ABSTRACT
Circular RNAs (CircRNAs) are reported to exert vital regulatory roles in the occurrence and development of various human malignancies, including non-small cell lung cancer (NSCLC). Bioinformatics methods identified the down-regulation of circSCN8A (circBase ID: hsa_circ_0026337) in NSCLC tissues. However, its biological functions and molecular mechanisms in NSCLC remain unknown. In this study, we found that circSCN8A expression was down-regulated in NSCLC tissues and cells. Low circSCN8A expression was positively associated with aggressive clinicopathological characteristics and poor prognosis in NSCLC patients. CircSCN8A suppressed cell proliferation, migration, invasion, and epithelial-mesenchymal transition (EMT) in vitro and blocked tumor growth in vivo. Moreover, circSCN8A promoted cell ferroptosis in NSCLC. Mechanistically, circSCN8A acted as a competing endogenous RNA (ceRNA) by sponging miR-1290 to enhance the expression of long-chain acyl-CoA synthetase-4 (ACSL4). Furthermore, the knockdown of ACSL4 or overexpression of miR-1290 reversed the effect of circSCN8A on facilitating ferroptosis and inhibiting cell proliferation and metastasis. In summary, circSCN8A represses cell proliferation and metastasis in NSCLC by regulating the miR-1290/ACSL4 axis to induce ferroptosis. Thus, circSCN8A may represent a promising therapeutic target against NSCLC.
Introduction
There are an estimated 2,206,771 lung cancer cases and 1,796,144 lung cancer deaths globally in 2020, representing 11.4% and 18.4% cancer incidence and mortality of in cancers, respectively [Citation1]. In addition to tobacco smoking, environmental and occupational exposures, chronic lung disease, and lifestyle factors are also associated with the development of lung cancer [Citation2]. Non-small cell lung cancer (NSCLC), including lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), and large cell carcinoma (LCC), occupies nearly 85% of lung tumors [Citation3]. Most patients with NSCLC are undiagnosed until this disease is late [Citation4]. Surgery, chemotherapy, radiotherapy, targeted therapy, and immunotherapy have been selectively applied to NSCLC treatments depending on the clinical situation, demonstrating the huge benefits for NSCLC patients [Citation5,Citation6]. However, the 5-year overall survival for NSCLC remains dismal due to distant metastasis, local recurrence, and drug resistance, from 68% in patients at stage IB to less than 10% in patients at stage IVA-IVB [Citation7]. To improve the survival outcomes in patients with NSCLC, there is an urgent need to investigate the pathological mechanisms underlying the progression of NSCLC and develop novel molecular targets.
Circular RNAs (circRNAs), generated by the back-splicing of pre-mRNA transcripts, are covalently closed single-stranded endogenous RNA molecules with no 5’-cap structure and 3’-poly(A) tail [Citation8]. There is accumulating evidence that circRNAs exert various biological functions by serving as transcriptional regulators, miRNA sponges, protein/peptide translation templates, as well as protein decoys, scaffolds, and recruiters [Citation9,Citation10]. Due to their high stability, abundance, and evolutionary conservation, and their presence in biological fluids, circRNAs have great potential as diagnostic biomarkers and pharmacological targets, opening up exciting possibilities for disease detection and treatment [Citation11]. With the development of high-throughput sequencing technology and bioinformatics, increasing circRNAs are found abundantly and differentially expressed in various human cancers [Citation12]. CircRNAs are implicated in the carcinogenesis and progression of multiple solid tumors and hematological malignancies [Citation13,Citation14]. CircRNAs are reported as tumor drivers or suppressors in lung cancer by regulating a series of pathophysiological events, such as cell proliferation, epithelial-mesenchymal transition (EMT), invasion, metastasis, apoptosis, autophagy, and chemoresistance [Citation15]. Based on the GEO microarray database, circSCN8A (circBase ID: hsa_circ_0026337) showed a decreased expression in NSCLC [Citation16]. However, the biological significance of circSCN8A in NSCLC remains elusive.
Ferroptosis is a novel defined form of regulated cell death driven by iron accumulation and lipid peroxidation, eventually causing mitochondrial damage and degradation of membrane integrity [Citation17]. Ferroptosis is intimately associated with the development and therapeutic responses of various human tumors [Citation18]. Recent studies demonstrate that non-coding RNAs (ncRNAs) play an important role in cancer progression by regulating ferroptosis [Citation19]. Ferroptosis-related circRNAs have been partially discovered by researchers, and circRNAs-based modulation of ferroptosis could be a promising approach to treat cancer [Citation20]. Here, an experimental study on the effect of circSCN8A on ferroptosis was also performed.
Among the different mechanisms circRNAs action in human cancers, competitive endogenous RNAs (ceRNAs), also known as miRNA “sponges” or “decoys”, is the best characterized [Citation21]. CircRNAs can competitively bind to miRNAs by base complementarity with miRNA response elements (MREs), thereby preventing miRNAs from binding to their target sites and increasing the expression of corresponding target mRNAs [Citation22]. There have been numerous reports about the circRNA-miRNA-mRNA regulation networks in lung cancer [Citation23]. For instance, circKEAP1 was significantly downregulated in LUAD and suppressed the progression of LUAD by sponging miR-141-3p to activate the KEAP1/NRF2 signal pathway [Citation24]. Circ_0003222 was highly expressed in NSCLC tissues and facilitated cell proliferation, migration, invasion, stemness-like properties, and chemoresistance by regulating miR-527/PHF21B axis [Citation25]. Nonetheless, it remains to be determined whether circSCN8A also participates in the malignant progression and ferroptosis of NSCLC in a ceRNA regulatory cascade.
Our study mainly focused on the biological roles of circSCN8A in NSCLC malignant progression and its relationship to ferroptosis. Decreased circSCN8A expression was observed in NSCLC tissues and cells. CircSCN8A overexpression suppressed cell proliferation and metastasis and induced ferroptosis in vitro. Moreover, circSCN8A repressed NSCLC tumorigenesis in vivo. Mechanistically, circSCN8A served as a sponge of miR-1290 to abate miRNA inhibition of target ACSL4, thus inhibiting NSCLC progression and accelerating ferroptosis. Our findings provide the experimental foundation for the application of circSCN8A to NSCLC treatment.
Materials and Method
Sample collection
A total of 64 patients who were histopathologically diagnosed as NSCLC the Affiliated Cancer Hospital of Zhengzhou University were enrolled in this study. None of the participants received preoperative adjuvant therapy. The clinicopathological characteristics of these patients are listed in . Tumor tissues and paired paracancerous tissues (3 cm away from the margin of neoplastic tissues), which were obtained by surgical resection, were quick-frozen and preserved at −80°C for further use. This study was permitted by the Research Ethics Committee of the Affiliated Cancer Hospital of Zhengzhou University and was performed in conformity to the Declaration of Helsinki. All patients agreed to the use of their lung tissues for experiment research and signed written informed consents.
Table 1. Correlation between circSCN8A expression and clinicopathological features in 64 patients with NSCLC.
Cell culture
Normal human bronchial epithelial cell line (BEAS-2B) and NSCLC cells (A549, H1299, NCI-H520, and SK-MES-1) were procured from the Cell Bank of Shanghai Institutes of Biological Sciences (Shanghai, China). BEAS-2B cells were cultured in BEGMTM BulletKit TM (Lonza, GA, USA). A549, H1299, and NCI-H520 cells were maintained in RPMI-1640 medium (Gibco, Waltham, MA, USA), and SK-MES-1 cells were cultured in Minimal Essential Medium (Gibco). Growth media for NSCLC cells was supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. Cell culture was performed in a 37°C incubator with 5% CO2. All cells were routinely tested for mycoplasma contamination. Cells were passaged every 2–3 days and exponentially growing cells were used for follow-up experiments.
