ABSTRACT
As a type of lung cancer, non-small cell lung cancer (NSCLC) has the characteristics of high mortality and high recurrence rate, which poses a great threat to human life and health. Due to the high risk of surgical treatment and the slow recovery of wounds, non-coding RNAs, especially lncRNAs are used as new potential clinical prognostic markers to prevent and treat cancer in advance. This study aims to explore the role of FAM138B in NSCLC and its possibility as a prognostic biomarker. Real-timequantitative polymerase chain reaction (RT-qPCR) was used to detect the expression and overexpression level of lncRNA FAM138B (FAM138B) in cells and tissues. The CCK-8, Transwell migration and invasion methods were performed to observe the cell transfection.The interaction between FAM138B and miR-105-5p was predicted by the bioinformatics tool starBase v2.0, and verified by the luciferase reporter gene experiment. Kaplan-Meier and Cox regression analyses were used to determine the prognostic significance of FAM138B in NSCLC. The expression of FAM138B is down-regulated in NSCLC cells and tissues. Overexpression of FAM138B can inhibit the expression level of miR-105-5p in NSCLC cells, and the ability of NSCLC cells to proliferate, migrate and invade is downregulated. FAM138B targets miR-105-5p, and there is a negative correlation between FAM138B and miR-105-5p. It is confirmed that FAM138B inhibits the progression of NSCLC by targeting miR-105-5p and can be a potential prognostic biomarker for NSCLC.
Introduction
As one of the most threatening malignant tumors to human health and life, the incidence and mortality of lung cancer have increased rapidly in recent decades [Citation1–3]. According to the latest data released in December 2020, lung cancer is still the leading cause of cancer deaths. There are an estimated 1,796,144 deaths from lung cancer worldwide, accounting for 18% of all cancer deaths [Citation4]. According to reports, the 5-year survival rate of lung cancer is less than 20%, the 1-year survival rate is less than 50%, and the recurrence rate is 50% within 5 years [Citation5,Citation6]. Lung cancers are categorized as small cell lung cancer or non-small cell lung cancer (NSCLC), Among them, NSCLC accounts for about 85% of all lung cancer cases, including squamous cell carcinoma, adenocarcinoma, and large cell carcinoma [Citation7,Citation8]. Studies have shown that about 75% of NSCLC patients are in the advanced stages when they are discovered and the 5-year survival rate is very low [Citation9]. So far, the molecular cause involved in NSCLC is not fully understood.
In the treatment of lung cancer, except to conventional surgery for the treatment of lung cancer, the combination of chemotherapy and radiotherapy is also an effective clinical method [Citation10]. However, the traditional lung cancer treatment cycle is long, with obvious side effects, pain and discomfort. In addition, advanced lung cancer patients have a short survival time and a high recurrence rate, it is very urgent to find new and effective prognostic biomarkers and treatment strategies to improve the treatment of lung cancer. Studies have shown that upregulation of lncRNA DANCR is linked to poor survival of patients with lung cancer, and miR-216a is down-regulated by DANCR, indicating that lncRNA DANCR interference has great potential to reduce the progression of aggressive lung cancer [Citation11]. Besides, lncRNA XLOC_009167 was screened as a novel diagnostic biomarker, which is up-regulated in lung cancer tissues and cell lines and can be used to distinguish lung cancer from benign lung diseases and healthy controls [Citation12], and research by Zhang et al. also showed that lncRNA TUC338 promotes lung cancer invasion by activating the MAPK pathway [Citation13]. Since Li et al confirmed through research that lncRNA FAM138B may be a prognostic biomarker of lung adenocarcinoma, its clinical and biological functions have not been studied yet [Citation14]. In summary, lncRNA FAM138B may be regarded as a latent prognostic biomarker for NSCLC, and its expression, function and interaction with miR-105-5p can be explored.
Therefore, under the premise of sequentially detecting the expression of FAM138B in NSCLC cells and tissues, this study further studied the effect of lncRNA FAM138B on miR-105-5p, and emphasized its potential as a prognostic biomarker for NSCLC.
