Abstract
Connexin43 (Cx43), a component of gap junctions, has a relatively large carboxy-terminal region with multiple proteomic interactions. Proteomic interactions with its cytoplasmic loop, however, are poorly defined. The goal of this study is to examine proteomic interactions involving the cytoplasmic loop (CL) of Cx43. The authors utilized various techniques, including glutathione-S-transferase (GST) pull-down, immunoblot analysis, two-dimensional (2D) gel electrophoresis, and mass spectrometry, to elucidate binding partners for Cx43-CL. The authors identified novel interactions with Cx43-CL involving α- and β-tubulin, myelin basic protein, and Purα. Because tubulin interacts with the C-terminus of Cx43 (Cx43-CT), the authors further investigated the nature of the interaction between β-tubulin and Cx43-CL. β-Tubulin binds with the full length of Cx43-CL with approximately one-fifth the affinity of the interaction between Cx43-CT and β-tubulin. This study demonstrates novel proteomic interactions involving Cx43-CL that may lead to a more complete understanding of trafficking and gating of gap junction channels.
INTRODUCTION
Gap junctions are relatively nonselective intercellular channels that allow for the passage of ions and small molecules between coupled cells (Saez et al. Citation2003). Gap junction channels are formed by joining two hemichannels from adjacent cells, each of which is comprised of a hexamer of connexin molecules. Connexin proteins are expressed in most mammalian tissues and play critical developmental and functional roles. For example, in the heart, gap junctions provide a low-resistance intercellular pathway that facilitates cardiac conduction (Gutstein et al. Citation2001). The most common connexin isoform in cardiac tissue is connexin43 (Cx43), which has three intracellular domains: the amino-terminal, the cytoplasmic loop (CL), and the carboxy-terminal (CT) (Saez et al. Citation2003).
Numerous proteins have been identified that interact with the carboxy terminus of Cx43 (Cx43-CT), including β-tubulin (Giepmans et al. Citation2001b), c-Src (Giepmans et al. Citation2001a), v-Src (Kanemitsu et al. Citation1997), ZO-1 (Chen et al. Citation2008; Giepmans and Moolenaar Citation1998; Sorgen et al. Citation2004; Toyofuku et al. Citation1998), and Drebrin (Butkevich et al. Citation2004). β-Tubulin has been shown to interact with the juxtamembrane domain of Cx43-CT (Giepmans et al. Citation2001b) and is thought to play a role in targeting Cx43 to points of cell-cell contact (Shaw et al. Citation2007). Localization of gap junctions in the membrane may also be influenced by interactions with ZO-1, which has been shown to bind the Cx43-CT by several groups independently (Chen et al. Citation2008; Giepmans and Moolenaar Citation1998; Sorgen et al. Citation2004; Toyofuku et al. Citation1998). The tyrosine kinase c-Src and Cx43-CT can interact via the SH3 domain of c-Src, likely resulting in phosphorylation of Y265 and, as a result, may regulate gap junctional intercellular communication (GJIC) (Giepmans et al. Citation2001a; Kanemitsu et al. Citation1997). An interaction between Cx43-CT and Drebrin may also modulate GJIC (Butkevich et al. Citation2004). Lastly, indirect binding between full-length Cx43 and β-catenin has been shown to occur after K258 (Maass et al. Citation2007), an association that may influence downstream signaling and negatively regulate the transcription of Cx43, as well as other β-catenin/Tcf–dependent transcriptional targets (Ai et al. Citation2000).
