ABSTRACT
Isoalantolactone (Iso) is a bioactive lactone isolated from the root of Inula helenium L, which has been reported to have many pharmacological effects. To investigate the role and mechanism of isoalantolactone in chronic myeloid leukemia (CML), we first investigated isoalantolactone’s anti-proliferative effects on imatinib-sensitive and imatinib-resistant CML cells by CCK8. Flow cytometry was used to detect isoalantolactone-induced cell apoptosis. Survivin was overexpressed in KBM5 and KBM5T315I cells using the lentivirus vector pSIN-3×flag-PURO. In KBM5 and KBM5T315I cells, shRNA was used to knockdown survivin. Cellular Thermal Shift Assay (CETSA) was used to detect the interaction between isoalantolactone and survivin. The ubiquitin of survivin induced by isoalantolactone was detected through immunoprecipitation. Quantitative polymerase-chain reaction (Q-PCR) and western blotting were used to detect the levels of mRNA and protein. Isoalantolactone inhibits the proliferation and promotes apoptosis of imatinib-resistant CML cells. Although isoalantolactone inhibits the proteins of BCR-ABL and survivin, it cannot inhibit survivin and BCR-ABL mRNA levels. Simultaneously, it was shown that isoalantolactone can degrade survivin protein by increasing ubiquitination. It was demonstrated that isoalantolactone-induced survivin mediated downregulation of BCR-ABL protein. It was also revealed that isoalantolactone triggered BCR-ABL protein degradation via caspase-3. Altogether, isoalantolactone inhibits survivin through the ubiquitin proteasome pathway, and mediates BCR-ABL downregulation in a caspase-3 dependent manner. These data suggest that isoalantolactone is a natural compound, which can be used as a potential drug to treat TKI-resistant CML.
1. Introduction
Chronic myeloid leukemia (CML) is a clonal myeloproliferative disorder characterized by a chromosome translocation that results in the formation of the BCR-ABL fusion gene, which encodes the constitutively activated BCR-ABL tyrosine kinase [Citation1–3]. The current treatment for CML (CML) with tyrosine kinase inhibitors (TKI) is effective but not curative in many patients, and it is frequently limited by drug resistance [Citation4,Citation5]. Survivin expression has been linked to increased tumor resistance to a variety of chemotherapy agents. Patients with undetectable survivin levels had cytogenetic complete remissions, whereas patients clinically resistant to imatinib had elevated survivin levels, similar to those seen in imatinib-resistant cell lines [Citation6]. The survivin protein levels were significantly reduced in an imatinib-sensitive CML cell line at the same time [Citation7–9]. Survivin plays an important role in the maintenance of CML drug resistance, but no natural compounds targeting survivin protein have been found to overcome TKI resistance in CML.
Natural compounds have medicinal potential and are a valuable source for the development of targeted regulation of BCR-ABL proteins. Diosmetin, a phytoestrogen, inhibits the phosphorylation of BCR-ABL and has a minor effect on BCR-ABL levels [Citation10]. By blocking the BCR-ABL signaling pathways, CD-200 induces apoptosis and has an anti-proliferative effect in BCR-ABL T315I cells, which are resistant to imatinib [Citation11]. Tambjamines are natural compounds isolated from marine invertebrates with a variety of pharmacological activities [Citation12–14]. T21, a novel indole-based tambjamine analog, has recently been identified as a promising antitumor agent that modulates the expression of apoptotic proteins, such as survivin [Citation15]. To that end, searching for candidate compounds targeting survivin downregulation among natural compounds may yield lead compounds to overcome TKI resistance. Isoalantolactone, a major sesquiterpene lactone isolated from the root of Inula helenium L. It is most commonly used in the treatment of chronic gastritis, gastrointestinal dysfunction, intercostal neuralgia, and chronic inflammation [Citation16–19]. In vitro studies have revealed that the drug has a potent anti-tumor activity in lung carcinoma, pancreatic carcinoma, prostate adenocarcinoma, and breast carcinoma by inducing apoptosis, autophagic cell death, or inhibiting migration and invasion [Citation20–24]. Isoalantolactone has been shown to induce apoptosis in human leukemia drug-resistant cell-line K562/A02 cells overexpressing the BCR-ABL oncogene by downregulating BCR-ABL and Bcl-2, as well as induced activation of Bax, cytochrome c, caspase-9, caspase-3, and PARP [Citation25]. It remains to be seen whether isoalantolactone can overcome TKI resistance by targeting the survivin protein.
To investigate the effect of isoalantolactone, we treated imatinib-sensitive and imatinib-resistant cells KBM5 and KBM5T315I with different concentrations of isoalantolactone and examined cell proliferation and apoptosis. We discovered that isoalantolactone-induced BCR-ABL degradation in a caspase-3 dependent manner via targeting survivin degradation. These findings suggest that isoalantolactone is a candidate natural compound that could be used to treat TKI-resistant CML.
2. Materials and methods
2.1. Chemicals
A stock concentration of 20 mM isoalantolactone was provided by Dr. Wu Ying-Li (Department of Pathophysiology, Shanghai Jiaotong University School of Medicine, Shanghai, China). It was isolated from the root of Inula helenium L. The batch number is Y-059-131027. HPLC is used as a test criterion. The pan-caspase inhibitor Z-VAD-FMK (catalog no. S7023; Selleck Chemicals), MG-132 (catalog no. s2619; Selleck Chemicals), CQ (catalog no. C6628; Sigma), 3 MA (catalog no. M9281; Sigma) was dissolved in DMSO and kept at −80°C.