Materials and methods
Research objects
136 patients treated in The Second Hospital of Jilin University were the research subjects, and the investigation time was from September 2015 to September 2016. No chemotherapy or other treatment methods were operated on the patients participating in the study. The NSCLC tissues and adjacent normal tissues collected during surgery are confirmed by a professional pathologist and then cryopreserved. This study used monthly telephone interviews, home visits or consultations with relatives of patients to conduct a five-year follow-up study on NSCLC to ensure the accuracy and authenticity of the research results. The ethics committee of The Second Hospital of Jilin University agreed and approved the study, and all participating patients were required to sign written informed consent. The correlation of the lncRNA FAM138B expression with clinical characteristics in NSCLC is shown in .
Table 1. Correlation of the lncRNA FAM138B expression with clinical characteristics in NSCLC.
Cultivation of cell lines and construction of overexpression vector
The BeNa Culture Collection (Suzhou, China) provides H1299, HCC95, NCI-H650, SK-MES-1 and BEAS-2B cells. All cells were cultured in a humidified incubator at 37°C and 5% CO2, and DMEM medium (Gibco, NY, USA) with 10% fetal bovine serum was used. In order to carry out the transfection experiment, the above-mentioned NSCLC cells were seeded and cultured in a 6-well plate. MiR-105-5p mimic, inhibition of miR-105-5p, and their several controls (mimic NC, inhibitor NC) were synthesized by Ribobio (Guangzhou, China). LncRNA FAM138B, FAM138B overexpression vector (pcDNA3.1-FAM138B) and pcDNA3.1 empty vector (pcDNA3.1) were constructed by GenePharma (Shanghai, China). Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) was used for transfection experiments. And after 48 hours, the cells were collected for follow-up operations.
RNA extraction and RT-qPCR analysis
First, TRIZOL reagent (Beyotime, Shanghai, China) was used to extract total RNA from tissues and cells. Secondly, reverse transcription of the obtained RNA into corresponding cDNA, including lncRNA FAM138B for reverse transcription using PrimeScript RT kit (TaKaRa, Dalian, China); reverse transcription of miR-105-5p using TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). Then, SYBR Green Master PCR mix (Applied Biosystems, Foster City, CA, USA) was used to detect gene expression through the ABI 7900 system (Applied Biosystems, Foster City, CA, USA). Finally, the data was calculated using the 2−ΔΔCt method, which needs to be normalized with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or small nuclear RNA U6 in advance.
Cell proliferation, migration and invasion assay
NSCLC cells H1299 and SK-MES-1 were cultured in a 96-well plate (2 × 104 cells/well) for 48 hours at 37°C and 5% CO2 in a humidified environment, and then use the Cell Count Kit-8 reagent (Sigma Aldrich, St Louis, Missouri) to determine the viability of the cells. In a 96-well plate, CCK-8 reagent was added at 0, 24, 48, and 72 hours and incubated for 1 hour. A microplate reader (Thermo Fisher Scientific) was used to detect the absorbance at 450 nm to analyze the proliferation of NSCLC cells. Each experiment was repeated three times.
In the cell migration experiment, serum-free DMEM medium was added to the upper chamber of a 24-well transwell chamber (Multiskan MK3, Thermo, Waltham, MA, USA), and DMEM medium containing 10% fetal bovine serum was added to the lower chamber. The transfected cells were transferred to the upper chamber for 24 hours and fixed with 4% paraformaldehyde (PFA) for 30 minutes, and then the cells were moved into the lower chamber. The cell invasion experiment is the same as the migration experiment, except that the matrix gel (200 mg/mL, BD Biosciences, Franklin Lakes, NJ, USA) needs to be coated 30 minutes in advance in the upper chamber. Finally, the cells were stained with crystal violet, and 5 areas were randomly photographed and recorded with a microscope (Olympus, Tokyo, Japan).
Dual luciferase reporter analysis
After using the online tool starBase v2.0 (http://starbase.sysu.edu.cn/) to determine the possible target genes, the binding sites between miR-105-5p and FAM138B were analyzed. The amplified FAM138B sequence with miR-105-5p binding site inserted into the dualluciferase reporter vector pGLO (Promega, Madison, WI, USA) was called wild type (WT-FAM138B) and mutant type (MUT-FAM138B). Then, the constructed WT-FAM138B, MUT-FAM138B and miR-105-5p were co-transfected into H1299 cells, and the luciferase activity was detected by the dualluciferase reporter gene.
