Abstract
The Drosophila innexin multigene family of gap junction encoding proteins consists of eight family members whose function in epithelial morphogenesis is mostly unknown. We have recently shown that innexin2 plays a crucial role in the organization of embryonic epithelia. Innexin2 protein accumulates in the epidermis in the apico-lateral membrane domain and colocalizes with core proteins of adherens junctions, such as DE-cadherin and Armadillo, the β -catenin homolog. Innexin2 localization is altered in both armadillo and DE-cadherin mutants Biochemical interaction studies point to a direct interaction of DE-cadherin and Armadillo with innexin2 suggesting a close link between gap junction and adherens junction biogenesis. We have used the Drosophila Schneider cell tissue culture system to further study the interaction of innexin2 with DE-cadherin. Our results provide evidence that DE-cadherin may be a key component to control trafficking, and localization of Innexin2 to the plasma membrane.
INTRODUCTION
Gap Junctions contain clusters of intercellular channels, which allow the direct exchange of ions and small molecules among neighboring cells, thus enabling cells and tissues to integrate signaling activities (Goodenough et al. Citation1996; Wei et al. Citation2004). A functional gap junction channel is formed by two hexameric hemichannels, one contributed by each of the opposing cells, which dock head-to-head in the extracellular space to form a double membrane-spanning intercellular channel (Segretain and Falk Citation2004; Martin and Evans Citation2004). Two major gene families have evolved to construct gap junction channels, the connexins in vertebrates and the innexins in invertebrates (Söhl and Willecke Citation2004; Bauer et al. Citation2005; Phelan Citation2005). Both gene families encode four-pass membrane domains with two extracellular loops, cytoplasmic N-and C-termini and a cytoplasmic loop domain. Although innexins and connexins are structurally and functionally analogous, they show virtually no sequence similarity to each other. Distantly related innexin homologs, named pannexins, have recently also been identified in mouse and humans (Panchin et al. Citation2000; Bruzzone et al. Citation2003; Panchin Citation2005).
Genome sequencing projects identified eight innexin genes in the fruit fly Drosophila, however, only a few mutants exist that have been molecularly and functionally characterized. It has been shown that innexin1 (ogre) and innexin8 (shakingB) are involved in the physiology of the giant fibre and the adult visual systems (Watanabe and Kankel Citation1990; Curtin et al. Citation2002; Krishnan et al. Citation1993; Phelan et al. Citation1996, Citation2005; Shimohigashi and Meinertzhagen Citation1998; Zhang et al. Citation1999; Jacobs et al. Citation2000); innexin4 (zero population growth) controls germ cell differentiation (Tazuke et al. Citation2002; Gilboa et al. Citation2003) and we have recently shown that during foregut morphogenesis, innexin2 is a Wingless/WNT target gene that coordinates epithelial morphogenesis (Bauer et al. Citation2002). Mutants for innexin2 (kropf), which lack both their maternal and zygotic innexin2 contributions fail to develop any epithelial tissues, and overexpression of innexin2 in the epidermis results in multilayering and irregular cell shapes suggesting that innexin2 is a key factor to control epithelial organization during morphogenesis of tissues and organs in the body (Bauer et al. Citation2004).
In the epidermis, innexin2 protein colocalizes with DE-cadherin and the β-catenin homolog Armadillo. Both proteins are core components of adherens junctions, which are multiprotein complexes mediating cell-cell adhesion and communication (Tepass Citation1999; Nagafuchi Citation2001, for reviews). Classic cadherins are major mediators of cell–cell adhesion. Their extracellular domain mediates calcium-dependent homophilic cell–cell adhesion, whereas their highly conserved intracellular domain is linked to the actin cytoskeleton (Tepass Citation1999). DE-cadherin possesses a highly conserved cytoplasmic domain that binds to the cytoplasmic Armadillo protein, thereby forming a cadherin-catenin complex. Armadillo, also serves as the transducer of the WNT signal/wingless signal (Tepass Citation1999; Nagafuchi Citation2001). Cadherins are required to maintain stable adhesion between epithelial cells (Larue et al. Citation1994; Riethmacher et al. Citation1995; Tepass et al. Citation1996; Uemura et al. Citation1996) but can also be used by migrating cells to adhere to a cellular substratum (Letourneau et al. Citation1990; Barami et al. Citation1994; Hazan et al. Citation2000; Li et al. Citation2001). In mutants for both zygotic armadillo and DE-cadherin, innexin2 localization is altered and in kropf mutants, components of adherens junctions are mislocalized (Bauer et al. Citation2004). Further evidence for a more direct interaction between innexin2 and adherens junction proteins was provided by yeast two hybrid analysis and coimmunoprecipitation experiments using embryonic extracts, which showed that innexin2 interacts via its cytoplasmic loop domain with the C-terminus of DE-cadherin (Bauer et al. Citation2004). These data suggest that the biogenesis of innexin2-containing gap junctions and of adherens junctions may be linked via the interaction of innexin2 and DE-cadherin/Armadillo. However, the molecular mechanism of how these proteins interact with each other is still rather elusive. We have used Drosophila tissue culture cells as a simpler model system to study the interactions of innexin2 and DE-cadherin. Our results provide evidence that DE-cadherin may be an important component to control trafficking and localization of innexin2 to the plasma membrane.
METHODS
Fly Strains
We used standard techniques for fly manipulation. The P allele kropf P16 that was used, is a transcript-null allele (Bauer et al. Citation2002).
Plasmids
pMT/V5-innexin2-GFP and pMT/V5-innexin2-Myc carry an innexin2 cDNA under the control of a copper-inducible promoter (pMT/V5 plasmid from; Invitrogen, Carlsbad, CA). innexin2-GFP cDNA was derived from pUAST-innexin2-GFP, which had been already used for the generation of transgenic flies (Bauer et al. Citation2004). innexin2-Mycwas generated by PCR reaction from LD11658 containing the entire innexin2cDNA using specific innexin2 primer pairs (innexin2 start-primer: 5′-GGAATTCCAAA ATGTTTGATGTCT TTGGGT -3′; innexin2 stop-primer containing the Myc-tag, which is printed in bold letters: 5′-CGCTCGAGCG GTTA CAGATCCTCTTCAGAGATGAGTTTC TGCTCGGCGTCGAAGGGCCGCTTG-3′). pR- mpRmHa3-DE-cadherin (DE-Cad wt) is also under control of a copper-inducible promoter, and was a kind gift of P. Rorth and A. Pacquelet (Pacquelet et al. Citation2003).