Cell transfection and treatment
To construct circSCN8A-overexpressing plasmid (circSCN8A), the full-length cDNA sequences of circSCN8A (exon 21 to exon 25 of SCN8A gene) were synthesized and cloned into pLCDH-ciR vector (Geneseed, Guangzhou, China). pLCDH-ciR vector (vector), containing a front and back circular frame to facilitate RNA cyclization, was used as a negative control. Small interfering RNA (siRNA) targeting the back-splicing junction sites of circSCN8A (si-circSCN8A), miR-1290 mimics (miR-1290), miR-1290 inhibitor (inh-miR-1290), and their corresponding negative control (si-NC/miR-NC/inh-NC) were designed by GenePharma (Shanghai, China). siRNA specially targeting ACSL4 (si-ACSL4) (GenePharma) was used to interfer the expression of ACSL4. RNA oligonucleotides and plasmids were transfected into A549 or SK-MES-1 cells at a final concentration of 20 nM by using Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA). At 6 h after transfection, the culture medium was refreshed. After incubation for 48 h under standard conditions, cells were subjected to qRT-PCR to determine the transfection efficiency. To observe cell death, Erastin, ferrostatin-1 (Fer-1), ZVAD-FMK, and necrostatin-1 (Nec-1) were purchased from Sigma-Aldrich St. Louis, MO, USA)
RNA and genomic DNA (gDNA) isolation
Total RNA extraction was performed in NSCLC tissues or cells by using TRIzol reagent (Invitrogen). The gDNA was extracted from NSCLC cells using a genomic DNA Isolation Kit (Sangon Biotech, Shanghai, China). Cytoplasmic and nuclear RNA extracts of NSCLC cells were prepared using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific, Rockford, IL, USA). All these experimental procedures were performed as per the manufacturer’s guide.
Quantitative real-time PCR (qRT-PCR) and qPCR
The total RNA was reversely transcribed into cDNA by using the PrimeScript RT kit (Takara, Dalian, China). The PCR products from cDNA and gDNA were examined by using electrophoresis on a 2% agarose gel. RT-PCR reactions were performed using Platinum SYBR Green qPCR Super Mix UDG kit (Invitrogen) on an ABI 7500 Fast Real-Time PCR System (Applied Biosystems, CA, USA) under the following conditions: 95°C for 20 s, 45 cycles at 95°C for 10 s and 60°C for 30 s. GAPDH and U6 were respectively used as internal references for circRNA/mRNA and miRNA. The relative expression levels of circSCN8A, miR-1290, and ACSL4 were calculated via the 2−ΔΔCt formula. The specific primer sequences were listed in Table S1.
RNase R and Actinomycin D treatment
The 3 μg of total RNA from NSCLC cells was incubated with or without 5 U/μg RNase R (Qiagen, Valencia, CA, USA) for 30 min at 37°C, followed by the qRT-PCR analysis of circSCN8A and SCN8A mRNA. NSCLC cells were cultured in a complete medium with or without 2 μg/ml actinomycin D (Sigma-Aldrich) for 4, 8, 12, and 24 h, followed by RNA extraction and qRT-PCR analysis.
CCK-8 assay
A549 and SK-MES-1 cells were inoculated into 96-well plates at a density of 2 × 103 cells per well. At 24, 48, 72, or 96 h after inoculation, 10 μl of CCK-8 solution (Beyotime, Shanghai, China) was added to each well. After 2 h of incubation at 37°C, the optical density at 450 nm was measured by a microplate reader (Biotek, Winooski, VT, USA) for each sample.
5-Ethynyl-2’-deoxyuridine (EdU) assay
Cell-Light™ EdU Apollo 567 In Vitro Kit (RiboBio, Guangzhou, China) was used to detect the proliferation ability of A549 and SK-MES-1 cells. Cells (4 × 103 cells/well) were seeded into 96-well plates and incubated for 24 h. After that, 50 μM EdU solution was added to each well for 2 h of incubation. Then, cells were fixed with 4% paraformaldehyde for 15 min, permeabilized with 0.5% Triton X-100 for 10 min, and stained with Apollo fluorescent dyes for 30 min. Finally, Hoechst 33,342 solution was added and incubated for 30 min to stain the cell nuclei. EdU-positive cells were photographed under a fluorescence microscope and counted with Image J software.
Colony formation assay
A549 and SK-MES-1 cells were placed into 6-well plates with a concentration of 500 cells per well. Then, cells were allowed to grow in an incubator at 37°C for 10 days. The culture medium was refreshed every 2 days. After being fixed with 4% paraformaldehyde for 20 min and stained with 0.1% crystal violet for 20 min, cells were put under an optical microscope to count the cell colonies.
Wound healing assay
A549 and SK-MES-1 cells (5 × 105 cells/well) were seeded into 6-well plates and cultured in an FBS-containing medium to form a confluent cell monolayer. A sterile 200-μL pipette tip was used to gently scratch an artificial gap on the monolayer. Detached cells were removed by PBS washing. Then, cells were cultured in a fresh serum-free medium for 24 h to allow migration. Images at 0 h and 24 h were captured under an inverted microscope to calculate the percentage of wound closure.
Transwell assay
Transwell inserts (Corning, New York, NY, USA) with 8-μm pore size membranes were used to assess cell migration and invasion. For migration assay, 800 μl medium containing 10% FBS was added to the lower chamber. A549 and SK-MES-1 cells (5 × 104 cells) in a 200 μL medium containing 1% FBS were plated into the upper chamber for 24 h of incubation at 37°C. Cells that migrated to the lower surface of membranes were fixed with 95% ethanol and stained with 0.1% crystal violet. Finally, representative images were acquired under a microscope, and migration cells were counted from three randomly selected fields. For cell invasion assay, the inserts were pre-coated with Matrigel matrix (BD Science, Sparks, MD, USA) at a 1:6 dilution, and then incubated at 37°C for 2 h. The remaining procedures were similar to those of the migration assay.
Western blot assay
Western blot analysis was performed according to a previous report [Citation26]. The antibodies used in this study were listed as follows: anti-E-cadherin (#3195), anti-N-cadherin (#13116), and anti-Vimentin (#5741) (Cell Signaling Technology, Danvers, MA, USA); anti-GPX4 (ab125066), anti-SLC7A11 (ab175186), and anti-ACSL4 (ab155282) (Abcam, Cambridge, MA, USA); anti-GAPDH (#2118) and anti-rabbit secondary antibody (#7074) (Cell Signaling Technology).
Cellular death assay
Cell Death Detection ELISA Plus kit (Roche, Basel, Switzerland) was used to determine the cellular death following the manufacturer’s instructions.
Detection of Fe2+, MDA, and GSH
An Iron Assay Kit (ab83366, Abcam) was used to determine the intracellular Fe2+ level. The MDA level was measured by using Lipid Peroxidation (MDA) Assay Kit (ab118970, Abcam). The intracellular GSH level was quantified by using GSH/GSSG Ratio Detection Assay Kit (ab138881, Abcam).
Measurement of reactive oxygen species (ROS)
The intracellular ROS level was detected by using a fluorescent probe 2’,7’-dichlorofluorescein-diacetate (DCFH-DA, Beyotime, Shanghai, China). A549 and SK-MES-1 cells were seeded into 6-well plates at a density of 5 × 105 cells/well and incubated for 24 h at 37°C and 5% CO2. Then, cells were incubated with a serum-free medium containing 10 µM DCFH-DA at 37°C for 30 min. After being washed with PBS 3 times, cells were subjected to a fluorescence microscope to analyze the fluorescence intensity.