In contrast to extensive work on the interactions of cellular proteins with Cx43-CT (Singh and Lampe Citation2003), there are only limited data available on proteomic interactions involving the cytoplasmic loop of Cx43 (Cx43-CL). Apart from a report suggesting that Cx43-CL physically interacts with Cx43-CT (Delmar et al. Citation2004) and the identification of a calmodulin-binding motif in the Cx43-CL (Zhou et al. Citation2007), there is little else in the literature describing protein-protein interactions involving Cx43-CL. Due to the relative paucity of data in this regard for Cx43-CL, we chose to focus our investigation on identifying possible novel binding partners for Cx43-CL. Using a nonbiased approach involving glutathione-S-transferase (GST) pull-downs, two dimensional (2D) gel electrophoresis, and mass spectrometry, we identified several novel proteomic interacting partners with Cx43-CL, including β-tubulin, myelin basic protein, and pur-α. Interestingly, β-tubulin, which has a well-defined interaction with Cx43-CT, binds with the full length of Cx43-CL at approximately one-fifth the affinity of the interaction between Cx43-CT and β-tubulin. Identification of novel interacting partners with Cx43-CL, such as those described in this study, may lead to a more complete understanding of the regulation and biology of the Cx43 gap junction channel.
METHODS
Cloning and PCR
We constructed GST-fusion constructs of truncated portions of Cx43-CL by polymerase chain reaction (PCR) and cloning. The truncation (TR) mutants included two proximal sequences, CL1 and CL2; two distal sequences, CL3 and CL4; and one section from the center of the loop, referred to as CL5 (). We also constructed a GST-fusion construct that joined CL1 and CL4 (CL1-4) in order to explore the extent to which CL-tubulin binding involves both the proximal and distal sections of the loop. All nucleotide sequences corresponding to regions of the Cx43 open reading frame were cloned into the pGEX-KG vector using restriction enzymes BamH1, Xba1, and/or Xho1. Inserts were ligated using the T4 DNA Ligase kit (Invitrogen, Carlsbad, CA). All constructs and plasmid insertions were verified by DNA sequencing and restriction digestion.
Figure 1. Relative positions and lengths of the various truncation mutants of connexin43 used in this study. Numbers refer to amino acid position in the connexin43 protein sequence. CLFL, full-length sequence of the connexin43 cytoplasmic loop; CTFL, full-length sequence of the connexin43 carboxy-terminus; CTTr, truncated connexin43 carboxy-terminus sequence.
Pull Down Assays with Glutathione-S-Transferase Fusion Products
GST pull-down experiments were performed as described elsewhere (Butkevich et al. Citation2004). All GST-Cx43 mutants were expressed in Escherichia coli bacterial cells (BL21) and induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). Cells were lysed in phosphate-buffered saline (PBS) with Complete Protease Inhibitor (Roche Pharmaceuticals) on ice for 1 min. The buffer included 1% (v/v) Triton X-100 (Sigma Aldrich) to ensure solubilization of membrane proteins. The samples were centrifuged and added to 1 ml of a 50% Glutathione Sepharose 4B bead slurry. Sepharose beads were then washed with cold PBS and centrifuged at 3000 rpm at 4°C.
Mouse brains were Dounce homogenized in cytomix buffer (120 mM KCl, 0.15 mM CaCl2, 10 mM KH2PO4, 25 mM HEPES, 2 mM EGTA, 5mM MgCl2, 2 mM ATP, and 5 mM glutathione; solution at pH 7.4). The homogenate was centrifuged at 50,000 rpm for 45 min at 4°C. The supernatant was set aside and the pellet was re-suspended in a 1% Triton X-100 solution, and centrifuged again at 50,000 rpm for 45 min at 4°C. The subsequent supernatant was taken as the fraction containing membrane-bound proteins.
Beads with bound GST were separated into two groups: one group of mouse brain lysates was exposed first to GST alone, and the other was used as a negative control. After exposure to GST alone, brain lysate samples were loaded onto beads with GST-bound fusion product overnight. Each sample was centrifuged at 9000 rpm for 3min, and the beads were washed with cytomix buffer. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (60 µl) containing 0.1% (v/v) β-mercaptoethanol (BME) was added to the beads. The beads and SDS-PAGE buffer were boiled for 2 min, centrifuged at 14,000 rpm for 3 min, and the liquid phase was collected to evaluate by gel electrophoresis.