Cell Culture and DNA Transfection
Drosophila SL2 cells were maintained at 25°C in Drosophila Schneider Medium (Invitrogen) supplemented with 10% fetal bovine serum (heat inactivated), and 1% penicillin/streptomycin (Sigma, St Louis, USA). Cells were subcultured to 3 × 106 cells/ml every week. Transfection was performed using Cellfectin reagent (Invitrogen) according to the manufacturer's instructions. Expression of pMT/V5-innexin2-GFP and pRmHa3 DE-cadherin was induced with 0.7 mM CuSO4 for 72 h. Stable SL2 cell lines were generated by transfection of pMT/V5-innexin2-GFP (innexin2-GFP cells) or pMT/V5-innexin2-Myc (innexin2-Myc cells) together with a pAcHygro (Invitrogen) vector (ratio of transfected cDNAs; 2: 1 μg) using Cellfectin. Stable transfected cells were selected in SL2 medium containing hygromycin at 300 μg/ml. Subsequently, the cells were cultured continuously in hygromycin containing medium. Most of the cells (usually >70%) in the stable cell lines expressed innexin2-GFP or innexin2-Myc.
Antibodies and Immunohistochemistry
Antibody staining of Drosophila embryos was performed as described earlier (Fuss and Hoch Citation1998; Bauer et al. Citation2004). SL2 cells were fixed in 4% paraformaldehyde for 10 min at room temperature (RT), washed twice with PBS containing 1% BSA (PBSB), and permeabilised with PBS supplemented with 0, 2% Triton X-100 for 5 min. Following two washes with PBS and two washes with PBSB, SL2 cells were stained with the corresponding primary antibodies in PBSB for 30 min at RT. After rinsing four times for 5 min each time with PBSB at RT, the samples were processed for indirect immunofluorescence by incubation with the corresponding secondary fluorescent antibody for 30 min at RT. After rinsing three times for 5 min each time with PBSB and one time with PBS, the samples were mounted in Vectashield (hard set) mounting medium with DAPI. The following stains and primary antibodies were used: rabbit anti-Innexin 1, 2, 3 (1:75); goat anti-DE-cadherin (1:50; Santa Cruz); mouse anti-Crumbs (1:10; Hybridoma Bank); rabbit anti-PDI (Phospho-Disulfide Isomerase; 1:100, Calbiochem) as ER marker; monoclonal anti-c-Myc 9E10 (1: 100, Santa Cruz); Alexa Fluor 488 Phalloidin (1:1000, Molecular Probes). Secondary antibodies used were: Alexa Fluor 488 (1:200; MoBiTec); Alexa Fluor 546 (1:200; MoBiTec); and Cy5 (1:100, Dianova).
Microscopy and Image Processing
Fluorescent and DIC (Differential Image Contrast) pictures were taken on a Zeiss Axiovert 200M microscope with ApoTome using Zeiss Axiovision software (Carl Zeiss AG Jena, Germany). Otherwise fluorescent images were recorded using a Leica TSP2 confocal microscope (Leica, Wetzlar, Germany). Images of multilabeled samples were acquired sequentially on separate channels. All images were processed with Adobe Photoshop software.
Immunoprecipitation and Immunoblotting
For immunoprecipitation reactions Drosophila innexin2-Myc cells, were homogenized 72 h after induction with CuSO4 in RIPA buffer (150 mM NaCl, 1% IGEPAL CA-630, 0.5% Sodium Deoxycholate, 0,1% SDS, 50 mM Tris (pH 8,0) with protease inhibitors (Roche)). Immunoprecipitation was performed using the Immunoprecipitation Starter Pack (Amersham Pharmacia Biotech) according to the manufacturer's instruction. To prevent unspecific binding of proteins to protein G Sepharose, preclearing of the SL2 cell lysate was performed in the presence of protein G Sepharose according the manufacturer's instructions. Cleared supernatant was incubated for 3 h with anti-innexin2 antibody or control IgG, followed by the addition 40 μl protein G-Sepharose to each mixture and incubation for 3 h. The beads were washed three times with excess RIPA-buffer and two times with excess PBS, resuspended in SDS-PAGE sample buffer, and boiled for 3 min. Samples were run on a 12.5% SDS-gel, and transferred to a PVDF-membrane (Immobilon P transfer-membrane; Millipore). The immunoblot membrane was blocked with nonfat dry milk in TBS plus 0.05% Tween (TBST) and then incubated with anti-DE-cadherin (1: 400, St Cruz). After washing, bound antibody was visualized with peroxidase–conjugated donkey antigoat IgG (1.15000) using the ECL system (Amersham Pharmacia).
RT-PCR
Total RNA was prepared from SL2 cells with the Qiagen RNAeasy Kit (Qiagen) according to the manufacturer's instructions. RT-PCR was performed as described by Zinke et al. (Citation2002) with minor modifications. Briefly, 1 μ g of total RNA was used for First strand cDNA reaction using oligo-d(T) primers (Amersham) and Super Script III reverse transcriptase (Gibco/Invitrogen) in a final volume of 20 μl at 50°C for 60 min. After inactivation of the Super Script III reverse transcriptase 80 μl H2O were added for a final volume of 100 μl. PCR was done on aliquots of this reaction in a total volume of 25 μl with GoTaq™ DNA Polymerase (Promega). As loading control we used actin 5C. The following innexin primers were used in the PCR reactions depicted in : innexin1: 5′-AAACTCGCTCATCACGTCCT-3′, 5′-TGCAGAAGCACGACTCACTC-3′; innexin2: 5′-CCGCCATCTCCTACTCCGAG-3′, 5′-TGTGG GCGCTATGCTCATC-3′; innexin3: 5′-CGGTCAT GTCTCCGTCTCCTT-3′, 5′-CTATTCACTGGTG GTTATCA-3′.