Bioinformatics analysis
Microarray data, including GSE112214 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE112214), GSE158695 [Citation27], and GSE101684 [Citation28] were used to analyze the differentially expressed circRNAs (DECs) in NSCLC. Microarray data (GSE32863 [Citation29], GSE33532 [Citation30], GSE19188 [Citation31], and GSE85841 [Citation32]) were downloaded to analyze the differentially expressed mRNAs in NSCLC. CircInteractome (https://omictools.com/circinteractome-tool) and CircBank (http://www.circbank.cn/) were used to predict the miRNAs that might bind to circSCN8A. miRDB (http://mirdb.org/) was employed to identify the miRNAs that might interact with ACSL4. Ferroptosis-related genes were obtained from the FerrDb (http://www.zhounan.org/ferrdb/) database.
Dual-luciferase reporter assay
The wild-type or mutant sequence of circSCN8A or ACSL4 containing the miR-1290 binding sites were subcloned to downstream of the Renilla gene in the psiCHECK-2 luciferase reporter plasmid (Promega, Madison, WI, USA). A549 and SK-MES-1 cells were inoculated in 24-well plates at a density of 4 × 104 cells/well 24 h before transfection. Then, cells were co-transfected with constructed luciferase plasmids and miR-NC or miR-1290 using Lipofectamine 3000 (Invitrogen). After 48 h, the luciferase activity was evaluated by a Dual-Luciferase Assay System (Promega) and normalized to Renilla luciferase activity.
Biotin-coupled miRNA capture
A549 and SK-MES-1 cells were transfected with biotinylated miRNA mimics (Biotin-miR-1290) or nonsense control (Biotin-NC) (Ribio, Guangzhou, China). After 24 h, cells were harvested, lysed, and incubated with streptavidin magnetic beads (Thermo Fisher Scientific) at 4°C overnight. Then, the beads were washed and eluted, and the bound RNAs were extracted with TRIzol reagent (Invitrogen). Finally, qRT-PCR analysis was performed to assess the abundance of circSNC8A or ACSL4 in immunoprecipitated RNA.
Animal study
Male BALB/c nude mice (3–4 weeks, 16–20 g) were purchased from Vital River Laboratory (Beijing, China) and housed under standard conditions. After acclimatization for one week, mice were randomly divided into two groups (n = 6/group). A549 cells infected with lentivirus circSCN8A (Lv-circSNC8A) or control (Lv-vector) were selected in the presence of puromycin (1 μg/ml). A549 cells (7 × 106) with Lv-circSNC8A or Lv-vector were suspended in 100 μL PBS and respectively injected into each mouse at the right flank. Tumor sizes were monitored at the indicated time points, and tumor volumes were calculated following the formula: volume (mm3) = (width2 × length)/2. After 31 days, the mice were killed, and the tumors were resected for further examination. All animal protocols were approved by the Institutional Animal Care and Use Committee of the Affiliated Cancer Hospital of Zhengzhou University and complied with the Guide for the Care and Use of Laboratory Animals.
Immunohistochemistry (IHC) assay
Xenografted tumors from mice were fixed with 4% paraformaldehyde, embedded in paraffin, and spliced into 4-μm slices. IHC was performed to detect the expression of Ki-67, E-cadherin, ACSL4, and GPX4 proteins. The detailed experimental procedures were performed according to a previous publication [Citation33]. The immunostaining images were photographed using a light microscope. The antibodies against Ki-67 (ab238020), E-cadherin (ab231303), ACSL4 (ab155282), and GPX4 (ab125066) were purchased from Abcam.
Statistical analysis
All experiments were repeated at least three times. Data were presented as mean ± standard deviation (SD). Statistical analyses were performed by two-tailed Student’s t-test or one-way analysis of variance with Dunnett’s multiple comparison test using the GraphPad Prism 8. Pearson’s correlation coefficients were used to perform gene correlation analysis. The association between circSCN8A expression and clinicopathological variables in NSCLC patients was analyzed by the Chi square test. The survival curves were constructed using the Kaplan-Meier method and compared with the log-rank test. P < 0.05 denotes a statistically significant difference (*P < 0.05, **P < 0.01, ***P < 0.001).
Results
CircSCN8A expression is down-regulated in NSCLC tissues and cells
We first searched for the differentially expressed circRNAs in NSCLC by analyzing 3 microarray data (GSE112214, GSE158695, and GSE101684). With the cutoff criteria of |log2Fold Change (FC)| ≥ 1 and P < 0.05, 149 DECs (16 up-regulated and 133 down-regulated), 185 DECs (84 up-regulated and 101 down-regulated), and 410 DECs (236 up-regulated and 174 down-regulated) were respectively identified from GSE112214, GSE158695, and GSE101684 (). There were 5 common down-regulated DECs (hsa_circ_0008234, hsa_circ_0026337, hsa_circ_0049271, hsa_circ_0029426, and hsa_circ_0043256) in these 3 datasets ( and Table S2). Among these 5 DECs, only hsa_circ_0026337 and hsa_circ_0029426 were not systematically investigated in NSCLC. By using qRT-PCR, we confirmed the decrease of hsa_circ_0026337 and hsa_circ_0029426 expression in ten pairs of NSCLC tissues and adjacent non-cancerous tissues (). Considering the more significant down-regulation of hsa_circ_0026337 in NSCLC, it was selected as the focus for our study. Hsa_circ_0026337, located on chr12 (chr12:52180325–52188425), was generated by the back-splicing of exon 21–25 of the host gene SCN8A () and its spliced mature sequence length is 853 bp (). To identify the circularization of circSCN8A, the convergent primers and divergent primers were designed to amplify linear SCN8A and circSCN8A based on cDNA and gDNA from A549 and SK-MES-1 cells. As shown in , circSCN8A could only be amplified by divergent primers in the cDNA, but not in the gDNA. To confirm the stability of circSCN8A, RNase R and Actinomycin D treatment assays were performed in A549 and SK-MES-1 cells. CircSCN8A was resistant to RNase R, while linear SCN8A mRNA expression was significantly reduced following RNase R treatment (). Actinomycin D assay also showed that circSCN8A had a longer half-life compared with the linear transcript (). Nucleocytoplasmic fractionation assays indicated that circSCN8A was mainly located in the cytoplasm (). qRT-PCR assays showed that circSCN8A expression was significantly lower in NSCLC tissues than that in adjacent paracancerous tissues (). Moreover, patients with low circSCN8A expression displayed larger tumor size, advanced TNM stage, and more lymph node metastasis (). Also, NSCLC patients with low circSCN8A expression showed a poorer overall survival than that with high circSCN8A expression (). Consistently, circSCN8A expression was decreased in NSCLC cells compared to normal bronchial epithelial cell line BEAS-2B (). These data suggested the potential involvement of circSCN8A in NSCLC progression and prognosis.
Figure 1. Expression and characterization of circSCN8A in NSCLC. (a) Volcano map showing the DECs in NSCLC according to the microarray data (GSE112214, GSE158695, and GSE101684). (b) Venn diagram showing five commonly down-regulated circRnas in GSE112214, GSE158695, and GSE101684. (c) the expression of hsa_circ_0026337 and hsa_circ_0029426 was determined by qRT-PCR in 10 pairs of NSCLC tissues and adjacent normal tissues. (d) Schematic diagram showing the circularization of hsa_circ_0026337 by back-splicing of SCN8A exons 21–25. (e) Detection of circSCN8A by agarose gel electrophoresis. (f) PCR was performed to amplify circSCN8A and its linear counterpart SCN8A by using convergent or divergent primers with cDNA or gDNA in NSCLC cells as the template. GAPDH acted as a control. (g and h) qRT-PCR analysis was used to assess the expression of circSCN8A and SCN8A mRNA in A549 and SK-MES-1 cells after treatment with RNase and actinomycin D. (i) Subcellular localization of circSCN8A in A549 and SK-MES-1 cells. (j) CircSCN8A expression in NSCLC tissues (n = 64) and corresponding paracancer tissues (n = 64). (k) Kaplan-Meier survival curve was plotted to evaluate the correlation between circSCN8A expression and overall survival of NSCLC patients. (l) CircSCN8A expression in NSCLC cells. *P <0.05, **P <0.01, ***P <0.001.