Two-Dimensional SDS-PAGE/IEF Analysis
Two-dimensional electrophoresis was performed according to the carrier ampholine method of isoelectric focusing (IEF) (O'Farrell Citation1975) by Kendrick Labs (Madison, WI) as follows: Isoelectric focusing was carried out in a glass tube of inner diameter 2.0 mm using 2% pH 3.5 to 10 ampholines (GE Healthcare, Piscataway, NJ) for 9600 V-h. An IEF internal standard, tropomyosin, was added to the sample. This protein migrates as a doublet with lower polypeptide spot of MW 33,000 and pI 5.2. The enclosed tube gel pH gradient plot for this set of ampholines was determined with a surface pH electrode.
After equilibration for 10 min in Buffer ‘O’ (10% glycerol, 50 mM dithiothreitol, 2.3% SDS, and 0.0625 M Tris, pH 6.8), each tube gel was sealed to the top of a stacking gel that overlaid a 10% acrylamide slab gel (0.75 mm thick). SDS slab gel electrophoresis was carried out for about 4 h at 15 mA/gel. The following proteins (Sigma Chemical, St. Louis, MO) were used as molecular weights standards: myosin (220,000), phosphorylase A (94,000), catalase (60,000), actin (43,000), carbonic anhydrase (29,000), and lysozyme (14,000). These standards appear as bands at the basic edge of the Coomassie-stained 10% acrylamide slab gel. Unique spots on the gel involving the protein of interest were removed and evaluated using mass spectrometry.
Mass Spectrometry
Gel spots were transferred to clean tubes, water was added to completely hydrate gels, and the plastic coating was removed with clean tweezers. Gel spots were prepared for digestion by washing twice with 100 µl 0.05M Tris, pH 8.5/30% acetonitrile for 20 min with shaking, then with 100% acetonitrile for 1 to 2 min. After removing the washes, the gel pieces were dried for 30 min in a Speed-Vac concentrator. Gels were digested by adding 0.06 µg modified trypsin (sequencing grade, Roche Molecular Biochemicals) in 13 to 15 µl 0.025 M Tris, pH 8.5. The tubes were placed in a heating block at 32°C and left overnight. Peptides were extracted with 2×50 µl 50% acetonitrile/2% trifluoroacetic acid (TFA); the combined extracts were dried and resuspended in matrix solution.
Matrix solution was prepared by making a 10 mg/ml solution of 4-hydroxy-α-cyanocinnamic acid in 50% acetonitrile/0.1% TFA and adding two internal standards, angiotensin and adrenocorticotropin hormone (ACTH) 7–38 peptide, to the matrix solution. The dried digest was dissolved in 3 µl matrix/standard solution and 0.5 µl was spotted onto the sample plate. When the spot was completely dried, it was washed twice with water. Matrix-assisted laser desorption/ionization (MALDI) mass spectrometric analysis was performed at the Columbia University Protein Core Facility (New York, NY) on a Voyager DE Pro in the positive linear mode, accelerating voltage 21000 V, grid voltage 95%, extraction delay 200 ns, laser intensity 1800 to 1900. The raw spectrum was smoothed using a Gaussian filter (19 points) and calibrated using two-point calibration with internal standards angiotensin and ACTH 7-38 peptide.
Peak lists from MALDI spectra were searched using the Mascot algorithm (www.matrixscience.com) with the following parameters: Database: NCBInr; Taxonomy: Mus musculus; Enzyme: trypsin 1 missed cleavage; Variable modifications: Propionamide-Cys, Met oxidation; Peptide tolerance for average MH+: 0.5 Da. Total score: 178; Expect: 2.2e-13; Sequence coverage: 72%.
Western Blotting and Coomassie Staining
Protein samples were run on 12% SDS-PAGE polyacrylamide gels and subsequently transferred to nitrocellulose paper. Monoclonal mouse antibodies (Sigma Aldrich) were used for detection of β-tubulin followed by appropriate secondary antibodies (Santa Cruz Biologicals).
Coomassie staining was performed by treating gels with 50 ml of staining solution (65% water, 25% methanol, 10% acetic acid, 0.05% Coomassie Brilliant Blue) for 1 h. Gels were then exposed to destaining solution (water with 50% methanol and 3% acetic acid) overnight.