Figure 1 Staining of DE-cadherin in Drosophila embryos, and mRNA expression levels of innexins 1, 2, and 3 in SL2 cells. (A) DE-cadherin (green) staining in a wildtype embryo. (B) Double staining of an innexin2 (kropf) mutant embryo monitoring expression of Crumbs (blue) and DE-cadherin (green). DE-cadherin is localized at the membrane in epithelial cells of wildtype embryos (A). In kropf embryos, the DE-cadherin signal is partially reduced and accumulates at the cortex or in the cytoplasm of epithelial cells. Most of the transmembrane protein Crumbs is still properly localized (B). (C, D) RT-PCR analysis of innexin 1, 2, and 3 mRNA expression level in SL2 cells. Actin 5C was used as loading control. (C) Innexins 1, and 3 are detectable after 30 PCR cycles. The mRNA innexin1 signal is slightly higher compared to the very weak innexin3 signal. Innexin2 clearly shows the highest expression level in SL2 cells compared to innexins 1, and 3. (E) iQ5 Real-Time PCR reaction using the iQ5 Real-Time PCR Detection System (BIO RAD) with innexin1, 2, and 3 primer pairs checked for primer-dimer artifacts and efficiencies confirmed the result depicted in (C) and (D); (SD: Standard deviation).
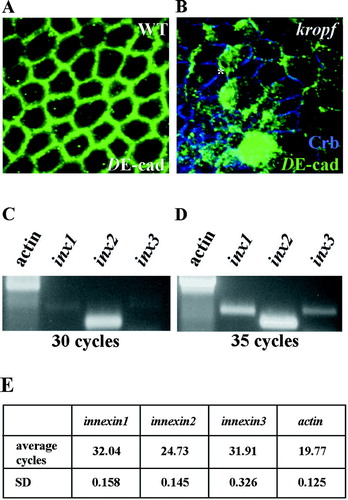
A second set of innexin1, 2, and 3 primer pairs was designed and tested for primer-dimer artefacts and efficiencies using the iQ5 Optical System Software (BIO RAD). Primer efficiencies determined for the different innexin primer pairs were: innexin1: 100%, (R2: 0.998); innexin2 : 98.3 %, (R2: 0.996); innexin3: 100%, (R2: 0.998).
PCR analysis with the second set of innexin1, 2, and 3 primer pairs was performed on aliquots of the reverse transcribed total RNA (described above) according to the manufacturer's instructions using the iQ5 Real-Time PCR Detection System (BIO RAD). PCR reactions were set up with the iQ™ SYBRR Green Supermix (BIO RAD), in a final volume of 25 μ l. The following innexin primers were used in the iQ5 Real-Time PCR reaction: innexin1: 5′-AATATCACGATCTGCACCCGC-3′; 5′-CCGA ATGGCGTACAGCTTGT-3′; innexin2: 5′-CCTAC TCCGAGCCCGTTCC-3′; 5′-TGCCCAGCTGA TAGAGCAGG-3′; innexin3: 5′-GATCGGTCCA GAAACACGACA-3′; 5′-GGAGATGGTGGCCA AGATGAT-3′.
RESULTS
In Drosophila melanogaster the apical epithelial region is defined by the subapical complex (SAC) and the zonula adherens (ZA). The SAC complex harbors the transmembrane protein Crumbs (Crb), the four-PDZ domain protein Discs Lost and the MAGUGK protein Stardust (Tepass, Citation1996; Bhat et al. Citation1999; Klebes and Knust Citation2000; Bachmann et al. Citation2001; Hong et al. Citation2001). DE-cadherin, α-catenin, and β-catenin/Armadillo are major ZA components (Tepass Citation1996). Within this apico-lateral region DE-cadherin and β-catenin/Armadillo colocalize with innexin2 (Bauer et al. Citation2004). In kropf mutants, DE-cadherin (compare and 1B) and β-catenin/ Armadillo (Bauer et al. Citation2004) are mislocalized. Moreover, in both zygotic armadillo and DE-cadherin mutants, the innexin2 localization is altered (Bauer et al. Citation2004). To further study the interaction of innexin2 with key proteins of the adherens junction complex such as DE-cadherin, we established a tissue culture system using Drosophila Schneider cells. Most of the established Schneider cell lines such as S2 (Schneider Citation1972), SL2 (Bruckner et al. Citation2000), Kc167 (Echalier and Ohanessian Citation1969), and S2R+ (Yanagawa et al. Citation1998) are derived from embryonic hemocytes. Whereas S2, SL2, and Kc167 cells are small and round, S2R+ cells are large, flat and strongly adherent to glass, and extracellular matrix (Kiger et al. Citation2003). As we have shown already that Schneider cell lines such as S2 and SL2 express innexin2 and that Armadillo, a component of the adherens junction complex and also a mediator of the WNT/Wg signalling pathway regulates innexin2 expression (Bauer et al. Citation2002), we started our analysis of innexin2 interactions with key proteins of the adherens junction complex in SL2 cells. This line is derived from a primary culture of 20–24-h-old embryos. SL2 cells are hemocyte-like and phagocytic cells. Under routine culture conditions, SL2 cells display a roughly spherical morphology (diameter of 10 μ m) and a thin rim of cytoplasm surrounds the nucleus ().
Figure 2 Localization of innexin 1, 2, and 3 within Drosophila SL2 cells. (A–C) Triple staining of SL2 cells using anti innexin 1, 2, and 3 (red), Phalloidin as actin marker (green), and Dapi as nuclear marker (blue). Innexin proteins are predominantly found in the cytoplasm of the SL2 cells. (D–F) Triple staining of innexin2-Myc cells displaying innexin2-Myc (green), PDI (red), an endoplasmic reticulum marker, and nuclear staining with Dapi (blue). (D) Most of innexin2-Myc is colocalized with the ER.