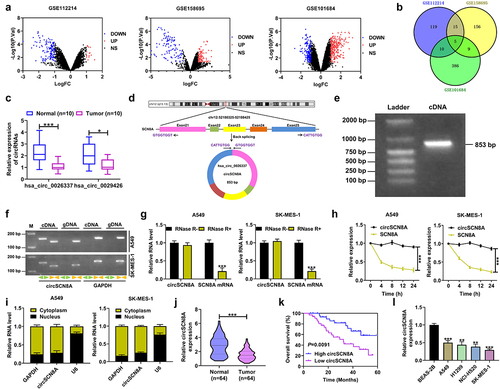
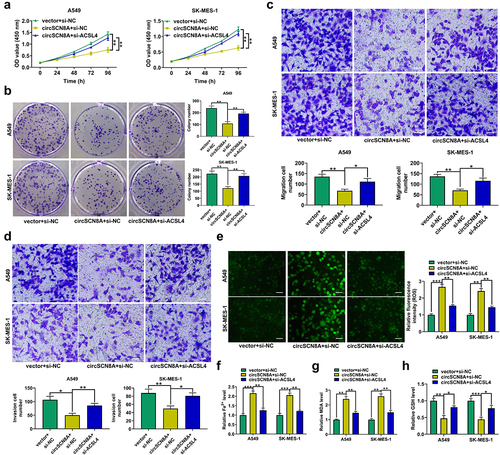
CircSCN8A inhibits cell proliferation and metastasis in NSCLC
To investigate the biological functions of circSCN8A in NSCLC, we overexpressed circSCN8A in A549 and SK-MES-1 cells (). CCK-8, colony forming, and EdU experiments demonstrated that overexpression of circSCN8A resulted in significant suppression of cell proliferation (). Wound healing assays showed a significant decrease in cell migration ability when circSCN8A was overexpressed (). As depicted by transwell assays, cell migration and invasion were significantly repressed due to circSCN8A overexpression (FandG). According to the data from western blot, increased E-cadherin expression and decreased N-cadherin and Vimentin expression were found in circSCN8A-overexpressing NSCLC cells (). Together, circSCN8A suppressed the aggressive phenotypes of NSCLC in vitro.
Figure 2. CircSCN8A inhibits the aggressive progression of NSCLC in vitro. (a) Transfection efficiency of circSCN8A-overexpressing plasmid in A549 and SK-MES-1 cells was determined by qRT-PCR. (b-d) CCK-8, colony formation, and EdU assays were performed to detect the proliferation ability of circSCN8A-overexpressing cells. (e) Wound healing assays were used to determine the effect of circSCN8A overexpression on cell migration. (f and g) Transwell assays were applied to determine the migration and invasion capability in cells with circSCN8A overexpression. (h) Western blot assays of EMT markers (e-cadherin, n-cadherin, and vimentin) in circSCN8A-overexpressing NSCLC cells. Scale bar: 50 μm. **P <0.01, ***P <0.001.
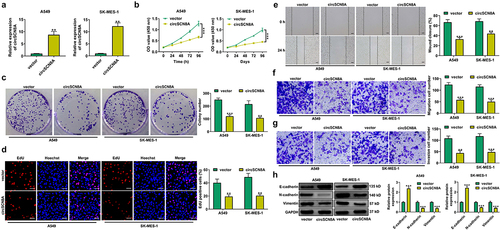
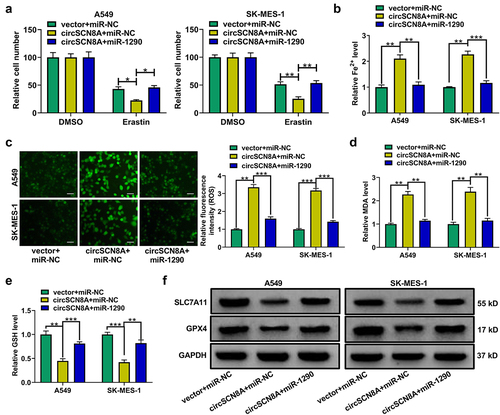
CircSCN8A induces ferroptosis in NSCLC cells
Next, we explored the effects of circSCN8A on cell death in NSCLC. As shown in AandB, cell death was significantly increased and cell viability was significantly suppressed by circSCN8A overexpression. To detect the cell death type caused by circSCN8A, circSCN8A-overexpressing NSCLC cells were treated with ZVAD-FMK (an apoptosis inhibitor), necrostatin-1 (Nec-1, a necroptosis inhibitor), or Ferrostatin-1 (Fer-1, a ferroptosis inhibitor). The results showed that circSCN8A-mediated cell death and growth inhibition was significantly reversed by Fer-1, but not ZVAD-FMK or Nec-1 (AandB), indicating that circSCN8A promoted cell death in NSCLC by inducing ferroptosis. To investigate the effect of circSCN8A on ferroptosis, circSCN8A-overexpressing cells were treated with erastin (a ferroptosis activator) or Fer-1. CCK-8 assays revealed that erastin-induced suppression of cell growth was reinforced by circSCN8A overexpression (). Moreover, circSCN8A overexpression increased Fe2+ (), ROS (), and malondialdehyde (MDA) () levels and decreased glutathione (GSH) content (), while these effects were greatly reversed by Fer-1 (). Western blot assays were also used to determine the effects of circSCN8A on the expression of ferroptosis-related proteins. SLC7A11 and GPX4 protein levels were suppressed by circSCN8A overexpression, while this effect was effectively weakened by Fer-1 (). These findings suggested that overexpressing circSCN8A could cause cells to undergo ferroptosis in NSCLC.
Figure 3. CircSCN8A induces ferroptosis in NSCLC cells. (a and b) CircSCN8A-overexpressing NSCLC cells were treated with ZVAD-FMK (10 μM), Nec-1 (10 μM), or Fer-1 (1 μM) for 24 h, followed by the determination of cell viability and cell death. (c and d) CircSCN8A-transfected NSCLC cells were treated with erastin (10 μM) or Fer-1 (1 μM) for 24 h, followed by CCK-8 assays of cell viability. (d-g) the levels of Fe2+ (d), ROS (e), MDA (f), and GSH (g) were measured in A549 and SK-MES-1 cells with circSCN8A overexpression. (h) Western blot assays were used to detect the protein expression of SLC7A11 and GPX4 in circSCN8A-overexpressing cells. Scale bar: 50 μm. *P <0.05, **P <0.01, ***P <0.001.