2.2. Cell culture.
Dr. Wu Ying-Li provided K562, K562R, KBM5, and KBM5T315I cells. At 37°C and 5% CO2, KBM5 cells were cultured in RPMI-1640 (catalog no. SH30809.01B; HyClone) with 10% FBS (catalog no. 10091148, Gibco; Thermo Fisher Scientific, Inc), 100 U penicillin, and 100 g/L streptomycin (catalog no. 15070063, Gibco; Thermo Fisher Scientific, Inc). KBM5T315I cells were cultured in 10 μM imatinib medium.
2.3. Cell proliferation assay
2000 cells in 100 μl were incubated in each well to assess cell viability and determine the study’s IC50 dose. The treatment group included 2.5, 5, 10, 20, 40, and 80 μmol/L isoalantolactone, while the control group included dimethyl sulfoxide (DMSO). After 12, 24, 36 and 48 h, 10 μl of CCK8 solution was added to each well of the plate, and the wells were incubated for 2 h. Each well’s absorbance at 450 nm was used to calculate the cell viability. The following equation calculated cell viability (%): = 100 × (mean OD of test wells/mean OD of control wells).
2.4. Flow cytometry
The apoptosis was determined by Annexin V-FITC/PI, Annexin V-Alexa Fluor 647/PI staining (we chose 647/PI staining for apoptosis detection because we transfected shRNA with a GFP tag). Briefly, the cells were treated with isoalantolactone for 24 h. Then, cells were centrifuged at 1,000 rpm for 5 min, and washed with a binding buffer. Following this, cells were incubated in working solution (100 μl binding buffer with 5 μl Annexin V-FITC/Alexa 647 and 5 μl PI) for 15 min in the dark.
2.5. Western blotting.
Cell pellets were lysed with 150 μl lysis buffer [50 mM Tris-HCl (pH6.8), 100 mM DTT, 2% SDS, 10% glycerol and ddH2O]. The cell lysates were denatured at 100°C for 10 min. The protein concentration of the cell lysate was measured using a spectrophotometer. The total protein (100 μg) of the cell lysate was resolved by SDS-PAGE, transferred to a nitrocellulose membrane, and blocked with 5% skim milk (catalog no. A600669, Sangon Biotech, China) for 1 h at room temperature. The membrane was incubated overnight at 4°C with the following appropriate primary antibodies: Anti-cleaved caspase-3 (catalog no. 9664, Cell Signaling Technology, Inc.), anti-caspase-3 (catalog no. ab184787, Abcam), anti-caspase-7 (catalog no. 9492, Cell Signaling Technology, Inc.), anti-c-Abl (catalog no. 2862, Cell Signaling Technology, Inc.), anti-cleaved PARP (catalog no. 5625, Cell Signaling Technology, Inc.), anti-survivin (catalog no. ab76424, Abcam) anti-Ub (catalog no.3932, Cell Signaling Technology, Inc) anti-Flag (catalog no.14793S, Cell Signaling Technology, Inc) anti-β-actin (catalog no. 4970, Cell Signaling Technology, Inc.). The membranes were then incubated with HRP-conjugated rabbit anti-mouse antibody (catalog no. 315-035-003, Jackson Immuno Research Laboratories, Inc.) for 1 h. ECL reagent (catalog no. WBKLS0500, EMD Millipore) was used to detect the signals.
2.6. RNA extraction and reverse transcription-quantitative PCR (RT-Qpcr)
Total RNA was extracted from cells using RNAiso Plus reagent (catalog no. 9109, Takara, Bio, Inc.) according to the manufacturer’s protocol. RNA (500 ng) was reverse transcribed to cDNA using the PrimeScript RT reagent kit (catalog no. RR047A, Takara, Bio, Inc.). qPCR analysis was conducted on a Roche Light Cycler480 machine with SYBR Green (catalog no. 11202ES03, Yeason). Relative expression was determined using the 2−ΔΔCq method with ACTB as the reference. The primers were synthesized as follows: BCR-ABL forward, 5´- CGG GAG CAG CAG AAG AAG TGT-3´ and reverse, 5´- CGA AAA GGT TGG GGT CAT TTT C-3´; Survivin forward, 5´-TGA ACT TCA GGT GGA TGA GGA GA −3´ and reverse, 5´-GTC TAA TCA CAC AGC AGT GGC AA −3´; and ACTB forward, 5´-CAT GTA CGT TGC TAT CCA GGC-3´, and reverse, 5´-CTC CTT AAT GTC ACG CAC GAT-3´. All experiments were performed three times independently.