Coimmunoprecipitation
HeLa cells were transiently transfected with PCS2 vector containing CT or CL with a cMyc tag (CL- or CT-cMyc, respectively) using Lipofectamine 2000 (Invitrogen). Three different samples were grown in culture (CL-cMyc, CT-cMyc, and vector alone), harvested, lysed, and immunoprecipitated with mouse monoclonal anti-β-tubulin antibodies (Sigma Aldrich, St. Louis, MO), followed by gel electrophoresis. Gels were blotted and probed with a polyclonal anti-cMyc antibody (Santa Cruz Biotechnology, Santa Cruz, CA).
RESULTS
Pull-Down Assay Suggests Novel Binding Partners for the Cytoplasmic Loop of Cx43
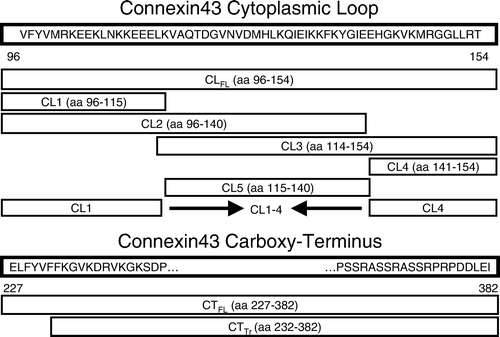
In order to detect novel proteins that interact with the cytoplasmic loop of Cx43, we generated a GST-CL full-length (CLFL) fusion construct and performed pull-down assays with and without mouse brain lysate (to serve as a negative control). After gel electrophoresis and Coomassie staining, multiple bands were observed in the lane containing protein eluted off GST- CLFL beads exposed to brain lysate, which were not seen in control lanes (). This suggests the presence of novel protein-protein interactions involving the cytoplasmic loop of Cx43.
Figure 2. Coomassie staining of protein samples obtained from glutathione-S-transferase (GST) pull-down assays. Products used in the assay included the full-length cytoplasmic loop (CLFL) sequence of Cx43 fused to GST and GST alone. These products were allowed to bind glutathione sepharose beads and were then eluted and collected (right-most two lanes) or exposed to mouse brain lysate prior to elution (left-most two lanes). Excess protein bands in the left-most lane containing eluate from the GST-CLFL beads exposed to brain lysate suggest novel proteomic interactions involving the Cx43-CLFL.
Novel Binding Partners for the Cytoplasmic Loop of Cx43 Are Identified by Two-Dimensional Gel Electrophoresis and Mass Spectrometry
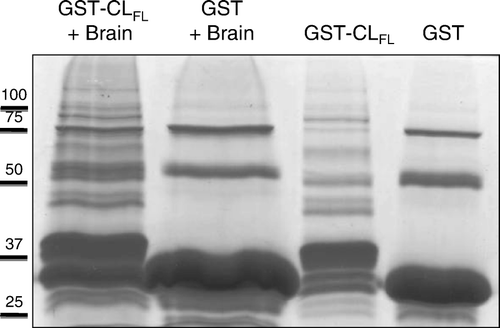
To identify potentially novel interacting proteins with the cytoplasmic loop of Cx43, we used two-dimensional gel electrophoresis of samples from GST pull-down assays, followed by mass spectrometry of isolated proteins. Five unique proteins were observed in 2D gels with samples eluted off GST-CLFL beads exposed to brain lysate, compared to control samples (). Using mass spectrometry, these five unique proteins were identified as myelin basic protein (MBP, isoforms 5 and 8), purine-rich single-stranded DNA-binding protein alpha (Purα), and α- and β-tubulin (, ). Thus, using pull-down assays with GST-CLFL fusion products, followed by 2D gel electrophoresis and mass spectrometry, we identified several novel proteomic interactions involving the cytoplasmic loop of Cx43.
Figure 3. Two-dimensional gel electrophoresis of GST pull-down assay involving Cx43-CLFL and matched controls. GST pull down assay with Cx43-CLFL GST fusion product followed by 2D gel electrophoresis (a) reveals five spots not seen in control experiments (b and c).