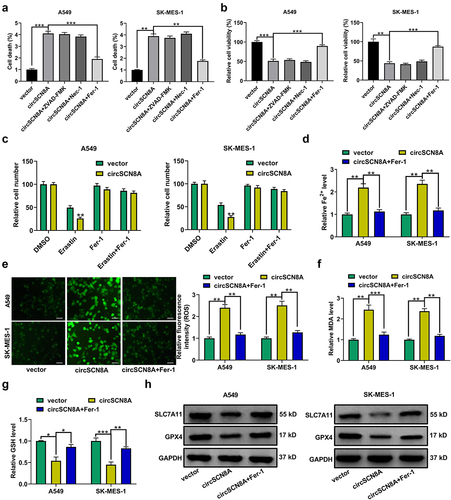
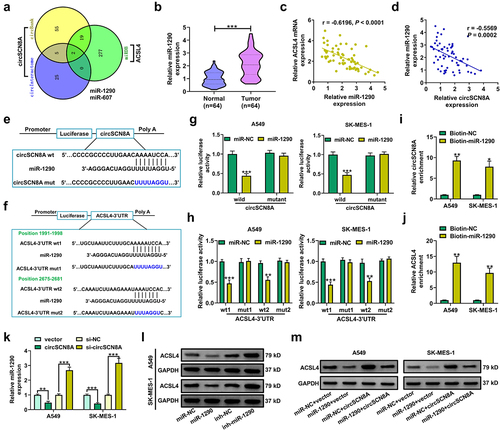
CircSCN8A promotes ACSL4 expression in NSCLC cells
Then, the molecular mechanisms by which circSCN8A regulates NSCLC progression and ferroptosis were further investigated. By analyzing the microarray data (GSE32863, GSE33532, GSE19188, and GSE85841), we found a total of 364 down-regulated mRNAs (). According to the intersection between down-regulated mRNAs in NSCLC and ferroptosis drivers from FerrDb database, we identified 6 candidate targets including EPAS1, ALOX5, CDO1, ACSL4, IL6, and ATF3 (). Then, qRT-PCR was used to determine the expression levels of these 6 mRNAs in A549 and SK-MES-1 cells with circSCN8A knockdown or overexpression. The results showed that the ACSL4 mRNA level was decreased by circSCN8A knockdown, and was promoted due to circSCN8A overexpression (CandD). Consistently, western blot assays demonstrated that circSCN8A knockdown inhibited ACSL4 protein expression (), and circSCN8A overexpression facilitated ACSL4 protein level (). TCGA analysis tools (UALCAN and starBase v2.0) showed that ACSL4 mRNA expression was abnormally down-regulated in NSCLC, including both LUAD and LUSC (Figure S1A and B). Compared with matched para-carcinoma tissues, ACSL4 mRNA level was decreased in tumor tissues from our cohort of NSCLC patients (). In addition, we observed a positive correlation between ACSL4 mRNA expression and circSCN8A expression in NSCLC tissues (). As presented by Kaplan-Meier Plotter, higher ACSL4 expression indicated a better prognosis for NSCLC patients (). By using western blot and IHC assays, we found lower ACSL4 protein expression in NSCLC tissues than that in adjacent normal tissues (JandK). These data suggested the promotive effect of circSCN8A on ACSL4 expression.
Figure 4. CircSCN8A facilitates ACSL4 expression in NSCLC cells. (a) Venn diagram showed the 364 down-regulated mRnas in NSCLC based on 4 datasets including GSE32863, GSE33532, GSE19188, and GSE85841. (b) the intersection between down-regulated mRnas in NSCLC and ferroptosis drivers from FerrDb database. (c and d) qRT-PCR assays of EPAS1, ALOX5, CDO1, ACSL4, IL6, and ATF3 mRNA expression in A549 and SK-MES-1 cells transfected with si-circSCN8A or circSCN8A. (e and f) Western blot assays of ACSL4 protein level in A549 and SK-MES-1 cells with circSCN8A knockdown or overexpression. (g) ACSL4 mRNA expression in 64 pairs of tumor tissues and adjacent non-cancerous samples. (h) Expression correlation between ACSL4 mRNA and circSCN8A in NSCLC tissues. (i) Kaplan-Meier plotter database presented the correlation between circSCN8A expression and prognosis in NSCLC patients. (j and k) Western blot and IHC assays were applied to assess the expression level of ACSL4 protein in NSCLC tissues and matched normal tissues. Scale bar: 100 μm. *P < 0.05, **P < 0.01, ***P < 0.001.
CircSCN8A suppresses NSCLC progression and induces ferroptosis by promoting ACSL4 expression
ACSL4 was previously reported as a tumor suppressor in LUAD by repressing tumor survival/invasiveness and inducing ferroptosis [Citation34]. To further explore whether circSCN8A exerted the anti-tumor roles in NSCLC by regulating ACSL4, we co-transfected circSCN8A and si-ACSL4 into A549 and SK-MES-1 cells. CircSCN8A-mediated inhibition of cell proliferation was significantly reversed by ACSL4 depletion (AandB). Also, knockdown of ACSL4 effectively attenuated the suppression of circSCN8A overexpression on cell migration and invasion (CandD). Moreover, circSCN8A-induced increase of ROS, Fe2+, and MDA levels and decrease in GSH content was greatly alleviated by ACSL4 down-regulation (). To sum up, circSCN8A inhibited cell malignant phenotypes and induces ferroptosis in NSCLC progression by facilitating ACSL4 expression.
Figure 5. CircSCN8A suppresses NSCLC progression and induces ferroptosis by promoting ACSL4 expression. A549 and SK-MES-1 cells were co-transfected with circSCN8A and si-ACSL4. (a and b) CCK-8 and colony formation assays of cell proliferation. (c and d) Transwell assays of cell migration and invasion. (e) DCFH-DA probe was used to detect the ROS level. (f-h) the commercial kits were used to determine the levels of Fe2+ (f), MDA (g), and GSH (h). Scale bar: 50 μm. *P <0.05, **P <0.01, ***P <0.001.
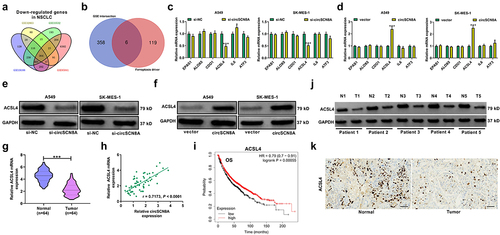
CircSCN8A increases ACSL4 expression by sponging miR-1290 in NSCLC cells
Considering the cytoplasmic localization of circSCN8A in NSCLC cells, we further explored whether circSCN8A promoted the ACSL4 expression through a ceRNA mechanism. With the help of bioinformatics tools (circBank, circInteractome, and miRDB), we discovered 2 miRNAs (miR-1290 and miR-607) containing the binding sites of both circSCN8A and ACSL4 (). miR-1290 is demonstrated as an oncogenic factor in multiple types of cancers, including lung cancer [Citation35]. Thus, it was selected as a focus to discuss. Higher expression of miR-1290 was found in tumor tissues than that in corresponding normal tissues from our cohort of NSCLC patients (). Interestingly, there was a negative correlation between ACSL4 and miR-1290 expression, as well as between miR-1290 and circSCN8A expression in NSCLC tissues (CandD). The binding sites between miR-1290 and circSCN8A or ACSL4 were shown in EandF. Then, the luciferase reporter assays and RNA pull-down experiments were performed to confirm the binding between miR-1290 and circSCN8A or ACSL4-3”UTR in A549 and SK-MES-1 cells. miR-1290 overexpression inhibited the luciferase activity of wild circSCN8A or ACSL4-3”UTR reporter but had no effects on the luciferase activity of mutant circSCN8A or ACSL4-3’UTR reporter (GandH). Moreover, circSCN8A and ACSL4 mRNA were significantly enriched by biotin-labeled miR-1290 compared with a negative control probe (IandJ). As demonstrated by qRT-PCR, miR-1290 expression was decreased in circSCN8A-overexpressing cells, while was increased in response to the knockdown of circSCN8A (). Western blot assays also revealed that ACSL4 protein expression was inhibited by miR-1290 overexpression, but was promoted due to miR-1290 inhibition (). Furthermore, circSCN8A-induced increase of ACSL4 protein level was reversed by the overexpression of miR-1290 (). Overall, the above results revealed that circSCN8A acted as a sponge for miR-1290 to promote the ACSL4 expression.