2.7. Transfection and viral infection
Dr. YingLi Wu provided the pSIN4-EF1alpha-IRES plasmids (Vector backbone identical to #61062, addgene). Survivin’s coding sequence was amplified by PCR using Jurkat CDNA as the template and the PrimeSTAR GXL DNA Polymerase Kit (#R050Q, TaKaRa, Japan). The following primers were used in the PCR: F: 5´ -GGG CTA GCT AGC TAG GAA TTC ATG GAC TAC AAA GAC GAT GAC GAC AAG GAC TAC AAA GAC GAT GAC GAC AAG GAC TAC AAA GAC GAT GAC GAC AAG ATG GGT GCC CCG ACG TTG-3´. R: 5´-GCC CTA GAT GCA TGC GGA TCC TCA ATC CAT GGC AGC CAG CTG-3´. 0.5 μg CDNA, 2 μl forward primers (10 μM), 2 μl reverse primers (10 μM), 4 μl dNTP Mixture, 10 μl 5 ×PrimeSTAR GXL Buffer (Mg2+ plus), 2 μl PrimeSTAR GXL DNA Polymerase (1.25 U μl), and RNase Free ddH2O were used in the PCR. Pre-denaturation at 98°C for 60 s, denaturation at 98°C for 10 s (30 cycles), annealing at 55°C for 15 s, and the extension at 68°C for 90 s were the reaction conditions. The PCR products were separated on a 1% agarose gel and purified using the TaKaRa MiniBEST Agarose Gel DNA Extraction Kit Ver.4.0 (#9762, TaKaRa, Japan). The restriction enzymes BamH I (#1605, TaKaRa, Japan) and EcoR I (#1611, TaKaRa, Japan) were used to digest the pSIN plasmids. The reaction was carried out in a 50 μl volume, with 1 μg pSIN plasmids, 1 μl BamH I, 1 μl EcoR I, 5 μl 10×QuickCut Buffer, and RNase Free ddH2O. The reaction conditions were as follows: 30 min of digestion at 37°C. After stopping the reaction with 0.8% agarose gel electrophoresis, the linearized pSIN plasmids were detected and purified using the TaKaRa MiniBEST Agarose Gel DNA Extraction Kit Ver.4.0. To obtain recombinant plasmids coding survivin, the ligation reaction of PCR products and linearized pSIN plasmids was catalyzed by the ClonExpress II One-Step Cloning Kit (#C11201, Vazyme, China). The reaction was carried out in a 20 μl volume with 160 ng linearized pSIN plasmids, 60 ng PCR products, 2 μl Exnase II, 4 μl 5×CE II Buffer, and RNase Free ddH2O. The reaction conditions were as follows: ligation at 37°C for 30 min, followed by 5 min on ice. The recombinant plasmids were amplified and DNA sequenced to confirm their existence (GENEWIZ, China) The Lipofectamine 3000 Transfection Kit (#L3000015, Invitrogen, USA) was used to transfect cells (50–60% cell confluent monolayer in one well of a 6 well plate) with pSIN or pSIN-Flag3-Survivin plasmids. The steps were as follows: Mix I is made up of 6 μl Lipofectamine 3000 Reagent and 250 μl OptiMEM Medium (#31985062, gibco). 2 μg pSIN/pSIN-Flag3-Survivin plasmids, 4 μl P3000 Reagent, and 250 μl OptiMEM Medium make up the Mix II solution. The above solutions I and II were thoroughly mixed and allowed to stand at room temperature for 5 min. The mixture was then added to one well of a 6-well plate containing 1.5 ml of normal medium. Transfected cells can then be used in subsequent experiments. All experiments were carried out three times independently and yielded similar results.
ShRNA sequences targeting survivin, caspase 3, caspase 7, and a negative control sequence were synthesized, annealed, and inserted into lentiviral pLL3.7 vectors. The following were the target sequences: survivin-1, 5´- GGC TGT TCC TGA GAA ATA A −3´; Survivin-2, 5´- GGA GAC AGA ATA GAG TGA T −3´; caspase-7–1, 5´-GGC AAA TGC ATC ATA ATA A-3´; caspase- 7–2, 5´-GCC CAT CAA TGA CAC AGAT-3´; caspase-3–1,5´-CCC TGG ACA ACA GTT ATA A-3´; caspase-3–2,5´-GCG TGA TGT TTC TAA AGA A-3´; and scrambled, 5´-GTT CTC CGA ACG TGT CAC GT-3´. 293T cells were transfected in Opti-MEM® I (Gibco; Thermo Fisher Scientific, Inc.) using Lipofectamine® 3000 (Thermo Fisher Scientific) once they were 70% confluent, according to the manufacturer’s protocol. The medium was changed to complete DMEM-H 8 h after transfection. Viral medium was created by co-transfecting 293T cells with shRNA expressing pLL3.7 lentiviral vectors as well as psPAX.2 and pMD2. G (Addgene, Inc.). In a 2:1:1 ratio, vectors were transfected. The viral particles were harvested 48 h after transfection, filtered through a 0.22 μm filter, and used for infection. KBM5 and KBM5-T315I cells were incubated with lentivirus-containing medium and Polybrene (10 g/ml). After 24 h, the culture medium was replaced with a complete medium, and cells expressing GFP were chosen.
2.8. Immunoprecipitation
Cells (70–80% cell confluent monolayer in a 6 cm cell culture dish) were washed twice with PBS buffer before being added to 1 ml RIPA lysis buffer (#P0013D, Beyotime Biotech, China) containing protease and phosphatase inhibitors (#B14001, Bimake). The cell lysate was then placed on ice for 60 min while being vibrated every 2 min. The cell lysate was then treated with an ultrasonic cell disruptor (power: 200 W; mode: work 4 s, stop 3 s, 5 times) (#DH92IIN, LAWSON, China). The supernatant of the cell lysate was collected and divided into two parts: one (350 μL) for the target protein immunoprecipitation assay and the other (200 μl) for total protein analysis. The immunoprecipitation assay protocol was as follows: Gently invert the resin to thoroughly resuspend it. Check that the ANTI-FLAG M2 Magnetic Beads (#M8823, Sigma) are in a uniform suspension in the bottle. 10 μl beads should be removed for use. Re-suspend the beads in 200 μl TBS to re-equilibrate them (50 mM Tris HCl, 150 mM NaCl, pH 7.4). To collect the beads, place the tube in an appropriate magnetic separator. Remove the storage buffer/TBS mixture and discard it. At 4°C, incubate the protein extract with the equilibrated beads for 34 h. Once the binding is finished, collect the magnetic beads by placing the tube in the appropriate magnetic separator and removing the supernatant. To remove all nonspecifically bound proteins, wash the resin beads with TBS buffer. The beads were then washed with TBS, and the protein binding to the beads was eluted with 30 μl SDS lysis buffer. Following that, the Western Blot protocol was carried out. All experiments were carried out three times independently and yielded similar results.