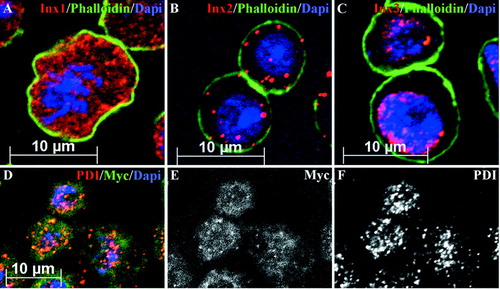
Towards further characterization of innexin expression levels in SL2 cells, we performed RT-PCR analysis using specific primers (see Methods) for the innexin genes 1, 2 and 3. All of the three innexin genes are expressed in SL2 cells (, ). The mRNA expression level is highest for innexin2 whereas innexin1 and 3 show a significantly lower expression. To exclude the possibility that different primer efficiencies of the different innexin primer pairs might influence RT-PCR analysis, we generated a second set of innexin primers. These primers were checked for primer-dimer artefacts and efficiencies (see Methods). We then used the tested primer pairs in repeat RT-PCR analysis of the relative abundance of innexins1, 2, and 3 in SL2 cells. As shown in , innexin2 expression is highest. An innexin2 signal is detected already after 25 PCR amplification cycles by the iQ5 Real-Time PCR Detection System (BIO RAD), whereas innexin1, and 3 signals are obtained after about 32 PCR amplification cycles. This experiment confirms the result depicted in and . To analyze the protein localization of endogenous innexin proteins in SL2 cells, we performed immunohistochemistry applying antibodies against the innexins 1, 2 and 3 (Bauer et al. Citation2001, Citation2003) in combination with DAPI for nuclear labeling, and phalloidin to visualize the actin cytoskeleton (). All three innexin proteins are localized within the cytoplasm of SL2 cells, with high level of expression around the nucleus (–); only a small fraction of the proteins is localized to the membrane. Coimmunostaining with the ER-marker Phospho-Disulfide-Isomerase (PDI; Lee et al. Citation2001; Urban et al. Citation2001) and an anti-Myc antibody in stable innexin2-Myc cells, indicates that most of the innexin2 protein is localized to the ER ( ). The cytoplasmic localization of endogenous innexins in SL2 cells is in contrast to their membrane localization in embryonic epithelia (Bauer et al. Citation2001, Citation2003, Citation2004) suggesting that key factors for trafficking, membrane insertion or for the stabilization of innexin hemichannels in the membranes may be lacking in SL2 cells. It is noteworthy in this context that SL2 cells are not polarized and also grow as single cells. Consistent with this, the cell adhesion molecule DE-cadherin is expressed in most of the SL2 cells at a low level (, ); in addition, we often find a large amount of the protein localized to the cytoplasm ().
Figure 3 Addition of DE-cadherin to Drosophihla SL2 cells alters the localization of innexin2. (A) DIC image of SL2 cells. (B) The same picture as depicted in (A) showing DE-cadherin (red) staining of SL2 cells. DE-cadherin is evenly distributed in the cytoplasm. Dapi (blue) was used as nuclear marker. (C) Different DE-cadherin levels of SL2 cells. Only a few cells express strongly DE-cadherin (red), whereas most of the SL2 cells show low DE-cadherin levels. (D, E) innexin2 localization before and after transfection of DE-cadherin. (D) Endogenous innexin2 in SL2 cells resides in the cytoplasm. (E) Addition of DE-cadherin results in an increased localization of innexin2 at the plasma membrane. Noteworthy, innexin2 is significantly enriched at cell-cell contact sites (arrows). (F–J) Distribution of innexin2-GFP and DE-cadherin in a single innexin2-GFP cell before and after the transfection of DE-cadherin. (F) Without addition of DE-cadherin innexin2-GFP is predominantly localized in the cytoplasm. (G–I) After transfection of DE-cadherin innexin2-GFP is predominantly and uniformly localized at the membrane. (G) Innexin2-GFP localization. (H) DE-cadherin distribution in the same cell as depicted in G. (I) Overlay of innexin2-GFP (G) and DE-cadherin (H). (J). 3-D view of the cell depicted in G–I rendered from different confocal sections. Note the high degree of DE-cadherin and innexin2-GFP colocalization. (K) Direct interaction of DE-cadherin and innexin2 in Drosophila SL2 cells. Innexin2-Myc cells either transfected (Lanes 1, 3, and 4) or not transfected with DE-cadherin (lane 2) were used for immunoprecipitation with an anti-innexin2 antibody (Lanes 1, 2, and 3). A control immunoprecipitation was performed using an IgG antibody (Lane 4). To accommodate the fact that innexin2-Myc cell extracts used for immunoprecipitation had either just some endogenous DE-cadherin or additionally transfected DE-Cadherin, endogenous DE-Cadherin and transfected DE-Cadherin are each identified by “+” and “+++”, respectively.
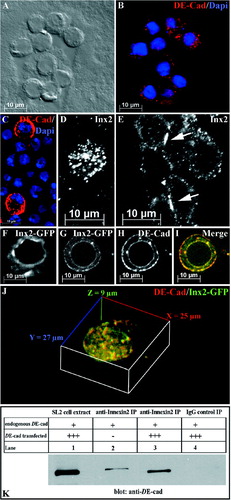
To test whether cell-to-cell contact may be required for correct innexin localization, we increased the amount of DE-cadherin in the cells by transfecting a DE-cad wt expression plasmid (see Methods). As has been previously observed, this causes SL2 cells to aggregate at a high rate (Pacquelet et al. Citation2003), although a considerable number of cells are still found that do not aggregate. Increasing the level of DE-cadherin, results in an altered localization of innexin2 protein. Whereas innexin2 is localized predominantly to the cytoplasm in untransfected cells (), it localizes to the membrane in transfected cells, which express high levels of DE-cadherin (, compare to D). Furthermore, we find a localized accumulation of innexin2 in membrane domains at cell-to-cell contact sites (arrows in ), which are highly reminiscent of gap junction plaques induced by overexpression of connexins in HeLa cells (Lauf et al. Citation2002; Gaietta et al. Citation2002).