Statistical analysis
SPSS software (IBM Corporation, Armonk, New York, USA) was used for data analysis and processing. The expression level of FAM138B was determined by using the ENCORI Pan-Cancer Analysis Platform, which has cancer gene expression data downloaded from the TCGA project through the Genomic data Commons data Portal, and taking lung adenocarcinoma (LUAD) as an example. To ensure the accuracy of the research results, all experiments were performed at least three times, and the data were expressed as mean ± standard deviation (SD). The statistical difference analysis of the data was carried out by Student’s t test, χ2 test or one-way analysis of variance (ANOVA) method, and then the analysis between variables was evaluated by Tukey test. Kaplan-Meier analysis and Cox regression analysis were used to express the prognostic probability of FAM138B. P < 0.05 is statistically significant.
Results
The expression of FAM138B in NSCLC cells and the proliferation and culture of NSCLC cells
The expression of FAM138B in NSCLC cell lines (H1299, HCC95, NCI-H650, SK-MES-1) was downregulated than that of normal adjacent cells BEAS-2B by RT-qPCR (). In order to further study the role and function of FAM138B in NSCLC cells, the expression of pcDNA3.1- FAM138B in H1299 and SK-MES-1 cells was detected by RT-qPCR, and the results are shown in . The content of FAM138B was significantly increased, indicating that the overexpression of pcDNA3.1-FAM138B was successfully constructed. As shown in , the proliferation ability of FAM138B in H1299 and SK-MES-1 cells was detected by CCK-8 method. Compared with the control group, the absorbance of pcDNA3.1-FAM138B at 450 nm decreased significantly. Similarly, in H1299 and SK-MES-1 cells, the overexpression of FAM138B vector pcDNA3.1-FAM138B showed a decrease in migration and invasion levels compared to the control group (). Through experiments in H1299 and SK-MES-1 cells, it can be confirmed that overexpression of FAM138B can inhibit the proliferation, migration and invasion of NSCLC cells.
Figure 1. The expression levels of FAM138B were detected by RT-qPCR. Compared with normal neighboring cells BEAS-2B, its expression level was reduced. **P <0.01, ***P <0.001. Data were analyzed by one-way ANOVA.
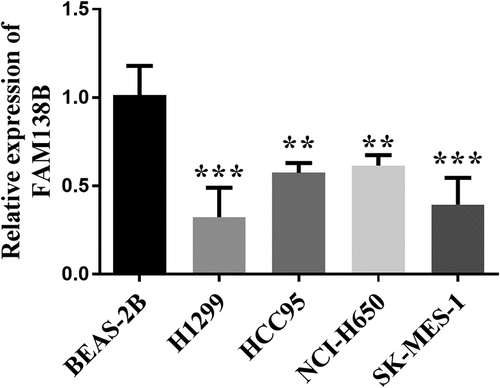
Figure 2. Overexpression of FAM138B and its inhibition of H1299 and SK-MES-1 cell proliferation, migration and invasion. (a) and (b) the relative expression of FAM138B in H1299 and SK-MES-1 cells transfected with pcDNA3.1-FAM138B was significantly up-regulated by RT-qPCR. (c) and (d) the CCK-8 method was used to detect the decreased proliferation ability of FAM138B in H1299 and SK-MES-1 cells. (e) to (h) Overexpression of FAM138B inhibited the migration and invasion levels of H1299 and SK-MES-1 cells compared with the control group. ***P <0.001. Data were analyzed by one-way ANOVA.