2.9. Cellular Thermal Shift Assay (CETSA)
×10 [Citation7] cells were cultured in 10 cm dishes for 24 h to produce a CETSA in HEK293 cell lysates. The cells were washed in PBS before being harvested in 1 ml of PBS [1% proteinase inhibitor cocktail]. After 3 min of incubation in Liquid Nitrogen and melting at room temperature, the process was repeated three times. The lysates were centrifuged for 30 min at 14,000 g at 4°C, and the supernatants were separated into two tubes. (1) The melt curve “CETSA” approach: The supernatants and 100 μM Isoalantolactone were incubated at room temperature for 30 min. Control cells were exposed to an equal volume of DMSO as a vehicle. Proteins from the two tubes were then divided equally into six 100 μl tubes and placed in the PCR instrument, which was set to the indicated temperatures. (2) The isothermal dose–response (ITDR) CETSA: The supernatants and Isoalantolactone were incubated with the indicated concentration (0 μM, 12.5 μM, 25 μM, 50 μM, 100 μM, 200 μM), and placed in the PCR instrument, 50°C 3 min, 25°C 3 min. The lysates were centrifuged at 14,000 g for 30 min at 4°C, and the supernatants were transferred to new tubes and incubated for 10 min at 100°C with an equal volume Lysis buffer. The supernatants were analyzed by Western blot.
2.10. Clinical samples collection
We collected bone marrow cells from four CML patients and treated them for 48 h with 20 μM isoalantolactone. All procedures were carried out in accordance with the ethical guidelines established by the Ethics Committee of Shanghai 9th People’s Hospital, Shanghai Jiao Tong University School of Medicine, and written informed consent was obtained.
2.11. Statistical analysis
The statistical analysis was performed using GraphPad Prism 8.0 software (GraphPad Software, Inc.) or SPSS 21.0 (IBM Corp.). Compared isoalantolactonens with the groups for cell viability and mRNA relative expression were assessed by one-way AVONA and Dunnett’s multiple comparison test. The apoptosis rates and protein expression were assessed by students’ t-test. P < 0.05 was considered to indicate a statistically significant difference.
3. Results
3.1. Isoalantolactone inhibits the proliferation and induces apoptosis of imatinib- resistant CML cells
Imatinib-sensitive cell lines (KBM5 and K562) and imatinib-resistant cell lines (KBM5T315I and K562R) were used for testing, as shown in Figures ( and Supplementary Figure S1A). The proliferation of KBM5, K562, KBM5T315I, and K562R cells was inhibited by isoalantolactone in a dose and time-dependent manner, with IC50 values of 8.58, 5.04, 15.13, and 19.32 μM at 48 h ( and Supplementary Figure S1B, C). When 20 μM isoalantolactone was added to KBM5, K562, KBM5T315I, and K562R cells, the amount of Annexin V+/PI+ (late apoptosis), and Annexin V-/PI+ (necrosis) stained cells were increased ( and Supplementary Figure S1D, E). This suggests isoalantoactone-induced apoptosis in both imatinib-sensitive and imatinib-resistant cells.
Figure 1. Isoalantolactone inhibits the proliferation and induces apoptosis of imatinib-sensitive and resistant CML cells.
3.2. Isoalantolactone downregulates BCR-ABL and survivin protein levels, but it can’t alter the mRNA levels of survivin and BCR-ABL in imatinib-resistant CML cells
We investigated whether isoalantolactone could affect survivin and BCR-ABL protein levels in imatinib-sensitive or imatinib-resistant CML cells. We discovered that isoalantolactone significantly reduced survivin and BCR-ABL protein levels in a dose and time-dependent manner (and Supplementary Figure S1F, G). Isoalantolactone was applied to imatinib-sensitive cells (KBM5 and K562) and imatinib-resistant cells (KBM5T315I and K562R) for 24 h at concentrations of 0, 10, 15, and 20 μM. The detection of cleaved-caspase-3 and cleaved-PARP indicated that isoalantolactone could induce apoptosis in imatinib-sensitive cells as well as imatinib-resistant cells ( and Supplementary Figure S1F, G). At the same time, we used clinical samples to show that 20 μM isoalantolactone could significantly inhibit survivin protein in patients with both early-stage and drug-resistant CML (Supplementary Figure S2A-C). However, no significant changes were identified in survivin and BCR-ABL mRNA levels (), indicating that the down-regulation of survivin and BCR-ABL induced by isoalantolactone was not at the transcriptional level.
Figure 2. Effect of isoalantolactone on survivin and BCR-ABL protein and mRNA levels in imatinib-sensitive and resistant CML cells.