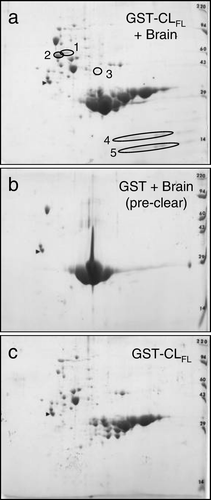
Figure 4. Mass spectrometry (MS) data acquired from analyzing spot 2 in Figure 3a. Identified peptide sequences (examples as shown) are consistent with β-tubulin.
Table 1. Proteins identified by 2-D gel electrophoresis and mass spectrometry
Highest Affinity Interaction of Cx43-CL with β-Tubulin Requires Full-Length CL
Because tubulin has a well-defined interaction with Cx43-CT (Giepmans et al. Citation2001b), we further investigated the nature of the interaction between β-tubulin and Cx43-CL. We initially theorized that binding of β-tubulin to Cx43-CL required one of two specific areas (CL1, CL4) on the loop sequence. A region in CL1 (Cx43 amino acids [aa] 102 to 110: KEEKLNKKE), the most proximal portion of the intracytoplasmic loop, shares homology with a tubulin binding region of microtubule-associated protein 1B (Map1B aa 723 to 731: KEEKEPKKE) (Noble et al. Citation1989). The second sequence of interest, CL4, is located at the distal end of the loop and has a high concentration of basic residues, which are thought to associate with microtubules (lysine, valine, proline, and glycine) (Giepmans et al. Citation2001b).
As expected, a truncated Cx43-CL GST fusion product (CL5) lacking sequences corresponding to both CL1 and CL4, showed no detectable affinity for β-tubulin. Surprisingly, however, affinity of the combined CL1 and CL4 GST fusion product (CL1 to CL4) with β-tubulin was substantially reduced compared to the Cx43-CLFL GST fusion product (a). We next investigated whether truncations of the Cx43-CL that were only missing the sequence corresponding to either CL1 or CL4 (CL3 and CL2, respectively) would bind tubulin with equal affinity to Cx43-CLFL. Interestingly, GST fusion products with either CL2 or CL3 each bound tubulin equally, but with substantially reduced affinity compared to Cx43-CLFL (b).
Figure 5. Comparisons of relative affinity for β-tubulin between Cx43-CLFL, portions of the CL of Cx43 and the carboxy terminus of Cx43 (Cx43-CTFL). (a) β-Tubulin abundance detected by Western blotting is shown in eluates from GST pull-down assays using GST fusions with Cx43-CLFL and truncated CL products, CL1-4 and CL5. Minimal β-tubulin is detected in the assay involving CL1-4 and no appreciable β-tubulin is seen in the eluate from the CL5 assay. (b) β-Tubulin abundance is substantially greater in eluates from assays involving the Cx43-CLFL GST fusion product than those involving either CL2 or CL3. (c) Eluates from GST pull down assays involving Cx43-CTFL contain more β-tubulin than those involving Cx43-CLFL. Shown also is an example of an eluate from an assay involving the GST construct without an insert. This control was performed for all of the experiments and demonstrated no tubulin signal. (d) Cx43-CTFL (lane 1) binds β-tubulin with fivefold greater affinity than a construct containing the sequence from the cytoplasmic tail of Cx43 in which the first five amino acids have been removed (CTTr) (lane 3). Lane 3 has 5X more protein loaded than lane 1.
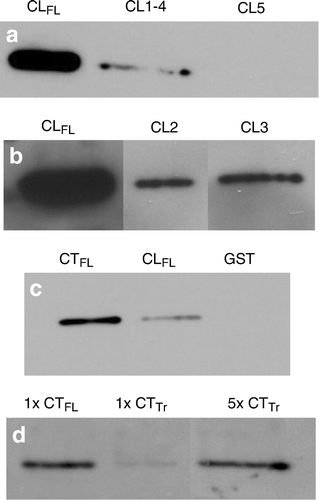
We next compared the tubulin binding affinity of Cx43-CLFL with that of Cx43-CTFL. Cx43-CTFL had substantially greater binding affinity for tubulin when compared to Cx43-CLFL (c).
