ABSTRACT
The development of genomic technologies over the past decades has enabled identification of genetic variants responsible of disease; occasionally however, protective rare variants emerged. Verweij et al have recently reported genetic variants in CIDEB gene that are protective from liver injury. Here, we briefly summarise the recent findings on the impact of CIDEB variants on liver disease, while emphasizing how phenotype-genotype studies tailored for the identification of “protective” mutations might direct development of prevention and therapeutic strategies for common diseases.
The next-generation sequencing techniques, including whole exome sequencing (WES) and whole genome sequencing, have boosted the identification of new and rare genetic variants linked to human disease. These substantial amounts of data of large patient cohorts have contributed to the generation of Biobanks, which often also provide detailed phenotypic and clinical data linked to medical records [Citation1–4]. In a recent article, published in The New England Journal of Medicine, Verweij and colleagues [Citation5] report the discovery of new allele variants associated with protection from liver injury. By performing WES of a cohort comprised of more than 500,000 patients belonging to the UK Biobank and Geisinger Health System MyCode databank, they aimed to find out new rare coding variants associated with liver status. Verweij et al. used a three-step approach to perform genetic association studies with liver status.
First, the identified rare gene coding variants associated with plasma levels of Alanine aminotransferase levels (ALT), an established biomarker of hepatocellular disease and injury [Citation6]. The second analysis stage aimed to corroborate the findings of stage one by performing association studies with the levels of Aspartate aminotransferase (AST). Third, they compared and estimated the association with liver disease using publicly available datasets comprising data from histopathological analyses, liver transcriptomics and in vitro experiments using cell lines. Through this multi-step approach, Verweij and colleagues could select rare variants in a subset of five genes that have a very strong correlation with liver phenotype, among which CIDEB.
CIDEB belongs to the Cell Death-inducing DNA Fragmentation Factor-like Effector (CIDE) family [Citation7], which regulates the metabolism of lipid droplets (LDs) inside adipocytes (CIDEA and CIDEC) [Citation8–10] and hepatocytes (CIDEB) [Citation11]. CIDEB plays an essential role in the regulation of fusion of LDs mediated the homodimerization between its N- and C-termini [Citation8]. Common liver injuries such as steatosis and nonalcoholic cirrhosis present dysregulation of the biochemical reactions involved in buildup and fusion of small LDs into large ones with a consequent accumulation of the latter. The protraction of this condition normally leads to liver failure which is rapidly becoming a major cause of liver transplantation [Citation12–14].
Rare germline variants, including missense and predicted loss of functin mutations were found in CIDEB coding sequence and showed a marked association with decreased content of hepatic lipids, lower level of ALT and AST, and thus a lower risk of developing liver failure or disease [Citation6]. Through the availability of transcriptomic analysis, the authors could observe the clinical relevance of two identified rare variants (K153* and c.336 + 1 G→A) which also correlate with decreased levels of CIDEB gene expression. To strengthen their finding, Verweij et al. assessed the effect of loss of CIDEB using hepatoma cell lines as model. Under resting conditions, loss of function of CIDEB did not result in altered size of LDs, but when treated with oleic acid, mimicking steatosis, cells depleted of CIDEB presented less accumulation of large LDs (). CIDEB not only has a direct effect on the maturation of intracellular large LDs but its reduction can also have a strong regulation of the lipid metabolism balance by regulating both enzymes involved in fatty acid synthesis among which acetyl-CoA carboxylase (ACC1), fatty acid synthase (FAS) and stearoyl-CoA desaturase-1 (SCD1), and transcription factors such as sterol-regulatory element binding protein-1C (SREBP1c). Studies using in vivo models have also provided evidence for a strong beta oxidation of fatty acid catabolic process, leading to a notable reduction of intracellular triglycerides level [Citation11].
Figure 1. CIDEB gene variants correlate with liver health.
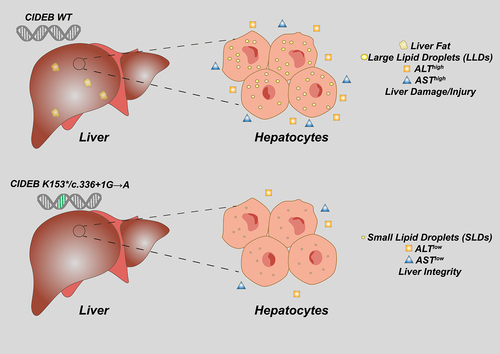
Overall, this study demonstrates that some rare nucleotide variants found in the human genome can have a strong protective effect against the development of diseases [Citation15,Citation16]. This is however not the first examples, G Protein-Coupled Receptor 75 (GPR75) and Proprotein Convertase Subtilisin/Kexin type 9 (PCSK9) mutations lower the risk of obesity and liver injury, respectively [Citation17,Citation18] and other cases have been found as well. “Beneficial” mutations represent precious information to instruct drug development. Often they are associated to attenuation of gene expression or protein activity [Citation19,Citation20]. Hence, they directly indicate potential drug targets for disease treatment or prevention. For instance, selective compounds downregulating CIDEB protein level can be developed, as observed for variants in PNPLA3 and HSD17B13, with the hope of reproducing the demonstrated effects on hepatic lipid metabolic processes and protective effects of liver integrity. Intriguingly, speculations might emerge on the capabilities of negative lifestyle, such as high-fat diet, to exert evolutionary pressure and select protective mutations. This however requires careful consideration of reasonable timescales of an evolutionary process. Moreover, it should be also taken into account that the discussed “environmental factors” are mainly influencing health and life expectancy in aged populations, and less the reproductive fitness of individuals; hence, they are less likely to impact evolution.