3.3. The survivin mediates the apoptosis and down-regulation of BCR-ABL protein in imatinib-resistant CML cells
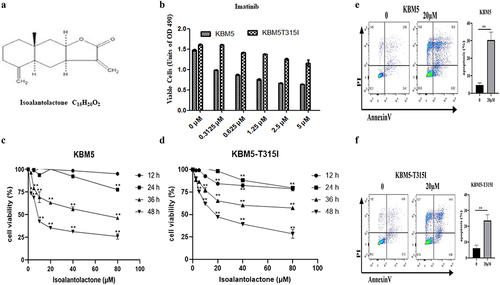
In imatinib resistant and sensitive CML cell lines, we established survivin knockdown cell lines and found that the BCR-ABL protein level of survivin knockdown cells was significantly reduced, which confirmed that the down-regulation of BCR-ABL was mediated by the reduction of survivin (). At the same time, apoptosis was significantly increased in CML cells that knocked down survivin (). These findings suggest that survivin is responsible for apoptosis and BCR-ABL protein downregulation in KBM5 and KBM5T315I cells.
Figure 3. The degradation of BCR-ABL protein in imatinib sensitive and resistant CML cells through down-regulation of survivin.
3.4. Isoalantolactone mediates imatinib resistance and sensitive CML cells apoptosis through down-regulation of survivin
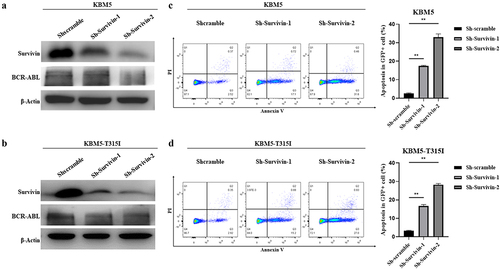
We constructed survivin overexpressed cell lines to investigate if isoalantolactone mediates imatinib resistance and sensitive cell-line apoptosis by down-regulating survivin. The overexpressed survivin in imatinib sensitive and resistant cells could resist the down-regulation of BCR-ABL protein induced by isoalantolactone (). Treating survivin overexpressed KBM5 and KBM5T315I cells with 20 μM isoalantolactone, the apoptosis was significantly inhibited (.
Figure 4. Isoalantolactone mediated imatinib resistance and sensitive CML cells apoptosis through down-regulation of survivin.
3.5. Isoalantolactone enhances ubiquitin-dependent degradation of survivin through direct interaction with surviving protein.
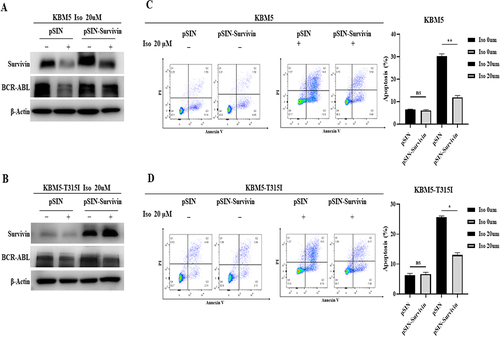
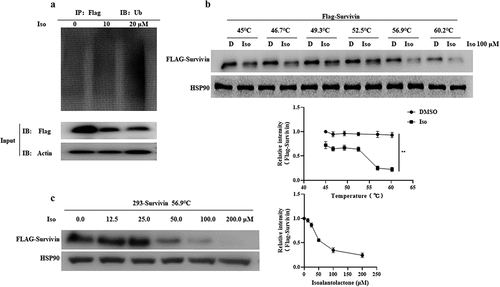
In order to understand the mechanism of survivin protein degradation in CML cells induced by isoalantolactone, both KBM5 and KBM5T315I cells were pretreated with the autophagy inhibitor 3-MA and the lysosomal inhibitor CQ in the presence or absence of isoalantolactone, which revealed no attenuation of survivin protein degradation, indicating that the autophagy pathway was not responsible for the isoalantolactone-induced survivin protein degradation (. Following that, KBM5 and KBM5T315I cells were treated with the proteasome inhibitor MG132 before being incubated with isoalantolactone. After treatment with MG132, survivin protein was restored, which means that the proteasome pathway may participate in the degradation of survivin . The immunoprecipitation test suggested that isoalantolactone may cause survivin degradation by increasing survivin ubiquitination (). However, it is unknown whether isoalantolactone directly targets survivin. We used CETSA to perform target validation to see if isoalantolactone directly binds to survivin. This method is based on the idea that ligand binding increases the thermal stability of a target protein. We discovered that when HEK293T cell lysates with overexpressed survivin were incubated with isoalantolactone, the thermal stability of survivin was reduced when compared to the control group, indicating that isoalantolactone may trigger survivin thermal stabilization via direct binding .
Figure 5. The proteasome pathway involved in isoalantolactone-induced down-regulation of survivin protein in imatinib-sensitive and resistant CML cells.
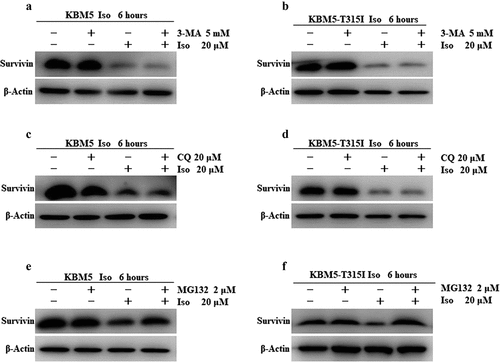
Figure 6. Isoalantolactone enhanced survivin protein ubiquitin modification by directly binding to survivin protein.