Figure 6. CircSCN8A serves as a sponge for miR-1290 to facilitate ACSL4 expression.(a) CircInteractome, circBank, and miRDB online databases were used to predict the potential miRnas that might bind to circSCN8A and ACSL4. (b) qRT-PCR analysis of miR-1290 expression in tumor tissues and adjacent normal tissues from NSCLC patients. (c and d) the correlation between miR-1290 expression and ACSL4 or circSCN8A in NSCLC tissues. (e) the predicted binding sites between miR-1290 and circSCN8A. (f) the putative binding sequences of miR-1290 on ACSL4-3”UTR. (g) the dual luciferase reporter assays were performed in A549 and SK-MES-1 cells after transfection with miR-NC or miR-1290 and wild or mutant circSCN8A reporter. (h) the luciferase activities were determined in A549 and SK-MES-1 cells co-transfected with miR-NC or miR-1290 and wild or mutant ACSL4-3”UTR reporter. (i and j) RNA pull-down assays were performed in A549 and SK-MES-1 cells by using a specific biotin-labeled miR-1290 probe, followed by the qRT-PCR to detect the enrichment of circSCN8A and ACSL4 mRNA. (K) qRT-PCR assays were performed to determine the effects of circSCN8A overexpression or knockdown on miR-1290 expression. (l) the ACSL4 protein expression was measured in A549 and SK-MES-1 cells with miR-1290 overexpression or knockdown. (m) the ACSL4 protein levels were determined in NSCLC cells transfected with miR-1290 or/and circSCN8A. *P <0.05, **P <0.01, ***P <0.001.
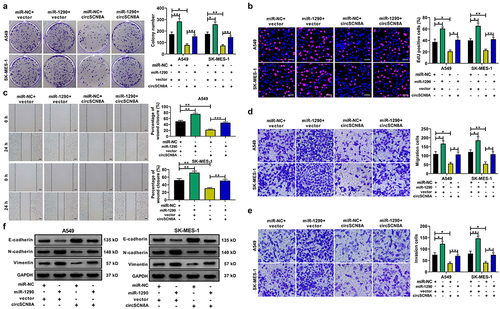
CircSCN8A suppresses cell proliferation and metastasis in NSCLC by sponging miR-1290
Subsequently, we further addressed whether the anti-tumor effects of circSCN8A in NSCLC was mediated by miR-1290. A549 and SK-MES-1 cells were transfected with miR-NC+vector, miR-1290+vector, miR-NC+circSCN8A, or miR-1290+circSCN8A. As presented in , overexpression of miR-1290 significantly promoted cell proliferation, migration, invasion, and EMT, suggesting the tumor-promotive role of miR-1290 in NSCLC. Moreover, circSCN8A-mediated suppression of cell proliferation, migration, invasion, and EMT was significantly abated by miR-1290 overexpression (). Based on the above results, we concluded that circSCN8A exerted a tumor-suppressive activity in NSCLC by down-regulating miR-1290.
Figure 7. CircSCN8A exerts a tumor-suppressive activity in NSCLC by sponging miR-1290.(a-e) A549 and SK-MES-1 cells were transfected with miR-NC+vector, miR-1290+vector, miR-NC+circSCN8A, or miR-1290+circscn8a. (a and b) Cell proliferation ability was assessed by using CCK-8 and colony formation assays. (c) Wound healing assay of cell migration. (d and e) Transwell assay of cell migration and invasion. (f) Western blot assay of E-cadherin, N-cadherin, and vimentin. Scale bar: 50 μm. *P <0.05, **P <0.01, ***P <0.001.
CircSCN8A induces ferroptosis in NSCLC cells by sponging miR-1290
We also investigated the effects of miR-1290 on ferroptosis in NSCLC cells. MiR-1290-overexpressing cells were treated with erastin or Fer-1. As shown in Figure S2A and B, miR-1290 overexpression abated erastin-induced growth inhibition of NSCLC cells. Overexpression of miR-1290 decreased intracellular levels of Fe2+ (Figure S2B), ROS (Figure S2C), MDA (Figure S2D) and increased GSH level (Figure S2E). Consistently, miR-1290 overexpression led to the increase of SLC7A11 and GPX4 protein levels (Figure S2F). These data suggested that miR-1290 induced NSCLC cells to resist ferroptosis. To explore whether circSCN8A-induced ferroptosis was associated with miR-1290, A549 and SK-MES-1 cells were co-transfected with circSCN8A and miR-1290. CCK-8 assays showed that circSCN8A overexpression aggravated erastin-induced inhibition of cell growth, while this effect was reversed by miR-1290 overexpression (). Moreover, circSCN8A-induced increase of Fe2+ (), ROS (), and malondialdehyde (MDA) () levels and decrease of glutathione (GSH) content () were greatly weakened by miR-1290 overexpression. Consistently, the decreased SLC7A11 and GPX4 protein levels mediated by circSCN8A overexpression was prominently restored due to the up-regulation of miR-1290 (). These results suggested that circSCN8A promoted ferroptosis in NSCLC cells by inhibiting miR-1290 expression.
Figure 8. CircSCN8A induces ferroptosis in NSCLC cells by inhibiting miR-1290. (a-f) A549 and SK-MES-1 cells were transfected with vector+mir-NC, circScn8a+mir-NC, or circScn8a+mir-1290. (a) Transfected NSCLC cells were treated with erastin (10 μM) for 24 h, followed by the CCK-8 assay of cell viability. (b-e) the levels of Fe2+ (b), ROS (c), MDA (d), and GSH (e) were measured in transfected cells. (f) Western blot assays of SLC7A11 and GPX4 protein expression in transfected cells. Scale bar: 50 μm. *P < 0.05, **P < 0.01, ***P < 0.001.
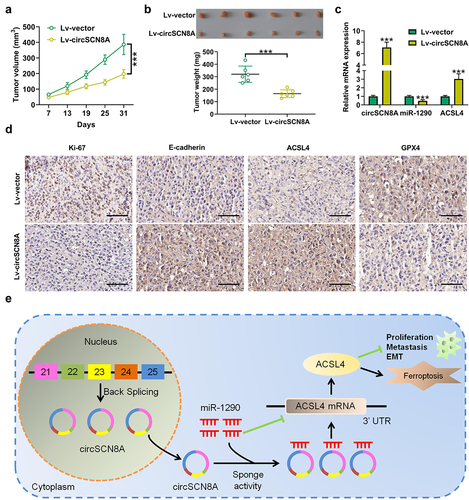
CircSCN8A represses the tumorigenesis of NSCLC in vivo
To further investigate the effects of circSCN8A on NSCLC tumor growth in vivo, A549 cells with Lv-circSNC8A or Lv-vector were subcutaneously injected into BALB/c nude mice. The xenograft tumor growth in the Lv-circSNC8A group was significantly slower than the control Lv-vector group (). Consistently, the weight of tumors from circSNC8A-overexpressing mice was lighter than that from control mice (). Then, these subcutaneous tumors were resected for further study. qRT-PCR indicated that circSNC8A and ACSL4 mRNA were higher, and miR-1290 expression was lower in circSNC8A-overexpressing tumors than in control tumors (). IHC assays revealed that overexpression of circSNC8A suppressed the expression of Ki-67 and GPX4 and promoted the expression of E-cadherin and ACSL4 (). These combined results indicated that the anti-tumor activity of circSNC8A in NSCLC in vivo were mediated by the miR-1290/ACSL4 axis.
Figure 9. CircSCN8A suppresses the growth of NSCLC cells in vivo. A549 cells transfected with Lv-circSCN8A or Lv-vector were subcutaneously injected into the right flanks of nude mice. (a) from the 7th day after cell inoculation, tumor growth was monitored every 6 days. (b) at 31 days after cell inoculation, tumors were excised for weight measurement. (c) the expression levels of circSCN8A, miR-1290, and ACSL4 mRNA in xenografted tumors. (d) IHC analysis of Ki-67, E-cadherin, ACSL4, and GPX4 in subcutaneous xenograft tumors. (e) Schematic diagram of the circScn8a/mir-1290/ACSL4 regulatory axis in NSCLC progression and ferroptosis. Scale bar: 100 μm. ***P <0.001.
Discussion
Although comprehensive therapy has brought unprecedented survival benefits for NSCLC patients, the overall cure and survival rates for NSCLC remain dismal, especially in metastatic disease [Citation36]. In lung cancer, a growing body of circRNAs are found to play vital roles in carcinogenesis, development, and response to different treatments [Citation37]. However, the potential effects of most circRNAs in NSCLC remain unclear.