To investigate whether it is merely cell aggregation that determines the membrane localization of innexin2, we analyzed DE-cadherin transfected innexin2-GFP cells that have not aggregated but still express DE-cadherin at a high level. In these cells, the cytoplasmic localization of innexin2-GFP is also altered. Both DE-cadherin and 2-GFP are predominantly localized to the membrane (–I) as compared to the distribution of 2-GFP in non-DE-cadherin transfected 2-GFP cells (). Noteworthy is also the high degree of 2-GFP and DE-cadherin colocalization in this cell (, and ). Coimmunoprecipitation experiments in innexin2-Myc cells transfected with DE-cadherin provided strong evidence for a direct interaction of DE-cadherin and innexin2 (). Recently our genetic and biochemical interaction studies in Drosophila embryos have already shown a direct interaction of DE-cadherin and innexin2 (Bauer et al. Citation2004). Recapitulating, these data suggest that DE-cadherin may control trafficking and innexin2 localization to the plasma membrane in SL2 cells.
DISCUSSION
The assembly of the junctional complement of a particular cell is a dynamic process that is essential for the proper differentiation of cells and tissues during development(Tepass and Hartenstein Citation1994). It has been demonstrated previously that the spatial association between gap and adherens junctions is an important factor in the maturation of vertebrate and invertebrate tissues (Angst et al. Citation1997; Tepass and Hartenstein Citation1994). Numerous studies in mammalian and avian tissue culture cells have provided evidence for the importance of cell adhesion contacts for gap junction formation (see Wei et al. Citation2004, for an extensive review). It was demonstrated in embryonic chick lens cultures, in adult rat cardiomyocytes, and in reaggregated Novikoff cells that N-cadherin is required for gap junction assembly (Frenzel and Johnson Citation1996; Hertig et al. Citation1996; Meyer et al. Citation1992; Ko et al. Citation2000) and a direct interaction of the Armadillo homologue β-catenin with connexin43 was shown to occur in cardiac myocytes (Ai et al. Citation2000). Similarly, tissue culture studies and the analysis of neural crest cell migration in the mouse embryo have shown that the assembly and function of gap junctions is directly controlled by E-and N-cadherins, respectively (Jongen et al. Citation1991; Musil et al. Citation1990; Fujimoto et al. Citation1997; Xu et al. Citation2001). Inhibition of cadherin function, such as in N-cadherin-deficient myocytes, can disrupt gap junction formation and inhibit cell-cell coupling, suggesting that localization of cadherin to cell-cell contact sites may be a prerequisite for gap junction formation (Meyer et al. Citation1992; Luo and Radice Citation2003). Conversely, inhibition of connexin43 can block adherens junction formation (Zuppinger et al. Citation2000). The formation of connexin channels and the development of junctional communication is also promoted by NCAM-mediated adhesion in neuroectoderm of the chick (Keane et al. 1998).
Also in Drosophila, the biogenesis of innexin2-containing gap junctions and of adherens junctions may be linked via the interaction of innexin2 and DE-cadherin/Armadillo. It was demonstrated in the embryonic epidermis that innexin2 protein colocalizes with the adherens junction proteins Armadillo and DE-cadherin. Kropf mutant embryos lacking both the maternal and zygotic innexin2 contribution fail to develop any epithelial tissues (Bauer et al. Citation2004). Embryos in which innexin2 has been extensively overexpressed are embryonic lethal and display defects such as multilayering and irregular cell shapes (Bauer et al. Citation2004). In mutants for both zygotic armadillo and DE-cadherin innexin2 localization is altered and in innexin2 overexpression experiments, Armadillo and DE-cadherin are organized into the innexin2 pattern (Bauer et al. Citation2004). Further evidence for a more direct interaction between innexin2 and adherens junction proteins was provided by yeast two hybrid analysis and coimmunoprecipitation experiments using embryonic extracts, which showed that innexin2 interacts via its cytoplasmic loop domain with the C-terminus of DE-cadherin (Bauer et al. Citation2004).
To study further the interaction of innexin2 with key proteins of the adherens junction complex such as DE-cadherin, we established a tissue culture system using Drosophila SL2 cells. Our RT-PCR experiments and our antibody staining show that innexin1, 2, and 3-mRNAs and -proteins are expressed in SL2 cells. However, the endogenous innexin proteins are localized within the cytoplasm of SL2 cells with high level of expression around the nucleus and only a small fraction of the proteins is localized to the membrane (). SL2 cells lack cell polarity and grow mostly as single cells. The cell adhesion molecule DE-cadherin is expressed in most of the SL2 cells at a quite low level (). Increasing the amount of DE-cadherin in the cells causes SL2 cells to aggregate and results in the displacement of innexin2 from the cytoplasm to the membrane (); innexin2 accumulates locally at cell-to-cell contact sites, a pattern which is highly reminiscent of gap junction plaques induced by overexpression of connexins in HeLa cells (Lauf et al. Citation2002; Gaietta et al. Citation2002). The membrane localization of innexin2 is also achieved in single DE-cadherin expressing cells (, , , , ), which are nonaggregated and lack cell-to-cell contact sites. innexin2 shows a uniform distribution in the membrane of these cells (, , , , ) suggesting that DE-cadherin may control trafficking of innexin2 to the plasma membrane in SL2 cells; cell-to-cell contact seems to be required for the accumulation of innexin2 in gap junction plaques (). It is known that E-cadherin is trafficked to and from the cell surface by exocytic and multiple endocytic pathways (Bryant and Stow Citation2005, for review).