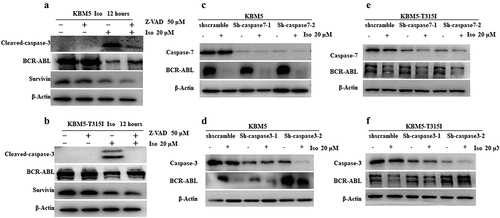
3.6. Isoalantolactone induced BCR-ABL protein degradation is caspase-3 dependent in imatinib sensitive and resistant cells
KBM5 and KBM5T315I cells were treated with caspase inhibitor Z-VAD-FMK alone or in combination with isoalantolactone to determine the mechanism of isoalantolactone-induced BCR-ABL degradation in CML cells, which revealed that the inhibitor Z-VAD-FMK blocked the caspase-3 activation induced by isoalantolactone . Furthermore, cotreatment with isoalantolactone and Z-VAD-FMK partially rescued BCR-ABL protein expression in KBM5 and KBM5T315I cells , indicating that isoalantolactone may downregulate BCR-ABL protein expression via caspase activation. However, this method had no effect on survivin protein expression. Caspase-3 and caspase-7 were knocked down in KBM5 and KBM5T315I cells to confirm that caspase activation contributes to isoalantolactone-induced BCR-ABL protein degradation. Caspase-7 knockdown could not rescue isoalantolactone-induced BCR-ABL protein degradation , whereas reducing caspase-3 could partially rescue isoalantolactone-induced BCR-ABL protein degradation . These findings suggest that isoalantolactone-induced BCR-ABL protein degradation in imatinib-resistant CML cells was partially dependent on caspase-3 activation.
Figure 7. The down-regulation of BCR-ABL protein induced by isoalantolactone depends on the activation of caspase-3 in imatinib-resistant and sensitive CML cells.
4. Discussion
LY2181308 is a single-strand antisense oligonucleotide that targets survivin by binding to and degrading its mRNA, preventing it from being translated into protein. In several tumor cell lines and human tumor xenografts, LY2181308 significantly reduced survivin mRNA and protein, as well as cell-cycle arrest, cell-death induction, and tumor growth inhibition [Citation26]. Another antisense oligonucleotide, SPC3042 (EZN-3042), was identified as a new agent with greater potency for survivin mRNA inhibition than previous antisense agents, including LY2181308 [Citation27]. SPC3042-induced cell cycle arrest, pronounced cellular apoptosis, and sensitization of prostate cancer cells to taxol treatment in vitro and in vivo. Survivin has emerged as an ideal target for oncology drug discovery, but no potential small molecule survivin inhibitor has been found [Citation28]. In this study, isoalantolactone inhibited proliferation and promoted apoptosis in both imatinib-sensitive cells (KBM5 and K562) and imatinib-resistant CML cell lines (KBM5T315I and K562R) in a dose- and time-dependent manner). We discovered that isoalantolactone inhibited survivin, which mediated BCR-ABL protein degradation in a caspase-3 dependent manner.
Whether CML cells are sensitive to imatinib or not, inhibition of survivin expression will lead to cell growth arrest and subsequent cell death [Citation6]. It suggests that suppressing survivin expression may have a therapeutic effect in patients with imatinib-resistant CML. We discovered that isoalantolactone induced the degradation of survivin and BCR-ABL, particularly in T315I mutated cells, but did not affect BCR-ABL and survivin at the mRNA level. The survivin protein changes can be reproducible in clinical samples. It was discovered that isoalantolactone mediated BCR-ABL degradation via survivin protein disruption, which may be the key reason for isoalantolactone’s ability to overcome BCR-ABL T315I cell resistance. On the other hand, more information about the interaction of isoalantolactone and survivin needs to be clarified.
The ubiquitin/proteasome pathway and the lysosomal pathway are two important intracellular protein degradation pathways [Citation29,Citation30]. However, in the current study, the proteasome inhibitor MG132, the autophagy inhibitor CQ, and the lysosomal inhibitor 3-MA were unable to reduce isoalantolactone-induced survivin and BCR-ABL protein degradation, indicating that these two pathways are not involved in isoalantolactone-induced survivin and BCR-ABL protein downregulation. The proteasome inhibitor MG132 can rescue isoalantolactone-induced survivin downregulation but not BCR-ABL protein, implying that the ubiquitin/proteasome pathway involved in survivin degradation.
Survivin overexpression in HEK293 cells treated with isoalantolactone resulted in increased survivin ubiquitination, implying that isoalantolactone can degrade survivin by increasing survivin ubiquitination. CETSA then observed isoalantolactone possibly binds and interacts with the surviving protein itself, presumably binding isoalantolactone. Survivin protein degradation may be aided by the intracellular ubiquitin proteasomal degradation pathway. This is analogous to M×106‘s direct interaction with survivin, which selectively degrades survivin via proteasome-mediated degradation [Citation31]. This demonstrates that isoalantolactone is directly targeted as an inhibitor to induce enhanced degradation of survivin protein, though it remains unknown how it interacts with survivin protein and how it increases the level of ubiquitin modification of survivin protein.