In the present study, we identified a down-regulated circRNA (hsa_circ_0026337, circSCN8A) in NSCLC from GEO datasets and further investigated its biological function and action mechanism. CircSCN8A exhibited a lower expression level in NSCLC tissues and cells. Clinical investigation demonstrated that circSCN8A expression was negatively correlated with the pathological states of NSCLC patients. Lower circSCN8A expression indicated a poorer prognosis. Functionally, circSCN8A overexpression inhibited NSCLC cell proliferation and metastasis in vitro. The subcutaneous tumor model in nude mice confirmed the anti-cancer activity of circSCN8A in vivo. All these evidence indicated the tumor-suppressive effect of circSCN8A in NSCLC. SCN8A expression was reduced in tumor tissues of colorectal carcinoma. There were also meaningful relationships between the tumor grade, tumor location, histopathological classification, and SCN8A expression [Citation38]. However, the expression of SCN8A in NSCLC and its correlation with patient prognosis are needed to be explored in detail.
Ferroptosis, an iron-dependent form of nonapoptotic cell death, is intimately correlated with tumorigenesis, invasion, and metastasis in cancers [Citation39]. CircRNAs are reported to regulate the ferroptosis of tumor cells [Citation40]. Here, we found the increase of ROS, Fe2+, and MDA levels and the decrease of GSH level in circSCN8A-overexpressing cells. As the key regulators in cancer cell ferroptosis, SLC7A11 and GPX4 were also suppressed by circSCN8A. These data suggested the inductive effects of circSCN8A on ferroptosis in NSCLC.
According to the GEO datasets and FerrDb database, ACSL4 was predicted as a candidate target of circSCN8A. Here, we found that ACSL4 expression was decreased in NSCLC tissues, consistent with a previous publication [Citation34]. ACSL4 is identified as a biomarker and contributor of ferroptosis in cancer cells [Citation41]. ACSL4 was reported as a ferroptosis inducer in several human cancers, such as cervical cancer [Citation42] and hepatocellular carcinoma [Citation43]. As previously described, LUDA patients with low ACSL4 expression had a worse prognosis. Moreover, ACSL4 inhibited cell survival, invasion, and migration and induced ferroptosis in LUDA [Citation34]. In this study, ACSL4 expression was positively associated with circSCN8A. Moreover, ACSL4 expression was increased by circSCN8A overexpression and reduced by circSCN8A knockdown. Furthermore, circSCN8A-mediated inhibition of cell proliferation, migration, invasion, and EMT and increase of ferroptosis was reversed by ACSL4 knockdown. These data suggested that circSCN8A repressed NSCLC progression and caused ferroptosis by increasing ASCL4 expression.
CircRNAs in the cytoplasm are inclined to exert roles by posttranscriptional regulation, such as miRNA sponging [Citation44]. In the current study, we found that circSCN8A was predominantly localized in the cytoplasm. Thus, circSCN8A is likely to exert regulatory effects in NSCLC through miRNA sponge activity. Through online databases, miR-1290 was identified as the common miRNA of circSCN8A and ACSL4. Biotinylated miRNA pull-down and luciferase reporter assays validated the binding between miR-1290 and circSCN8A or ACSL4. In mechanistic experiments, circSCN8A acted as a sponge for miR-1290 to up-regulate ACSL4 expression. miR-1290 exerts an oncogenic function by promoting tumor initiation, growth, survival, angiogenesis, and metastasis in the vast majority of cancers [Citation45]. In NSCLC, serum and exosomal miR-1290 was revealed as a potential diagnostic and prognostic biomarker [Citation46,Citation47]. MiR-1290 was demonstrated as a tumor-promoting factor in NSCLC. For example, overexpression of miR-1290 promoted cell proliferation and invasion in NSCLC by targeting IRF2 [Citation48]. MiR-1290 overexpression contributed to tumor growth and metastasis in NSCLC by repressing MT1 G expression [Citation49]. In this study, we also found that miR-1290 overexpression facilitated NSCLC cell proliferation, migration, invasion, and EMT. Moreover, miR-1290 overexpression induced ferroptosis in NSCLC cells. Furthermore, circSCN8A-mediated suppression of cell malignant phenotypes and promotion of ferroptosis was greatly abrogated by the up-regulation of miR-1290. These findings suggested that circSCN8A inhibited NSCLC progression and induced ferroptosis by sponging miR-1290. Combining all these results together, we drew a conclusion that circSCN8A inhibited cell proliferation and metastasis and triggered ferroptosis in NSCLC by sponging miR-1290 to increase ACSL4 expression ().
Conclusion
In summary, our present study demonstrated that circSCN8A was down-regulated in NSCLC cells and tissues. Low expression of circSCN8A was associated with unfavorable clinicopathological features of NSCLC patients. CircSCN8A overexpression suppressed cell proliferation and metastasis and induced ferroptosis. Mechanistically, circSCN8A served as a sponge for miR-1290 to up-regulate ACSL4 expression. Our findings proposed circSCN8A as a ferroptosis inducer in NSLCL to repress cell proliferation and metastasis by miR-1290/ACSL4 axis. These data highlight the potential of circSCN8A as a prognostic biomarker and therapeutic target for NSCLC. Further animal and clinical experiments are required for applying circSCN8A to ferroptosis-inducing therapy in NSCLC.
Author contributions
Baoxing Liu and Wenqun Xing contribute to concept design and experiment implementation. Baoxing Liu and Haibo Ma are in charge of draft preparation and data interpretation. Xingyu Liu are responsible for chart plotting and statistical analysis. All authors approved the final submitted version.
Supplemental Material
Download MS Word (433.5 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384101.2022.2154543
Additional information
Funding
References
- Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: gLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. DOI:10.3322/caac.21660
- Bade BC, Dela Cruz CS. Lung Cancer 2020: epidemiology, etiology, and prevention. Clin Chest Med. 2020;41(1):1–24.
- Relli V, Trerotola M, Guerra E, et al. Abandoning the notion of non-small cell lung cancer. Trends Mol Med. 2019;25(7):585–594. DOI:10.1016/j.molmed.2019.04.012
- Kocher F, Hilbe W, Seeber A, et al. Longitudinal analysis of 2293 NSCLC patients: a comprehensive study from the TYROL registry. Lung Cancer. 2015;87:193–200.
- Duma N, Santana-Davila R, Molina JR. Non–small cell lung cancer: epidemiology, screening, diagnosis, and treatment. Mayo Clin Proc. 2019;94(8):1623–1640.
- Arbour KC, Riely GJ. Systemic therapy for locally advanced and metastatic non–small cell lung cancer: a review. JAMA. 2019;322(8):764–774.
- Goldstraw P, Chansky K, Crowley J, et al. The IASLC lung cancer staging project: proposals for revision of the TNM stage groupings in the forthcoming (Eighth) edition of the TNM classification for lung cancer. J Thorac Oncol. 2016;11(1):39–51. DOI:10.1016/j.jtho.2015.09.009
- Ebbesen KK, Hansen TB, Kjems J. Insights into circular RNA biology. RNA Biol. 2017;14(8):1035–1045.
- Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21(8):475–490.
- Zhou WY, Cai ZR, Liu J, et al. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer. 2020;19(1):172. DOI:10.1186/s12943-020-01286-3
- Verduci L, Tarcitano E, Strano S, et al. CircRnas: role in human diseases and potential use as biomarkers. Cell Death Dis. 2021;12(5):468. DOI:10.1038/s41419-021-03743-3
- Bach DH, Lee SK, Sood AK. Circular RNAs in cancer. Mol Ther Nucleic Acids. 2019;16:118–129.