During trafficking to and from the cell surface, cadherin–catenin complexes are dynamically assembled and disassembled. β-catenin binds to E-cadherin early in the biosynthetic pathway and the two proteins are trafficked and sorted as a complex for delivery to the epithelial-cell surface, as demonstrated in Madin–Darby canine kidney cells (Chen et al. Citation1999; Miranda et al. Citation2003). Since E-cadherin is intimately involved in the establishment and maintenance of epithelial-cell polarity, it is sorted and delivered in a polarized manner to the lateral cell surface. Putative sorting signals have been characterized within the cytoplasmic tail of E-cadherin (Bryant and Stow Citation2005, for review). The direct interactions of innexin2 with DE-cadherin, which we have shown previously by yeast two hybrid and immunoprecipitation analysis, (Bauer et al. Citation2004) suggest the possibility that innexin2 may be part of the cadherin-catenin complexes during trafficking. Experiments in rat cardiomyocytes have indicated that the formation of a catenin-ZO1-connexin43 complex is required for connexin43 transport to the plasma membrane during the assembly of gap junctions (Wu et al. Citation2003). Furthermore, it was recently shown that the mammalian connexin43α 1 coassembles in a multiprotein complex containing N-cadherin and various N-cadherin associated proteins in NIH3T3 cells (Wei et al. Citation2005). By using siRNA knock down, it was demonstrated that cell surface expression of connexin43α 1 requires N-cadherin and conversely, that N-cadherin surface expression requires connexin43α 1. Pulse chase labeling and biotinylation experiments indicated that in the absence of N-cadherin, connexin43α 1 trafficking is blocked, suggesting that the intracellular coassembly of connexins and cadherin is required for gap junction and adherens junction formation (Wei et al. Citation2005). Thus in summary, there seems an intimate linkage between the assembly of connexin-and innexin-containing gap junctions and adherens junctions in both vertebrates and in Drosophila. Our results suggest that also in the Drosophila epidermis, common trafficking routes of adherens junction and gap junction proteins within cells may ensure the positioning of innexin-containing hemichannels in membrane domains close to adherens junctions.
ACKNOWLEDGMENTS
We thank P. Rorth and A. Paquelet for the pRmHa3-DE-cadherin construct, F. Eckardt for technical assistance, and F. Josten for his assistance with the microscopes. Also we want to thank Prof. Waldemar Kolanus for helpful comments. The work was supported by a DFG grant to M.H. (SFB 645).
REFERENCES
- Ai Z, Fischer A, Spray D C, Brown A MC, Fishman G I. Wnt-1 regulation of connexin 43 in cardiac myocytes. J Clin Invest 2000; 105: 161–171, [PUBMED], [CSA]
- Angst A D, Khan L U, Severs N J, Whitely K, Rothery S, Thompson R P, Magee A I, Gourdie R G. Dissociated spatial patterning of gap junctions and cell adhesion junctions during postnatal differentiation of ventricular myocardium. Circ Res 1997; 80: 88–94, [PUBMED], [CSA]
- Bachmann A, Schneider M, Grawe F, Theilenberg E, Knust E. Drosophila Stardust is a partner of Crumbs in the control of epithelial cell polarity. Nature 2001; 414: 638–643, [PUBMED], [CSA]
- Barami K, Kirschenbaum B, Lemmon V, Goldman S A. N-cadherin and Ng-CAM/8D9 are involved serially in the migration of newly generated neurons into the adult songbird brain. Neuron 1994; 13: 567–582, [PUBMED], [CSA]
- Bhat M A, Izaddoost S, Lu Y, Cho K O, Choi K W, Bellen H J. Discs lost a novel multi-PDZ domain protein establishes and maintains epithelial polarity. Cell 1999; 96: 833–845, [PUBMED], [CSA]
- Bauer R, Lehmann C, Fuss B, Eckardt F, Hoch M. The Drosophila gap junction channel gene innexin 2 controls foregut development in response to Wingless signalling. J Cell Sci 2002; 115: 1859–1867, [PUBMED], [CSA]
- Bauer R, Lehmann C, Hoch M. Gastrointestinal development in the Drosophila embryo requires the activity of Innexin gap junction channel proteins. Cell Commun Adhes 2001; 8: 307–310, [PUBMED], [CSA]
- Bauer R, Lehmann C, Martini J, Eckardt F, Hoch M. Gap junction channel protein innexin 2 is essential for epithelial morphogenesis in the Drosophila embryo. Mol Biol Cell 2004; 15: 2992–3004, [PUBMED], [CSA]
- Bauer R, Loer B, Ostrowski K, Martini J, Weimbs A, Lechner H, Hoch M. Intercellular communication: The Drosophila innexin multiprotein family of gap junction proteins. Chem Biol 2005; 12: 515–526, [PUBMED], [CSA]
- Bauer R, Martini J, Lehmann C, Hoch M. Cellular distribution of innexin 1 and 2 gap junctional channel proteins in epithelia of the Drosophila embryo. Cell Commun Adhes 2003; 10: 1–225, [CSA]
- Bruzzone R, Hormuzdi S G, Barbe M T, Herb A, Monyer H. Pannexins a family of gap junction proteins expressed in brain. PNAS 2003; 100: 13644, [PUBMED], [CSA]
- Bruckner K, Perez L, Clausen H, Cohen S. Glycosyltransferase activity of fringe modulates Notch-Delta interactions. Nature 2000; 27: 411–415, [CSA]
- Bryant D M, Stow J L. Nuclear translocation of cell-surface receptors: lessons from fibroblast growth factor. Traffic 2005; 6: 947–954, [PUBMED], [CSA]
- Chen Y T, Stewart D B, Nelson W J. Coupling assembly of the E-cadherin–β-catenin complex to efficient endoplasmic reticulum exit and basal–lateral membrane targeting of E-cadherin in polarized MDCK cells. J Cell Biol 1999; 144: 687–699, [PUBMED], [CSA]