To determine whether the Cx43-CT, in contrast to the Cx43-CL, required a specific motif for binding of β-tubulin, we focused on the juxtamembrane (JM) region of the CT. Other investigators have previously reported a putative tubulin binding domain within the 35–amino acid JM region of the Cx43-CT (Giepmans et al. Citation2001b). We compared a truncated CT construct (deletion of residues 227 to 231; CTTr) to CTFL and found that CTFL had a fivefold increase in binding affinity for β-tubulin. This suggests that residues 227 to 231 (ELFYV) located in the JM region of the Cx43 molecule are critical for tubulin binding (d).
Cx43-CL and Cx43-CT Immunoprecipitate with β-Tubulin
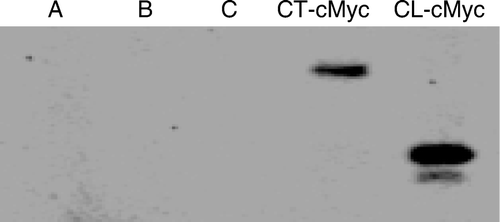
We performed an immunoprecipitation assay using a cMyc tag to verify the presence of an interaction between β-tubulin and both Cx43-CL and Cx43-CT. As shown in , both Cx43-CL-cMyc and Cx43-CT-cMyc immunoprecipitated with β-tubulin. Note that because this experiment was not normalized for molar concentrations of tagged constructs, the results should be considered qualitative rather than quantitative. However, these results suggest that β-tubulin physically interacts with both the Cx43-CL and Cx43-CT.
Figure 6. Immunoprecipitation of Cx43-CT-cMyc and Cx43-CL-cMyc fusion products with anti-β-tubulin antibodies. Polyacrylamide gel electrophoresis was performed to separate proteins, followed by probing with anti-cMyc antibodies. Controls are shown in lanes A (PCS2 vector–transfected HeLa cell lysate, immunoprecipitated with anti-β-tubulin), B (PCS2 vector containing CT-cMyc-transfected HeLa cell lysate, not exposed to anti-β-tubulin), and C (PCS2 vector–containing CL-cMyc-transfected HeLa cell lysate, not exposed to anti-β-tubulin).
DISCUSSION
In this study, we identified several novel binding partners of the cytoplasmic loop of Cx43, including α- and β-tubulin, Purα, and myelin basic protein. To our knowledge, this is the first study of an unbiased proteomic screen for binding partners of the cytoplasmic loop of Cx43.
We focused on a novel interaction between the Cx43-CL and β-tubulin, which has previously been shown to interact with the Cx43-CT. β-Tubulin was found to associate with the CL segment of Cx43, as illustrated by GST pull-down assays and via immunoprecipitation. Our pull-down assays using various portions of the loop demonstrated that the affinity between Cx43-CL and β-tubulin is not based upon a specifically identified motif, but rather is greatest in the presence of the entire cytoplasmic loop sequence. The interaction between Cx43-CL and β-tubulin may play an accessory yet significant role in the trafficking, localization and gating of Cx43 gap junction channels.
In contrast to the interaction between Cx43-CL and β-tubulin, the interaction between the CT portion of Cx43 and β-tubulin has been well characterized. The binding site of β-tubulin with the Cx43-CT has previously been identified in the juxtamembrane region, between residues 228 and 263 (Giepmans et al. Citation2001b), an area containing multiple basic residues. Another protein that contains regions that bind specifically to β-tubulin is Map1B. In the case of Map1B, β-tubulin binds specifically at two regions of the enzyme, which are known to have multiple copies of the sequences KKEE and KKE (Noble et al. Citation1989). Interestingly, Vaillant et al. demonstrated tubulin binding to regions of Map1a that contain basic KKE repeats, as well as flanking regions without these domains (Vaillant et al. Citation1998). Thus, basic KKE repeats may not be absolutely required for tubulin binding. Despite a proximal region of the Cx43-CL containing KKEE and KKE motifs, as well as a concentration of basic putative microtubule-binding residues at the distal end of the Cx43-CL, our studies demonstrate that the binding between β-tubulin and the Cx43-CL requires the entire length of the CL.