FAM138B targets miR-105-5p in NSCLC
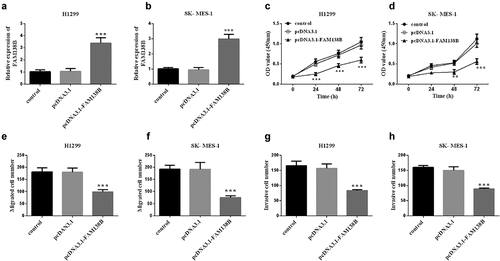
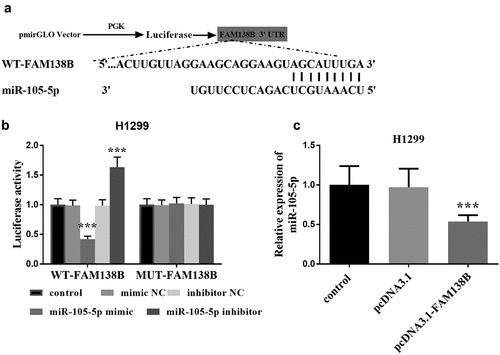
Taking H1299 cells in NSCLC as an example, we studied the targeting effect of FAM138B on miR-105-5p and the effects of the two on NSCLC. In , miR-105-5p and FAM138B form multiple base pairs, and the binding sites between miR-105-5p and FAM138B are shown, which can be known from bioinformatics analysis. By detecting the luciferase activity in H1299 cells, it can be seen that, as shown in , WT-FAM138B and MUT-FAM138B have different performance results. The luciferase activity of H1299 cells after transfection with MUT-FAM138B was not affected by the expression of miR-105-5p. However, transfection of WT-FAM138B significantly reduced the luciferase activity of H1299 cells due to the high expression of miR-105-5p. Similarly, inhibiting the expression of miR-105-5p would also significantly increase the luciferase activity of H1299 cells. In , the expression of miR-105-5p in H1299, pcDNA3.1-FAM138B was significantly decreased compared with the control group, which can be known by RT-qPCR.
Figure 3. Bioinformatics and dual-luciferase report analysis. (a) Bioinformatics analysis showed the binding site between miR-105-5p and FAM138B 3‘UTR. (b) Luciferase activity was examined in H1299 cell cotransfected with miR-105-5p mimic, miR-105-5p inhibitor, mimic NC or inhibitor NC and WT-FAM138B or MUT-FAM138B. (c) RT-qPCR analysis showed that overexpression of FAM138B would inhibit the expression of miR-105-5p in H1299 cells. ***P <0.001. Data were analyzed by one-way ANOVA.
Rescue experiments
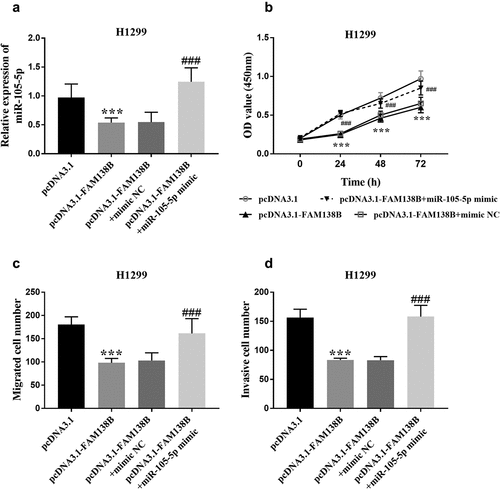
To further verify the relationship between FAM138B and miR-105-5p, H1299 cells were co-transfected with pcDNA3.1, pcDNA3.1-FAM138B, pcDNA3.1-FAM138B+mimic NC or pcDNA3.1-FAM138B+miR-105- 5p mimic. As shown in , pcDNA3.1-FAM138B+miR-105-5p mimic increased the relative expression of miR-105-5p compared with pcDNA3.1-FAM138B. shows that pcDNA3.1-FAM138B+miR-105-5p mimic counteracted the inhibitory effect of overexpression of FAM138B on the proliferation of H1299 cells. Similarly, pcDNA3.1-FAM138B+miR-105-5p mimic reversed the effects of overexpression FAM138B-mediated inhibition on the migration and invasion progression of H1299 cells ().
Figure 4. Rescue experiments confirmed that FAM138B targets miR-105-5p. (a) pcDNA3.1-FAM138B+miR-105-5p mimic increased the relative expression of miR-105-5p. (b)-(d) pcDNA3.1-FAM138B+miR-105-5p mimic reversed the effects of overexpression FAM138B-mediated inhibition on the proliferation, migration and invasion progression of H1299 cells. ***P <0.001, ###P <0.001. Data were analyzed by one-way ANOVA.