- Curtin K D, Zhang Z, Wyman R J. Gap junction proteins expressed during development are required for adult neural function in the Drosophila optic lamina. J Neurosci 2002; 22: 7088, [PUBMED], [CSA]
- Echalier G, Ohanessian A. C R Acad. Sci. Hebd. Seances Acad. Sci. D. 1969; 31: 1771–1773, [CSA]
- Frenzel E M, Johnson R G. Gap junction formation between cultured embryonic lens cells is inhibited by antibody to N-cadherin. Dev Biol 1996; 179: 1–16, [PUBMED], [CSA]
- Fujimoto K, Nagafuchi A, Tsukita S KA, Ohokuma A, Shibata Y. Dynamics of connexins E-cadherin and α -catenin on cell membranes during gap junction formation. J Cell Sci 1997; 110: 311–322, [PUBMED], [CSA]
- Fuss B, Hoch M. Drosophila endoderm development requires a novel homeobox gene which is a target of Wingless and Dpp signaling. Mech Dev 1998; 79: 83–87, [PUBMED], [CSA]
- Gaietta G, Deerinck T J, Adams S R, Bouwer J, Tour O, Laird D W, Sosinsky G E, Tsien R Y, Ellisman M H. Multicolor and electron microscopic imaging of connexin trafficking. Science 2002; 19: 503–507, [CSA]
- Gilboa L, Forbes A, Tazuke S I, Fuller M T, Lehmann R. Germ line stem cell differentiation in Drosophila requires gap junctions and proceeds via an intermediate state. Development 2003; 130: 6625–6634, [PUBMED], [CSA]
- Goodenough D A, Goliger J A, Paul D L. Connexins connexons and intercellular communcation. Annu Rev Biochem 1996; 65: 475–602, [PUBMED], [CSA]
- Hazan R B, Phillips G R, Qiao R F, Norton L, Aaronson S A. Exogenous expression of N-cadherin in breast cancer cells induces cell migration invasion and metastasis. J Cell Biol 2000; 21: 779–790, [CSA]
- Hertig C M, Eppenberger-Eberhardt M, Koch S, Eppenberger H M. N-cadherin in adult rat cardiomyocytes in culture. I. Functional role of N-cadherin and impairment of cell-cell contact by a truncated N-cadherin mutant. J Cell Sci 1996; 109: 1–10, [PUBMED], [CSA]
- Hong Y, Stronach B, Perrimon N, Jan L Y, Jan Y N. DrosophilaStardust interacts with Crumbs to control polarity of epithelia but not neuroblasts. Nature 2001; 414: 634–638, [PUBMED], [CSA]
- Jacobs K, Todman M G, Allen M J, Davies J A, Bacon J P. Synaptogenesis in the giant-fibre system of Drosophila: Interaction of the giant fibre and its major motorneuronal target. Development 2000; 127: 5203–5212, [PUBMED], [CSA]
- Jongen W MF, Fithgerald D J, Asamoto M, Piccoli C, Slaga T J, Gros D, Takeichi M, Yamasaki H. Regulation of connexin 43-mediated gap junctional intercellular communication by Ca2+ in mouse epidermal cells is controlled by E-cadherin. J Cell Biol 1991; 114: 545–555, [PUBMED], [CSA]
- Keane R W, Mehta P P, Rose B, Honig L S, Loewenstein W R, Rutishauser U. Neural differentiation NCAM-mediated adhesion and gap junctional communication in neuroectoderm. A study in vitro. J Cell Biol 1988; 106: 1307–1319, [PUBMED], [CSA]
- Kiger A A, Baum B, Jones S, Jones M R, Coulson A, Echeverri C, Perrimon N. A functional genomic analysis of cell morphology using RNA interference. J Biol 2003; 2: 27, [PUBMED], [CSA]
- Klebes A, Knust E. A conserved motif in Crumbs is required for E-cadherin localisation and zonula adherens formation in Drosophila. Current Biology 2000; 10: 76–85, [PUBMED], [CSA]
- Ko K, Arora P, Lee W, McCulloch C. Biochemical and functional characterization of intercellular adhesion and gap junctions in fibroblasts. Cell Phys 2000; 279: C147, [CSA]
- Krishnan S N, Frei E, Swain G P, Wyman R J. Passover: A gene required for synaptic connectivity in the giant fibre system of Drosophila. Cell 1993; 73: 967, [PUBMED], [CSA]
- Larue L, Ohsugi M, Hirchenhain J, Kemler R. E-cadherin null mutant embryos fail to form a trophectoderm epithelium. Proc Natl Acad Sci USA 1994; 16: 8263–8267, [CSA]
- Lauf U, Giepmans B NG, Lopez P, Braconnot S, Chen S -C, Falk M M. Dynamic trafficking and delivery of connexons to the plasma membrane and accretion to gap junctions in living cells. PNAS 2002; 99: 10446–10451, [PUBMED], [CSA]
- Lee S, Gilula N B, Warner A E. Gap junctional communication and compaction during preimplantation stages of mouse development. Cell 1987; 51: 851–60, [PUBMED], [CSA]
- Lee J R, Urban S, Garvey C F, Freeman M. Regulated intracellular ligand transport and proteolysis control EGF signal activation in Drosophila. Cell 2001; 107: 161–171, [PUBMED], [CSA]
- Letourneau P C, Shattuck T A, Roche F K, Takeichi M, Lemmon V. Nerve growth cone migration onto Schwann cells involves the calcium-dependent adhesion molecule N-cadherin. Dev Biol 1990; 138: 430–442, [PUBMED], [CSA]
- Li G, Satyamoorthy K, Herlyn M. N-cadherin-mediated intercellular interactions promote survival and migration of melanoma cells. Cancer Res 2001; 61: 3819–3825, [PUBMED], [CSA]
- Luo Y, Radice G L. Cadherin-mediated adhesion is essential for myofibril continuity across the plasma membrane but not for assembly of the contractile apparatus. J Cell Sci 2003; 116: 1471–1479, [PUBMED], [CSA]
- Martin P E, Evans W H. Incorporation of connexins into plasma membranes and gap junctions. Cardiovasc Res 2004; 62: 378–387, [PUBMED], [CSA]
- Meyer R A, Laird D W, Revel J P, Johnson R G. Inhibition of gap junction and adherens junction assembly by connexin and A-CAM antibodies. J Cell Biol 1992; 119: 179–189, [PUBMED], [CSA]
- Miranda K C, Joseph S R, Yap A S, Teasdale R D, Stow J L. Contextual binding of p120ctn to E-cadherin at the basolateral plasma membrane in polarized epithelia. J Biol Chem 2003; 278: 43480–43488, [PUBMED], [CSA]
