ_0004904 could be a potential biomolecule to improve the clinical management of PE.
Supplemental Material
Download Zip (1.1 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384101.2022.2124616
Additional information
Funding
References
- Rana S, Lemoine E, Granger JP, et al. Preeclampsia: pathophysiology, challenges, and perspectives. Circ Res. 2019;124(7):1094–1112. DOI:10.1161/CIRCRESAHA.118.313276
- Han C, Chen Y-Y, Dong J-F. Prothrombotic state associated with preeclampsia. Curr Opin Hematol. 2021;28(5):323–330.
- Chourdakis E, Oikonomou N, Fouzas S, et al. Preeclampsia emerging as a risk factor of cardiovascular disease in women. High Blood Press Cardiovasc Prev. 2021;28(2):103–114. DOI:10.1007/s40292-020-00425-7
- Staff AC. The two-stage placental model of preeclampsia: an update. J Reprod Immunol. 2019;134-135:1–10.
- Lee CQ, Gardner L, Turco M, et al. What is trophoblast? A combination of criteria define human first-trimester trophoblast. Stem Cell Reports. 2016;6:257–272.
- Flint EJ, Cerdeira AS, Redman CW, et al. The role of angiogenic factors in the management of preeclampsia. Acta Obstet Gynecol Scand. 2019;98(6):700–707. DOI:10.1111/aogs.13540
- Xu M, Xie F, Tang X, et al. Insights into the role of circular RNA in macrophage activation and fibrosis disease. Pharmacol Res. 2020;156:104777.
- Li C, Li Q. Circular RNA circ_0111277 serves as ceRNA, targeting the miR-424-5p/nfat5 axis to regulate the proliferation, migration, and invasion of trophoblast cells in preeclampsia. Reprod Sci. 2021;29(3):923–935.
- Jiang M, Lash GE, Zhao X, et al. CircRNA-0004904, CircRNA-0001855, and PAPP-A: potential novel biomarkers for the prediction of preeclampsia. Cell Physiol Biochem. 2018;46(6):2576–2586. DOI:10.1159/000489685
- Dai W, Liu X. Circular RNA 0004904 promotes autophagy and regulates the fused in sarcoma/vascular endothelial growth factor axis in preeclampsia. Int J Mol Med. 2021;47. DOI:10.3892/ijmm.2021.4944
- Wang N, Li R, Xue M. Potential regulatory network in the PSG10P/miR-19a-3p/il1rap pathway is possibly involved in preeclampsia pathogenesis. J Cell Mol Med. 2019;23(2):852–864.
- Li FH, Han N, Wang Y, et al. Gadd45a knockdown alleviates oxidative stress through suppressing the p38 MAPK signaling pathway in the pathogenesis of preeclampsia. Placenta. 2018;65:20–28.
- Yang Y, Shang H. Silencing lncRNA-DGCR5 increased trophoblast cell migration, invasion and tube formation, and inhibited cell apoptosis via targeting miR-454-3p/gadd45a axis. Mol Cell Biochem. 2021;476:3407–3421.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408.
- Li B, Wang Z, Yang F, et al. miR449a5p suppresses CDK6 expression to inhibit cardiomyocyte proliferation. Mol Med Rep. 2021;23:14.
- Huppertz B. The critical role of abnormal trophoblast development in the etiology of preeclampsia. Curr Pharm Biotechnol. 2018;19(10):771–780.
- Ridder A, Giorgione V, Khalil A, et al. Preeclampsia: the relationship between uterine artery blood flow and trophoblast function. Int J Mol Sci. 2019;20(13):3263. DOI:10.3390/ijms20133263
- Gomes VCL, Sones JL. From inhibition of trophoblast cell invasion to proapoptosis: what are the potential roles of kisspeptins in preeclampsia? Am J Physiol Regul Integr Comp Physiol. 2021;321(1):R41–R48.
- Ping Z, Ai L, Shen H, et al. Identification and comparison of circular RNAs in preeclampsia. PeerJ. 2021;9:e11299.
- Zhang Y, Yang H, Zhang Y, et al. circCRAMP1L is a novel biomarker of preeclampsia risk and may play a role in preeclampsia pathogenesis via regulation of the MSP/RON axis in trophoblasts. BMC Pregnancy Childbirth. 2020;20(1):652. DOI:10.1186/s12884-020-03345-5
- Zhang Y, Cao L, Jia J, et al. CircHIPK3 is decreased in preeclampsia and affects migration, invasion, proliferation, and tube formation of human trophoblast cells. Placenta. 2019;85:1–8.
- Shen XY, Zheng LL, Huang J, et al. CircTRNC18 inhibits trophoblast cell migration and epithelial–mesenchymal transition by regulating miR-762/grhl2 pathway in pre-eclampsia. RNA Biol. 2019;16(11):1565–1573. DOI:10.1080/15476286.2019.1644591
- Wang H, Xiao Y, Wu L, et al. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-000911/mir-449a pathway in breast carcinogenesis. Int J Oncol. 2018;52(3):743–754. DOI:10.3892/ijo.2018.4265
- Chen F, Wang X, Fu S, et al. Circular RNA circ-CDYL sponges miR-1180 to elevate yes-associated protein in multiple myeloma. Exp Biol Med (Maywood). 2020;245(11):925–932. DOI:10.1177/1535370220918191
- Su MT, Tsai PY, Wang CY, et al. Aspirin facilitates trophoblast invasion and epithelial-mesenchymal transition by regulating the miR-200-ZEB1 axis in preeclampsia. Biomed Pharmacother. 2021;139:111591.
- Yang X, Meng T. miR-215-5p decreases migration and invasion of trophoblast cells through regulating CDC6 in preeclampsia. Cell Biochem Funct. 2020;38(4):472–479.
- Li W, Yu N, Fan L, et al. Circ_0063517 acts as ceRNA, targeting the miR-31-5p-ETBR axis to regulate angiogenesis of vascular endothelial cells in preeclampsia. Life Sci. 2020;244:117306.
- Wang H, Luo C, Wu X, et al. Circular RNA hsa_circ_0081343 promotes trophoblast cell migration and invasion and inhibits trophoblast apoptosis by regulating miR-210-5p/dlx3 axis. Reprod Biol Endocrinol. 2021;19(1):123. DOI:10.1186/s12958-021-00795-0
- Zhu H, Niu X, Li Q, et al. Circ_0085296 suppresses trophoblast cell proliferation, invasion, and migration via modulating miR-144/E-cadherin axis. Placenta. 2020;97:18–25.