The α-tubulin monomer was among the other novel binding partners we discovered for Cx43-CL. This interaction is not wholly surprising, due to the similarity and strong dimeric association between α-tubulin and β-tubulin monomers. Interestingly, the carboxy-terminal region of heag2, a potassium channel protein, associates with α-tubulin and is thought to play a role in the trafficking of this channel to the membrane (Bracey et al. Citation2008).
Purα, another binding partner of Cx43-CL, is a transcription factor that has been implicated in cell growth and malignancies such as retinoblastoma and prostate cancer. It is highly conserved between species and is known to bind single- and double-stranded DNA in a sequence-specific manner, preferring single purine-rich sequences such as GGN. For instance, Purα is known to regulate the transcription of myelin basic protein (Gallia et al. Citation2000).
Myelin basic protein (MBP), another of the proteins we observed to associate with Cx43-CL, plays a major role in the structural composition of central nervous system (CNS) myelin. MBP commonly associates with negatively charged cytosolic membrane lipids and has the ability to bind both membrane and actin filaments, linking the two together. The association between MBP and CL may possibly serve to help localize the Cx43 channel within the membrane (Boggs Citation2006).
In previous studies, a physical interaction was demonstrated between the CT and CL of Cx43. Delmar and colleagues determined that this interaction occurred between residues 119 and 144 of the Cx43-CL and was pH dependent (Delmar et al. Citation2004). Of note, Cx43 CL-CT binding is increased at a pH of 5.8 to 6.5 and decreased at pH 7.4 to 8.0. It is likely that we did not observe a similar interaction between CL and CT because all of our solutions were prepared at pH 7.4.
An important limitation of our study is that experiments investigating the interaction between Cx43 and β-tubulin were performed with overexpressed, isolated domains of the Cx43 molecule and, as such, must be interpreted with caution. However, a known interaction between the Cx43-CT and β-tubulin (Giepmans et al. Citation2001b), at a higher affinity than the interaction between the Cx43-CL and β-tubulin, would complicate interpretation of studies utilizing full-length Cx43 that would otherwise provide reassurance of a true interaction between the Cx43-CL and β-tubulin. Future studies investigating an interaction between full-length Cx43 and the other potential binding partners of the Cx43-CL in this paper will help to strengthen the hypothesis that the Cx43-CL may mediate interactions between Cx43 and other cellular proteins.
In summary, we have demonstrated that the cytoplasmic loop of Cx43 interacts with β-tubulin with relatively greatest affinity in the presence of the entire length of the CL. In addition, we have identified novel interactions between Cx43-CL and MBP, as well as the transcription factor Purα. β-Tubulin also interacts with Cx43-CT and our data suggest that the first five residues of the juxtamembrane region of the CT are critical for that interaction. These findings suggest that novel complex proteomic interactions with the Cx43 molecule may play important roles in the biology and trafficking of gap junction channels.
Acknowledgements
This work was supported by NIH grant HL081336 and a Grant-in-Aid from the American Heart Association (DEG).