Caspase activation has been shown in the literature to downregulate BCR-ABL protein [Citation32,Citation33]. In this study, we discovered that isoalantolactone activated caspase-3 and increased cleaved-caspase-3 accumulation. PARP cleavage was induced after caspase activation [Citation34]. Furthermore, caspase inhibitors were found to reduce isoalantolactone-induced BCR-ABL degradation. These findings suggest that isoalantolactone can inhibit BCR-ABL protein expression by activating caspases. Caspase-3 and caspase-7 are thought to be involved in protein degradation, according to previous research [Citation35]. For example, activated caspase-3 promotes FTase/GGTase-subunit cleavage, resulting in the accumulation of unpropylated G-proteins. WT1 protein can also be degraded by Caspase-3 [Citation36]. Furthermore, caspase-7 has been shown to be capable of cleaving TNFRI [Citation37]. Caspase-3 and caspase-7 are also involved in BCR-ABL degradation induced by p53 [Citation4]. To investigate the molecular mechanism of isoalantolactone-induced BCR-ABL degradation, we knocked down caspase-3 expression in KBM5 and KBM5T315I cells and discovered that caspase-3 knockdown can partially rescue isoalantolactone-induced BCR-ABL degradation. Caspase 7 inhibition did not restore BCR-ABL degradation. This could be due to the fact that caspase-3 and caspase-7 have different activities toward substrate proteins. Caspase-3 has been reported to be more promiscuous than caspase-7 and to be the primary executioner caspase during the demolition phase of apoptosis [Citation35]. However, the detailed molecular mechanism of BCR-ABL fusion protein degradation by activated caspase-3 remains unknown.
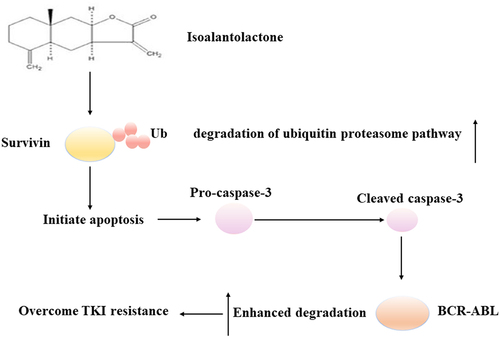
TKI resistance during CML treatment is urgently needed in the search for new anti-resistance strategies. According to current reports, most studies are focused on the discovery of TKI that can either downregulate BCR-ABL transcription or inhibit tyrosine kinase activity. We propose that isoalantolactone inhibits survivin by the ubiquitin proteasome pathway and mediates BCR-ABL downregulation in a caspase-3 dependent manner (). This provides a potential candidate compound as well as a hint for potential new strategies to overcome different types of TKI resistance.
Figure 8. The schematic diagram of the effect and mechanism of isoalantolactone in the process of overcoming imatinib resistance.
Authors’ contributions
Shan-Shan Yin: Conceptualization, Methodology, Writing – original draft preparation. Chen Chen: Conceptualization, Methodology, Writing – original draft preparation. Zhen Liu: Methodology. Shan-Ling Liu: Methodology, Resources. Chao Zhang: Supervision, Project administration. Quan-Wu Zhang: Supervision, Project administration. Feng-Hou Gao: Supervision, Project administration, Writing – review and editing, Conceptualization.
Ethics approval and consent to participate
All procedures were carried out in accordance with the ethical guidelines established by the Ethics Committee of Shanghai 9th People’s Hospital, Shanghai Jiao Tong University School of Medicine, and written informed consent was obtained.
List of abbreviations
| CML | = | Chronic myeloid leukemia |
| TKI | = | Tyrosine Kinase Inhibitors |
| IC50 | = | half maximal inhibitory concentration |
| CESTA | = | Cellular Thermal Shift Assay |
Supplemental Material
Download Zip (1.7 MB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.
Supplementary Material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384101.2023.2209963.
Additional information
Funding
References
- Melo JV, Barnes DJ. Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat Rev Cancer. 2007;7:441–453.
- Rangatia J, Bonnet D. Transient or long-term silencing of BCR-ABL alone induces cell cycle and proliferation arrest, apoptosis and differentiation. Leukemia. 2006;20:68–76.
- Shet AS, Jahagirdar BN, Verfaillie CM. Chronic myelogenous leukemia: mechanisms underlying disease progression. Leukemia. 2002;16:1402–1411.
- Di Bacco AM, TG C. P53 expression in K562 cells is associated with caspase-mediated cleavage of c-ABL and BCR-ABL protein kinases. Br J Haematol. 2002;117:588–597.
- Saglio G, Fava C. BCR-ABL1 mutation not equal ponatinib resistance. Blood. 2016;127:666–667.
- Carter BZ, Mak DH, Schober WD, et al. Regulation of survivin expression through Bcr-Abl/MAPK cascade: targeting survivin overcomes imatinib resistance and increases imatinib sensitivity in imatinib-responsive CML cells. Blood. 2006;107:1555–1563.
- Badran A, Yoshida A, Wano Y, et al. Expression of the antiapoptotic gene survivin in chronic myeloid leukemia. Anticancer Res. 2003;23:589–592.
- Conte E, Stagno F, Guglielmo P, et al. Survivin expression in chronic myeloid leukemia. Cancer Lett. 2005;225:105–110.
- Mori A, Wada H, Nishimura Y, et al. Expression of the antiapoptosis gene survivin in human leukemia. Int J Hematol. 2002;75:161–165.
- Liu Y, Shao Z, Liao Y, et al. Targeting SKP2/Bcr-Abl pathway with Diosmetin suppresses chronic myeloid leukemia proliferation. Eur J Pharmacol. 2020;883:173366.
- Fang Z, Jung KH, Yan HH, et al. CD-200 induces apoptosis and inhibits Bcr-Abl signaling in imatinib-resistant chronic myeloid leukemia with T315I mutation. Int J Oncol. 2015;47:253–261.