The prognostic effect of FAM138B in NSCLC
In , FAM138B expression was significantly decreased in LUAD samples compared with normal samples, and the sample data were obtained from starbase database. The expression of FAM138B in NSCLC tissues was also decreased compared to normal tissues (). Lymph node metastasis was associated with low expression of FAM138B (). The expression of FAM138B in NSCLC patients with TNM stage (III-IV) was lower than that in stage I-II (). Meanwhile, as shown in , through the χ2 test and data analysis, it can be seen that the downregulated expression of FAM138B is only related to the lymph node metastasis (P = 0.008) and TNM stage (P < 0.001), but not related to patient age, gender, tumor size, smoking status, and differentiation. shows the survival probability of lncRNA FAM138B high expression group within 5 years was higher than that of lncRNA FAM138B low expression group (log-rank P = 0.001). In order to evaluate the prognostic value of FAM138B, multivariate Cox analysis of clinical characteristics and overall survival shows that FAM138B can be used as an independent prognostic factor for NSCLC. Where in , HR = 3.077, 95% CI = 1.373–6.893, P = 0.006. The above studies confirmed that lncRNA FAM138B can be used as an independent prognostic factor of NSCLC, and the high expression of lncRNA FAM138B will have a positive impact on patients with NSCLC. According to , through the analysis of the correlation between FAM138B and miR-105-5p, the results showed a negative correlation, again verifying the following hypothesis that overexpression of FAM138B can inhibit the expression ability of miR-105-5p in NSCLC. According to the expression of lncRNA FAM138B in NSCLC tissues, all 136 NSCLC patients were divided into lncRNA FAM138B high expression group (n = 65) and lncRNA FAM138B low expression group (n = 71).
Figure 5. The expression of FAM138B in NSCLC tissues and the survival analysis of patients with high or low expression of FAM138B by Kaplan-Meier method. (a) the expression of FAM138B was significantly reduced in LUAD samples compared to normal samples provided by the starbase database. (b) the expression of FAM138B decreased in NSCLC tissues compared to normal tissues. (c) Lymph node metastasis was associated with low expression of FAM138B. (d) the expression of FAM138B in NSCLC patients with TNM stage (III-IV) was lower than that in stage I-II. (e) Within five years, the survival probability of NSCLC patients with high expression of FAM138B was higher than that of low expression of FAM138B (log-rank P = 0.001). (f) the expression of FAM138B and miR-105-5p was negatively correlated (r = −0.6916, P <0.0001). ***P <0.001. Data were analyzed by Student’s t-test. The correlation coefficient was determined using Pearson correlation. Survival analysis was performed using the log-rank test.
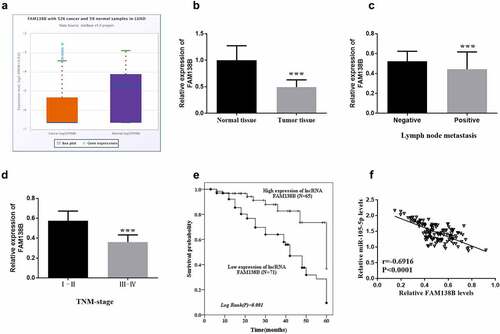
Table 2. Multivariate Cox analysis of clinical characteristics in relation to overall survival.
Discussion
With the worldwide industrialization, urbanization and environmental pollution, lung cancer has become a global problem and one of the most common cancers [Citation15]. Lung cancer is a complex disease composed of diverse histological and molecular types with clinical relevance [Citation16]. A large number of studies have demonstrated that long non-coding RNA (lnc RNA) is a novel type of cancer regulatory factor that controls basic biochemical and cellular processes [Citation11,Citation17]. In recent years, the regulatory effect of lncRNA on cancer has attracted the attention of more and more researchers.