- Musil L S, Cunningham B A, Edelman G M, Goodenough D A. Differential phosphorylation of the gap junction protein connexin 43 in junctional communication-competent and -deficient cell lines. J Cell Biol 1990; 111: 2077–2088, [PUBMED], [CSA]
- Nagafuchi A. Molecular architecture of adherens junctions. Curr Opin Cell Biol 2001; 13: 600–603, [PUBMED], [CSA]
- Pacquelet A, Lin L, Rorth P. Binding site for p120/delta-catenin is not required for Drosophila E-cadherin function in vivo. J Cell Biol 2003; 160: 313–319, [PUBMED], [CSA]
- Panchin Y, Kelmanson I, Matz M, Lukyanov K, Usman N, Lukyanov S. A ubiquitous family of putative gap junction molecules. Curr Biol 2000; 10: 473–474, [CSA]
- Panchin Y V. Evolution of gap junction proteins—the pannexin alternative. J Exp Biol 2005; 208: 1415–1419, [PUBMED], [CSA]
- Phelan P, Nakagawa M, Wilkin M B, Moffat K G, O'Kane C J, Davies J A, Bacon J P. Mutations in shaking-B prevent electrical synapse formation in the Drosophila giant fibre system. J Neurosci 1996; 16: 1101–1113, [PUBMED], [CSA]
- Phelan P. Innexins: Members of an evolutionarily conserved family of gap-junction proteins. Biochim Biophys Acta 2005; 1711: 225–245, [PUBMED], [CSA]
- Riethmacher D, Brinkmann V, Birchmeier C. A targeted mutation in the mouse E-cadherin gene results in defective preimplantation development. Proc Natl Acad Sci USA 1995; 92: 855–859, [PUBMED], [CSA]
- Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. Embryol Exp Morphol 1972; 27: 353–365, [CSA]
- Segretain D, Falk M. Regulation of connexin biosynthesis assembly gap junction formation and removal. Biochim Biophys Acta 2004; 23: 3–21, [CSA]
- Shimohigashi M, Meinertzhagen I A. The shaking B gene in Drosophila regulates the number of gap junctions between photoreceptor terminals in the lamina. J Neurobiol 1998; 35: 105, [PUBMED], [CSA]
- Söhl G, Willecke K. Gap junctions and the connexin protein family. Cardiovasc Res 2004; 62: 228–232, [CSA]
- Tazuke S I, Schulz C, Gilboa L, Fogarty M, Mahowald A P, Guichet A, Ephrussi A, Wood C G, Lehmann R, Fuller M T. A germline-specific gap junction protein required for survival of differentiating early germ cells. Development 2002; 129: 2529–2539, [PUBMED], [CSA]
- Tepass U. Genetic analysis of cadherin function in animal morphogenesis. Curr Opin Cell Biol 1999; 11: 540–548, [PUBMED], [CSA]
- Tepass U, Hartenstein V. The development of cellular junctions in the Drosophila embryo. Dev Biol 1994; 161: 563–596, [PUBMED], [CSA]
- Tepass U, Gruszynski-DeFeo E, Haag T A, Omatyar L, Török T, Hartenstein V. Shotgun encodes Drosophila E-cadherin and is preferentially required during cell rearrangement in the neuroectoderm and other morphogenetically active epithela. Genes Dev 1996; 10: 672–685, [PUBMED], [CSA]
- Uemura T, Oda H, Kraut R, Hayashi S, Kataoka Y, Takeichi M. Zygotic Drosophila E-cadherin expression is required for processes of dynamic epithelial cell rearrangement in the Drosophila embryo. Genes Dev 1996; 10: 659–671, [PUBMED], [CSA]
- Urban S, Lee J R, Freeman M. Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell 2001; 19: 173–182, [CSA]
- Van d er, Heyden M A, Rook M B, Hermans M M, Rijksen G, Boonstra J, Defize L H, Destree O H. Identification of connexin 43 as a functional target for Wnt signalling. J Cell Sci 1998; 111: 1741–1749, [CSA]
- Wang Y, Rose B. An inhibition of gap-junctional communication by cadherins. J Cell Sci 1997; 110: 301–309, [PUBMED], [CSA]
- Watanabe T, Krankel D R. Molecular cloning and analysis of l(1)ogre a locus of Drosophila melanogaster with prominent effects on postembryonic development of the central nervous system. Genetics 1990; 126: 1033–1044, [PUBMED], [CSA]
- Wei C -J, Xu X, Lo W. Connexins and cell signalling in development and disease. Annu Rev Cell Dev Biol 2004; 4: 811–838, [CSA]
- Wei C -J, Francio R, Xu X, Lo C W. Connexin43 associated with an N-cadherin-containing multiprotein complex is required for gap junction formation in NIH3T3 cells. J Biol Chem 2005; 280: 19925–19936, [PUBMED], [CSA]
- Willecke K, Eiberger J, Degen J, Echardt D, Romualdi A, Guldenagel M, Deutsch U, Sohl G. Structural and functional diversity of connexin genes in the mouse and human genome. Biol Chem 2002; 383: 725–737, [PUBMED], [CSA]
- Wu J C, Tsai R Y, Chung T H. Role of catenins in the development of gap junctions in rat cardiomyocytes. J Cell Biochem 2003; 88: 823–835, [PUBMED], [CSA]
- Xu X, Li W E, Huang G Y, Meyer R, Chen T, Luo Y, Thomas M P, Radice G L, Lo C W. Modulation of mouse neural crest cell motility by N-cadherin and connexin 43 gap junctions. J Cell Biol 2001; 154: 217–229, [PUBMED], [CSA]
- Yanagawa S, Lee J S, Ishimoto A. J Biol Chem 1998; 27: 32353–32359, [CSA]
- Zhang Z, Curtin K D, Sun Y A, Wyman R J. Nested transcripts of gap junction gene have distinct expression patterns. J Neurobiol 1999; 40: 288, [PUBMED], [CSA]
- Zinke I, Schutz C S, Katzenberger J D, Bauer M, Pankratz M J. Nutrient control of gene expression in Drosophila: Microarray analysis of starvation and sugar-dependent response. EMBO J 2002; 21: 6162–6173, [PUBMED], [CSA]
- Zuppinger C, Schaub M C, Eppenberger H M. Dynamics of early contact formation in cultured adult rat cardiomyocytes studied by N-cadherin fused to green fluorescent protein. J Mol Cell Cardiol 2000; 32: 539–555, [PUBMED], [CSA]