- Franks A, Egan S, Holmstrom C, et al. Inhibition of fungal colonization by Pseudoalteromonas tunicata provides a competitive advantage during surface colonization. Appl Environ Microbiol. 2006;72:6079–6087.
- Kancharla P, Kelly JX, Reynolds KA. Synthesis and structure-activity relationships of tambjamines and B-Ring functionalized prodiginines as potent antimalarials. J Med Chem. 2015;58:7286–7309.
- Pinkerton DM, Banwell MG, Garson MJ, et al. Antimicrobial and cytotoxic activities of synthetically derived tambjamines C and E - J, BE-18591, and a related alkaloid from the marine bacterium Pseudoalteromonas tunicata. Chem Biodivers. 2010;7:1311–1324.
- Martinez-Garcia D, Perez-Hernandez M, Korrodi-Gregorio L, et al. The natural-based antitumor compound T21 decreases survivin levels through potent STAT3 inhibition in lung cancer models. Biomolecules. 2019;9:361.
- Ding YH, Song YD, Wu YX, et al. Isoalantolactone suppresses LPS-induced inflammation by inhibiting TRAF6 ubiquitination and alleviates acute lung injury. Acta Pharmacol Sin. 2019;40:64–74.
- Lu N, Lv Q, Sun X, et al. Isoalantolactone restores the sensitivity of gram-negative enterobacteriaceae carrying MCR-1 to carbapenems. J Cell Mol Med. 2020;24:2475–2483.
- Tripathi YB, Tripathi P, Upadhyay BN. Assessment of the adrenergic beta-blocking activity of Inula racemosa. J Ethnopharmacol. 1988;23:3–9.
- Xu R, Peng Y, Wang M, et al. Intestinal absorption of isoalantolactone and alantolactone, two sesquiterpene lactones from radix inulae, using Caco-2 cells. Eur J Drug Metab Pharmacokinet. 2019;44:295–303.
- Jin C, Zhang G, Zhang Y, et al. Isoalantolactone induces intrinsic apoptosis through p53 signaling pathway in human lung squamous carcinoma cells. PLoS ONE. 2017;12:e0181731.
- Khan M, Ding C, Rasul A, et al. Isoalantolactone induces reactive oxygen species mediated apoptosis in pancreatic carcinoma PANC-1 cells. Int J Biol Sci. 2012;8:533–547.
- Li Y, Ni ZY, Zhu MC, et al. Antitumour activities of sesquiterpene lactones from Inula helenium and Inula japonica. Z Naturforsch C J Biosci. 2012;67:375–380.
- Li Z, Qin B, Qi X, et al. Isoalantolactone induces apoptosis in human breast cancer cells via ROS-mediated mitochondrial pathway and downregulation of SIRT1. Arch Pharm Res. 2016;39:1441–1453.
- Rasul A, Di J, Millimouno FM, et al. Reactive oxygen species mediate isoalantolactone-induced apoptosis in human prostate cancer cells. Molecules. 2013;18:9382–9396.
- Cai H, Meng X, Li Y, et al. Growth inhibition effects of isoalantolactone on K562/A02 cells: caspase-dependent apoptotic pathways, S phase arrest, and downregulation of Bcr/Abl. Phytother Res. 2014;28:1679–1686.
- Carrasco RA, Stamm NB, Marcusson E, et al. Antisense inhibition of survivin expression as a cancer therapeutic. Mol Cancer Ther. 2011;10:221–232.
- Hansen JB, Fisker N, Westergaard M, et al. SPC3042: a proapoptotic survivin inhibitor. Mol Cancer Ther. 2008;7:2736–2745.
- Dang CV, Reddy EP, Shokat KM, et al. Drugging the ‘undruggable’ cancer targets. Nat Rev Cancer. 2017;17:502–508.
- Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17:528–542.
- Zientara-Rytter K, Subramani S. The roles of ubiquitin-binding protein shuttles in the degradative fate of ubiquitinated proteins in the ubiquitin-proteasome system and autophagy. Cells. 2019;8:40.
- Zhao G, Wang Q, Wu Z, et al. Ovarian primary and metastatic tumors suppressed by survivin knockout or a novel survivin inhibitor. Mol Cancer Ther. 2019;18:2233–2245.
- Lan X, Zhao C, Chen X, et al. Platinum pyrithione induces apoptosis in chronic myeloid leukemia cells resistant to imatinib via DUB inhibition-dependent caspase activation and Bcr-Abl downregulation. Cell Death Dis. 2017;8:e2913.
- Shi X, Chen X, Li X, et al. Gambogic acid induces apoptosis in imatinib-resistant chronic myeloid leukemia cells via inducing proteasome inhibition and caspase-dependent Bcr-Abl downregulation. Clin Cancer Res. 2014;20:151–163.
- Duriez PJ, Shah GM. Cleavage of poly(ADP-ribose) polymerase: a sensitive parameter to study cell death. Biochem Cell Biol. 1997;75:337–349.
- Walsh JG, Cullen SP, Sheridan C, et al. Executioner caspase-3 and caspase-7 are functionally distinct proteases. Proc Natl Acad Sci U S A. 2008;105:12815–12819.
- Ruan J, Gao S, Yang J, et al. WT1 protein is cleaved by caspase-3 in apoptotic leukemic cells. Leuk Lymphoma. 2018;59:162–170.
- Ethell DW, Bossy-Wetzel E, Bredesen DE. Caspase 7 can cleave tumor necrosis factor receptor-I (p60) at a non-consensus motif, in vitro. Biochim Biophys Acta. 2001;1541:231–238.