Genotype–phenotype association studies represent a powerful tool for the identification of novel “beneficial” mutations. There is however lower chance of finding mutations that are protective rather than causative of diseases [Citation21]. The selection of the cohorts for these studies tends to have bias for individuals that carry causative mutations. Furthermore, the protective effects of these rare variants come out under environmental stress conditions, such as high fat diet; hence, the result of a mutation or variant should be always considered in a context-dependent manner [Citation22]. However, a relevant limitation is that such comprehensive studies often do not consider variants in non-coding regions, as they are generally excluded in exome sequencing analyses [Citation23]. Indeed, non-coding mutations in cis- and trans-acting modifier elements, such as promoter or enhancer, can also modulate the expression of proximity genes [Citation20]. Although, Verweij et al. could corroborate their findings using an in vitro model, the molecular mechanism underpinning liver injury remains poorly described. Hence, an integration of gene-phenotype studies with a deeper dissection of the pathways deregulated in such pathologies might be a prerequisite for the development of tailored therapies.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Backman JD, Li AH, Marcketta A, et al. Exome sequencing and analysis of 454, 787 UK Biobank participants. Nature. 2021;599:628–634.
- Wang Q, et al. Rare variant contribution to human disease in 281, 104 UK Biobank exomes. Nature. 2021;597:527–532.
- Ganini C, Amelio I, Bertolo R, et al. Global mapping of cancers: the cancer genome atlas and beyond. Mol Oncol. 2021;15(11):2823–2840.
- Amelio I, Bertolo R, Bove P, et al. Cancer predictive studies. Biol Direct. 2020; 15(1).
- Verweij N, Haas ME, Nielsen JB, et al. Germline Mutations in CIDEB and protection against liver disease. N Engl J Med. 2022;387(4):332–344.
- Kwo PY, Cohen SM, Lim JK. ACG clinical guideline: evaluation of abnormal liver chemistries. Am J Gastroenterol. 2017;112(1):18–35.
- Xu W, Wu L, Yu M, et al. Differential roles of cell death-inducing DNA Fragmentation Factor-α-like Effector (CIDE) proteins in promoting lipid droplet fusion and growth in subpopulations of hepatocytes. J Biol Chem. 2016;291(9):4282–4293.
- Barneda D, et al. The brown adipocyte protein CIDEA promotes lipid droplet fusion via a phosphatidic acid-binding amphipathic helix, 2015; 1–24. DOI:10.7554/eLife.07485
- Sun Z, Gong J, Wu H, et al. Perilipin1 promotes unilocular lipid droplet formation through the activation of Fsp27 in adipocytes. Nat Commun. 2014; 5(1).
- Li X, Ye J, Zhou L, et al. Opposing roles of cell death-inducing DFF45-like effector B and perilipin 2 in controlling hepatic VLDL lipidation. J Lipid Res. 2012;53(9):1877–1889.
- Li JZ, Ye J, Xue B, et al. Cideb regulates diet-induced obesity, liver steatosis, and insulin sensitivity by controlling lipogenesis and fatty acid oxidation. Diabetes. 2007;56(10):2523–2532.
- Sepanlou SG, Safiri S, Bisignano C, et al. The global, regional, and national burden of cirrhosis by cause in 195 countries and territories, 1990–2017: a systematic analysis for the global burden of disease study 2017. Lancet Gastroenterol Hepatol. 2020;5(3):245–266.
- Sheka AC, Adeyi O, Thompson J, et al. Nonalcoholic steatohepatitis: a review. JAMA. 2020;323(12):1175–1183.
- Noureddin M, Vipani A, Bresee C, et al. NASH leading cause of liver transplant in women: updated analysis of indications for liver transplant and ethnic and gender variances. Am J Gastroenterol. 2018;113(11):1649–1659.
- Müller M, Forbes SJ, Bird TG. Spotlight beneficial noncancerous mutations in liver disease. xx. 2019;35(7):475–477.
- Ng SWK, Rouhani FJ, Brunner SF, et al. Convergent somatic mutations in metabolism genes in chronic liver disease. Nature. 2021;598:473–478.
- Lebeau PF, Wassef H, Byun JH, et al. Chaperones GRP78 and GRP94 and protects against liver injury the loss-of-function PCSK9 Q152H variant increases ER chaperones GRP78 and GRP94 and protects against liver injury. J Clin Investig. 2021; 131(2).
- Genomics H, et al. Sequencing of 640,000 exomes identifies GPR75 variants associated with protection from obesity. Science. 2021;373:6550.
- Kozlitina J, Cheng X, Li AH, et al. A protein-truncating hsd17b13 variant and protection from chronic liver disease. N Engl J Med. 2018;378(12):1096–1106.
- Harper AR, Nayee S, Topol EJ. Protective alleles and modifier. Nat Rev Genet. 2015;16(12):689–701.
- Nelson MR, Tipney H, Painter JL, et al. The support of human genetic evidence for approved drug indications. Nat Genet. 2015;47(8):856–860.
- Peters A, Nawrot TS, Baccarelli AA. Ll Hallmarks of environmental insults. Cell. 2021;184(6):1455–1468.
- Wells A, Heckerman D, Torkamani A, et al. Ranking of non-coding pathogenic variants and putative essential regions of the human genome. Nat Commun. 2019; 10(1).