Recent studies have shown that ZFHX3-MT can be used as a new predictive marker for NSCLC treatment, and it is positively correlated with known immunotherapy response biomarkers [Citation18]. In addition, it has also been confirmed that lncRNA DLEU2 is a new oncogene and can be used as a potential new target for NSCLC treatment [Citation19]. Xu et al also confirmed that in order to inhibit cell proliferation of NSCLC, LncRNA MIR503HG can be used to down-regulate Cyclin D1 to induce cell cycle arrest [Citation20]. LncRNA can be used as a tumor suppressor to play a role in NSCLC, and its clinical potential for NSCLC treatment has also been confirmed [Citation21]. In short, the role and function of LncRNA FAM138B in NSCLC can be taken as the research direction of this time to explore the influence of FAM138B and miR-105-5p on the progress of NSCLC.
The content of FAM138B in NSCLC cells and tissues is reduced, which is derived from this study. In addition, in NSCLC cells, overexpression of miR-1298 [Citation22], miR-3180-3p [Citation23], miR-545 [Citation24] and miR-186 [Citation25] will inhibit cell proliferation, migration and invasion. A large number of experiments are consistent with the results of this study that overexpression of FAM138B can inhibit the proliferation, migration and invasion of NSCLC cells. Studies have shown that in NSCLC patient specimens, linc00673 is negatively correlated with miR-150-5p, and overexpression of miR-150-5p inhibits the expression of lin00673 [Citation26]. Dong et al demonstrated that the expression of miR-105-5p could accelerate tumorigenesis as a biomarker for early diagnosis of NSCLC patients [Citation27]. Furthermore, the expression of miR-105-5p was shown to be altered in lung squamous cell carcinoma [Citation28]. This study also confirmed that miR-105-5p is negatively regulated by FAM138B, and high expression of FAM138B caused a significant decrease in the expression of miR-105-5p in NSCLC. In previous literature, miR-105-5p not only affected the immunogenicity of gastric cancer cells by regulating PD-L1 [Citation29], but also had a positive effect on the treatment of esophageal squamous cell carcinoma by mediating SPARCL1 and FAK/Akt signaling pathways [Citation30]. Therefore, we have reason to speculate that the miR-105-5p target gene may have an impact on the progression of NSCLC, which will also become the focus of follow-up research. Further studies have shown that the decreased expression of FAM138B in NSCLC is associated with lymph node metastasis and TNM staging is related. Xu et al found that the expression of miR-18a is closely related to clinical TNM staging, tumor differentiation and regional lymph node metastasis [Citation31]; and, Liu et al also confirmed that the decreased expression of Per1/2/3 in non-small cell lung cancer also involves high TNM staging, Lymph node metastasis [Citation32]. LncRNA CCDC144NL-AS1 is an independent prognostic factor of NSCLC, and patients with overexpression of lncRNA CCDC144NL-AS1 have a poor prognosis, which was confirmed by Zhang et al [Citation33]. The exploration of overexpression of lncRNA dle2 shows that lncRNA dle2 can also be an independent prognostic factor for poor survival of NSCLC patients [Citation34]. Meanwhile 14-3-3-3 zeta has also been confirmed as a biomarker for predicting the prognosis of patients with early NSCLC [Citation35]. In the same way, lncRNA FAM138B can be used as an independent prognostic factor of NSCLC, and lncRNA FAM138B can be a potential biomarker of NSCLC, which is proved by this experiment.
In order to ensure the accuracy and authenticity of the results of t
his experimental study, all the above experimental steps were repeated at least three times. At the same time, telephone return visits, on-site home visits, communication with the patients’ relatives, and consultations with the patients themselves were used to continuously conduct NSCLC patients within five years. Research to ensure that relevant information can be grasped in time, and the latest data can be summarized and updated. Admittedly, there are still some defects in this study, such as high treatment cost and relatively single target, insufficient attention has been paid to the mutated lncRNA FAM138B, and a large number of clinical experiments are needed to further confirm, so more in-depth participation is needed to realize the application.
In short, this study confirmed that lncRNA FAM138B targets miR-105-5p, and when FAM138B is highly expressed, it inhibits the progression of NSCLC, indicating that lncRNA FAM138B can be used as a potential prognostic marker for NSCLC.
Supplemental Material
Download PDF (715.9 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The corresponding author can provide relevant data for this study.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384101.2022.2154556.