- Kristensen LS, Hansen TB, Venø MT, et al. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565. DOI:10.1038/onc.2017.361
- Kristensen LS, Jakobsen T, Hager H, et al. The emerging roles of circRnas in cancer and oncology. Nat Rev Clin Oncol. 2022;19(3):188–206. DOI:10.1038/s41571-021-00585-y
- Chen HH, Zhang TN, Wu QJ, et al. Circular RNAs in lung cancer: recent advances and future perspectives. Front Oncol. 2021;11:664290.
- Zhang Q, Kang L, Li X, et al. Bioinformatics analysis predicts hsa_circ_0026337/mir-197-3p as a potential oncogenic ceRNA network for non-small cell lung cancers. Anticancer Agents Med Chem. 2022;22(5):874–886. DOI:10.2174/1871520621666210712090721
- Tang D, Chen X, Kang R, et al. Ferroptosis: molecular mechanisms and health implications. Cell Res. 2021;31(2):107–125. DOI:10.1038/s41422-020-00441-1
- Chen X, Kang R, Kroemer G, et al. Broadening horizons: the role of ferroptosis in cancer. Nat Rev Clin Oncol. 2021;18(5):280–296. DOI:10.1038/s41571-020-00462-0
- Balihodzic A, Prinz F, Dengler MA, et al. Non-coding RNAs and ferroptosis: potential implications for cancer therapy. Cell Death Differ. 2022;29(6):1094–1106. DOI:10.1038/s41418-022-00998-x
- Zhang X, Wang L, Li H, et al. Crosstalk between noncoding RNAs and ferroptosis: new dawn for overcoming cancer progression. Cell Death Dis. 2020;11(7):580. DOI:10.1038/s41419-020-02772-8
- Verduci L, Strano S, Yarden Y, et al. The circRna–microRNA code: emerging implications for cancer diagnosis and treatment. Mol Oncol. 2019;13(4):669–680. DOI:10.1002/1878-0261.12468
- Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRnas to regulate and control human cancer progression. Mol Cancer. 2018;17(1):79. DOI:10.1186/s12943-018-0827-8
- Liang ZZ, Guo C, Zou MM, et al. circRNA-miRNA-mRNA regulatory network in human lung cancer: an update. Cancer Cell Int. 2020;20(1):173. DOI:10.1186/s12935-020-01245-4
- Wang Y, Ren F, Sun D, et al. CircKEAP1 suppresses the progression of lung adenocarcinoma via the miR-141-3p/keap1/nrf2 Axis. Front Oncol. 2021;11:672586.
- Li C, Zhang J, Yang X, et al. Hsa_circ_0003222 accelerates stemness and progression of non-small cell lung cancer by sponging miR-527. Cell Death Dis. 2021;12(9):807. DOI:10.1038/s41419-021-04095-8
- Rugamba A, Kang DY, Sp N, et al. Silibinin regulates tumor progression and tumorsphere formation by suppressing PD-L1 expression in non-small cell lung cancer (NSCLC) cells. Cells. 2021;10(7):1632. DOI:10.3390/cells10071632
- Li B, Zhu L, Lu C, et al. circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nat Commun. 2021;12(1):295. DOI:10.1038/s41467-020-20527-z
- Zhao J, Li L, Wang Q, et al. CircRNA expression profile in early-stage lung adenocarcinoma patients. Cell Physiol Biochem. 2017;44(6):2138–2146. DOI:10.1159/000485953
- Selamat SA, Chung BS, Girard L, et al. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res. 2012;22(7):1197–1211. DOI:10.1101/gr.132662.111
- Meister M, Belousov A, Xu EC, et al. Intra-tumor heterogeneity of gene expression profiles in early stage non-small cell lung cancer. J Bioinf Res Stud. 2014;1:1.
- Hou J, Aerts J, den Hamer B, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS ONE. 2010;5(4):e10312. DOI:10.1371/journal.pone.0010312
- Yan H, Guan Q, He J, et al. Individualized analysis reveals CpG sites with methylation aberrations in almost all lung adenocarcinoma tissues. J Transl Med. 2017;15(1):26. DOI:10.1186/s12967-017-1122-y
- Liu T, Zhu J, Du W, et al. AKT2 drives cancer progression and is negatively modulated by miR-124 in human lung adenocarcinoma. Respir Res. 2020;21(1):227. DOI:10.1186/s12931-020-01491-0
- Zhang Y, Li S, Li F, et al. High-fat diet impairs ferroptosis and promotes cancer invasiveness via downregulating tumor suppressor ACSL4 in lung adenocarcinoma. Biol Direct. 2021;16(1):10. DOI:10.1186/s13062-021-00294-7
- Kalhori MR, Soleimani M, Arefian E, et al. The potential role of miR-1290 in cancer progression, diagnosis, prognosis, and treatment: an oncomiR or onco-suppressor microRNA? J Cell Biochem. 2022;123(3):506–531. DOI:10.1002/jcb.30191
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553(7689):446–454.
- Feng B, Zhou H, Wang T, et al. Insights into circRnas: functional roles in lung cancer management and the potential mechanisms. Front Cell Dev Biol. 2021;9:636913.
- Igci YZ, Bozgeyik E, Borazan E, et al. Expression profiling of SCN8A and NDUFC2 genes in colorectal carcinoma. Exp Oncol. 2015;37(1):77–80. DOI:10.31768/2312-8852.2015.37(1):77-80
- Shi Z, Zhang L, Zheng J, et al. Ferroptosis: biochemistry and Biology in Cancers. Front Oncol. 2021;11:579286–579286. DOI:10.3389/fonc.2021.579286.
- Xie B, Guo Y. Molecular mechanism of cell ferroptosis and research progress in regulation of ferroptosis by noncoding RNAs in tumor cells. Cell Death Discov. 2021;7(1):101.
- Yuan H, Li X, Zhang X, et al. Identification of ACSL4 as a biomarker and contributor of ferroptosis. Biochem Biophys Res Commun. 2016;478(3):1338–1343. DOI:10.1016/j.bbrc.2016.08.124
- Xiaofei J, Mingqing S, Miao S, et al. Oleanolic acid inhibits cervical cancer Hela cell proliferation through modulation of the ACSL4 ferroptosis signaling pathway. Biochem Biophys Res Commun. 2021;545:81–88.
- Lu Y, Chan YT, Tan HY, et al. Epigenetic regulation of ferroptosis via ETS1/miR-23a-3p/acsl4 axis mediates sorafenib resistance in human hepatocellular carcinoma. J Exp Clin Cancer Res. 2022;41(1):3. DOI:10.1186/s13046-021-02208-x
- Jarlstad OM, S Kristensen L, Hon CC. Circular RNAs as microRNA sponges: evidence and controversies. Essays Biochem. 2021;65(4):685–696.
- Guz M, Jeleniewicz W, Cybulski M. An Insight into miR-1290: an Oncogenic miRNA with Diagnostic Potential. Int J Mol Sci. 2022;23(3):1234.
- Mo D, Gu B, Gong X, et al. miR-1290 is a potential prognostic biomarker in non-small cell lung cancer. J Thorac Dis. 2015;7(9):1570–1579. DOI:10.3978/j.issn.2072-1439.2015.09.38
- Wu Y, Wei J, Zhang W, et al. Serum exosomal miR-1290 is a potential biomarker for lung adenocarcinoma. Onco Targets Ther. 2020;13:7809–7818.
- Jin JJ, Liu YH, Si JM, et al. Overexpression of miR-1290 contributes to cell proliferation and invasion of non small cell lung cancer by targeting interferon regulatory factor 2. Int J Biochem Cell Biol. 2018;95:113–120.
- Zhang WC, Chin TM, Yang H, et al. Tumour-initiating cell-specific miR-1246 and miR-1290 expression converge to promote non-small cell lung cancer progression. Nat Commun. 2016;7(1):11702. DOI:10.1038/ncomms11702