Additional information
Funding
References
- Li X, Yuan N, Lin L, et al. Targeting cysteine-rich angiogenic inducer-61 by antibody immunotherapy suppresses growth and migration of non-small cell lung cancer. Exp Ther Med. 2018 Aug;16(2):730–738.
- Dang X, Zhao W, Li C, et al. Impact of COL6A4P2 gene polymorphisms on the risk of lung cancer: a case-control study. PLoS ONE. 2021;16(5):e0252082. DOI:10.1371/journal.pone.0252082
- Ding CZ, Guo XF, Wang GL, et al. High glucose contributes to the proliferation and migration of non-small cell lung cancer cells via GAS5-TRIB3 axis. Biosci Rep. 2018 Jan 24;38(2). DOI:10.1042/BSR20171014
- Yu F, Xiao R, Li X, et al. Combined effects of lung disease history, environmental exposures, and family history of lung cancer to susceptibility of lung cancer in Chinese non-smokers. Respir Res. 2021 Jul 23;22(1):210. DOI:10.1186/s12931-021-01802-z
- Wen P, Chidanguro T, Shi Z, et al. Identi?cation of candidate biomarkers and pathways associated with SCLC by bioinformatics analysis. Mol Med Rep. 2018 Aug;18(2):1538–1550.
- Monkam P, Qi S, Xu M, et al. CNN models discriminating between pulmonary micro-nodules and non-nodules from CT images. Biomed Eng Online. 2018 Jul 16;17(1):96. DOI:10.1186/s12938-018-0529-x
- Collins LG, Haines C, Perkel R, et al. Lung cancer: diagnosis and management. Am Fam Physician. 2007 Jan 1;75(1):56–63.
- Molina JR, Yang P, Cassivi SD, et al. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008 May;83(5):584–594.
- Sun W, Zhang L, Yan R, et al. LncRNA DLX6-AS1 promotes the proliferation, invasion, and migration of non-small cell lung cancer cells by targeting the miR-27b-3p/gspt1 axis. Onco Targets Ther. 2019;12:3945–3954.
- Wei T, Cheng S, Fu XN, et al. miR-219a-5p enhances the radiosensitivity of non-small cell lung cancer cells through targeting CD164. Biosci Rep. 2020 Jul 31;40(7). DOI:10.1042/BSR20192795
- Zhen Q, Gao LN, Wang RF, et al. LncRNA DANCR promotes lung cancer by sequestering miR-216a. Cancer Control. 2018 Jan-Mar;25(1):1073274818769849.
- Jiang N, Meng X, Mi H, et al. Circulating lncRNA XLOC_009167 serves as a diagnostic biomarker to predict lung cancer. Clin Chim Acta. 2018 Nov;486:26–33.
- Zhang YX, Yuan J, Gao ZM, et al. LncRNA TUC338 promotes invasion of lung cancer by activating MAPK pathway. Eur Rev Med Pharmacol Sci. 2018 Jan;22(2):443–449.
- Li X, Li B, Ran P, et al. Identification of ceRNA network based on a RNA-seq shows prognostic lncRNA biomarkers in human lung adenocarcinoma. Oncol Lett. 2018 Nov;16(5):5697–5708.
- Mao Y, Yang D, He J, et al. Epidemiology of lung cancer. Surg Oncol Clin N Am. 2016 Jul;25(3):439–445.
- Rodriguez-Canales J, Parra-Cuentas E, Wistuba II. Diagnosis and molecular classification of lung cancer. Cancer Treat Res. 2016;170:25–46.
- Loewen G, Jayawickramarajah J, Zhuo Y, et al. Functions of lncRNA HOTAIR in lung cancer. Journal of Hematology & Oncology. 2014 Dec 10;7(1):90. DOI:10.1186/s13045-014-0090-4
- Zhang J, Zhou N, Lin A, et al. ZFHX3 mutation as a protective biomarker for immune checkpoint blockade in non-small cell lung cancer. Cancer Immunol Immunother. 2021 Jan;70(1):137–151.
- Zhou Y, Shi H, Du Y, et al. lncRNA DLEU2 modulates cell proliferation and invasion of non-small cell lung cancer by regulating miR-30c-5p/sox9 axis. Aging. 2019 Sep 20;11(18):7386–7401. DOI:10.18632/aging.102226
- Xu S, Zhai S, Du T, et al. LncRNA MIR503HG inhibits non-small cell lung cancer cell proliferation by inducing cell cycle arrest through the downregulation of cyclin D1. Cancer Manage Res. 2020;12:1641–1647.
- Ginn L, Shi L, Montagna M, et al. LncRnas in non-small-cell lung cancer. Noncoding RNA. 2020 Jun 30;6(3):25. DOI:10.3390/ncrna6030025
- Chen W, Lu Q, Li S, et al. microRNA-1298 inhibits the malignant behaviors of breast cancer cells via targeting ADAM9. Biosci Rep. 2020 Dec 23;40(12). DOI:10.1042/BSR20201215
- Chen T, Liu Y, Chen J, et al. Exosomal miR-3180-3p inhibits proliferation and metastasis of non-small cell lung cancer by downregulating FOXP4. Thoracic Cancer. 2021 Feb;12(3):372–381.
- Cui J, Pan G, He Q, et al. MicroRNA-545 targets ZEB2 to inhibit the development of non-small cell lung cancer by inactivating Wnt/β-catenin pathway. Oncol Lett. 2019 Sep;18(3):2931–2938.
- Cui G, Cui M, Li Y, et al. MiR-186 targets ROCK1 to suppress the growth and metastasis of NSCLC cells. Tumour Biol. 2014 Sep;35(9):8933–8937.
- Lu W, Zhang H, Niu Y, et al. Long non-coding RNA linc00673 regulated non-small cell lung cancer proliferation, migration, invasion and epithelial mesenchymal transition by sponging miR-150-5p. Mol cancer. 2017 Jul 11;16(1):118. DOI:10.1186/s12943-017-0685-9
- Dong X, Chang M, Song X, et al. Plasma miR-1247-5p, miR-301b-3p and miR-105-5p as potential biomarkers for early diagnosis of non-small cell lung cancer. Thoracic Cancer. 2021 Feb;12(4):539–548.
- Sun F, Yang X, Jin Y, et al. Bioinformatics analyses of the differences between lung adenocarcinoma and squamous cell carcinoma using the cancer genome atlas expression data. Mol Med Rep. 2017 Jul;16(1):609–616.
- Miliotis C, Slack FJ. miR-105-5p regulates PD-L1 expression and tumor immunogenicity in gastric cancer. Cancer Lett. 2021 Oct 10;518:115–126. DOI:10.1016/j.canlet.2021.05.037.
- He B, Zhang K, Han X, et al. Extracellular vesicle-derived miR-105-5p Promotes Malignant Phenotypes of Esophageal Squamous Cell Carcinoma by targeting SPARCL1 via FAK/AKT signaling pathway. Front genet. 2022;13:819699.
- Xu X, Zhu S, Tao Z, et al. High circulating miR-18a, miR-20a, and miR-92a expression correlates with poor prognosis in patients with non-small cell lung cancer. Cancer Med. 2018 Jan;7(1):21–31.
- Deng F, Yang K. Current status of research on the period family of clock genes in the occurrence and development of cancer. J Cancer. 2019;10(5):1117–1123.
- Zhang L, Chi B, Chai J, et al. LncRNA CCDC144NL-AS1 serves as a prognosis biomarker for non-small cell lung cancer and promotes cellular function by targeting miR-490-3p. Mol Biotechnol. 2021 Jun 11;63(10):933–940. DOI:10.1007/s12033-021-00351-6
- Wu W, Zhao Y, Gao E, et al. LncRNA DLEU2 accelerates the tumorigenesis and invasion of non-small cell lung cancer by sponging miR-30a-5p. J Cell Mol Med. 2020 Jan;24(1):441–450.
- Zhao Y, Qiao W, Wang X, et al. 14-3-3ζ/TGFβR1 promotes tumor metastasis in lung squamous cell carcinoma. Oncotarget. 2016 Dec 13;7(50):82972–82984. DOI:10.18632/oncotarget.12690