References
- Ai Z, Fischer A, Spray DC, Brown AM, Fishman GI. Wnt-1 regulation of connexin43 in cardiac myocytes. J Clin Invest. 2000; 105: 161–171
- Boggs J. M. Myelin basic protein: A multifunctional protein. Cell Mol Life Sci 2006; 63: 1945–1961
- Bracey K., Ju M., Tian C., Stevens L., Wray D. Tubulin as a binding partner of the heag2 voltage-gated potassium channel. J Membr Biol 2008; 222: 115–125
- Butkevich E., Hulsmann S., Wenzel D., Shirao T., Duden R., Majoul I. Drebrin is a novel connexin-43 binding partner that links gap junctions to the submembrane cytoskeleton. Curr Biol 2004; 14: 650–658
- Chen J., Pan L., Wei Z., Zhao Y., Zhang M. Domain-swapped dimerization of ZO-1 PDZ2 generates specific and regulatory connexin43-binding sites. EMBO J 2008; 27: 2113–2123
- Delmar M., Coombs W., Sorgen P., Duffy H. S., Taffet S. M. Structural bases for the chemical regulation of Connexin43 channels. Cardiovasc Res 2004; 62: 268–275
- Gallia G. L., Johnson E. M., Khalili K. Puralpha: a multifunctional single-stranded DNA- and RNA-binding protein. Nucleic Acids Res 2000; 28: 3197–3205
- Giepmans B. N., Hengeveld T., Postma F. R., Moolenaar W. H. Interaction of c-Src with gap junction protein connexin-43. Role in the regulation of cell-cell communication. J Biol Chem 2001a; 276: 8544–8549
- Giepmans B. N., Moolenaar W. H. The gap junction protein connexin43 interacts with the second PDZ domain of the zona occludens-1 protein. Curr Biol 1998; 8: 931–934
- Giepmans B. N., Verlaan I., Hengeveld T., Janssen H., Calafat J., Falk M. M., Moolenaar W. H. Gap junction protein connexin-43 interacts directly with microtubules. Curr Biol 2001b; 11: 1364–1368
- Gutstein D. E., Morley G. E., Tamaddon H., Vaidya D., Schneider M. D., Chen J., Chien K. R., Stuhlmann H., Fishman G. I. Conduction slowing and sudden arrhythmic death in mice with cardiac-restricted inactivation of connexin43. Circ Res 2001; 88: 333–339
- Kanemitsu M. Y., Loo L. W., Simon S., Lau A. F., Eckhart W. Tyrosine phosphorylation of connexin 43 by v-Src is mediated by SH2 and SH3 domain interactions. J Biol Chem 1997; 272: 22824–22831
- Maass K., Shibayama J., Chase S. E., Willecke K., Delmar M. C-terminal truncation of connexin43 changes number, size, and localization of cardiac gap junction plaques. Circ Res 2007; 101: 1283–1291
- Noble M., Lewis S. A., Cowan N. J. The microtubule binding domain of microtubule-associated protein MAP1B contains a repeated sequence motif unrelated to that of MAP2 and tau. J Cell Biol 1989; 109: 3367–3376
- O'Farrell P. H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem 1975; 250: 4007–4021
- Saez J. C., Berthoud V. M., Branes M. C., Martinez A. D., Beyer E. C. Plasma membrane channels formed by connexins: Their regulation and functions. Physiol Rev 2003; 83: 1359–1400
- Shaw R. M., Fay A. J., Puthenveedu M. A., von Zastrow M., Jan Y. N., Jan L. Y. Microtubule plus-end-tracking proteins target gap junctions directly from the cell interior to adherens junctions. Cell 2007; 128: 547–560
- Singh D., Lampe P. D. Identification of connexin-43 interacting proteins. Cell Commun Adhes 2003; 10: 215–220
- Sorgen P. L., Duffy H. S., Sahoo P., Coombs W., Delmar M., Spray D. C. Structural changes in the carboxyl terminus of the gap junction protein connexin43 indicates signaling between binding domains for c-Src and zonula occludens-1. J Biol Chem 2004; 279: 54695–54701
- Toyofuku T., Yabuki M., Otsu K., Kuzuya T., Hori M., Tada M. Direct association of the gap junction protein connexin-43 with ZO-1 in cardiac myocytes. J Biol Chem 1998; 273: 12725–12731
- Vaillant A. R., Muller R., Langkopf A., Brown D. L. Characterization of the microtubule-binding domain of microtubule-associated protein 1A and its effects on microtubule dynamics. J Biol Chem 1998; 273: 13973–13981
- Zhou Y., Yang W., Lurtz M. M., Ye Y., Huang Y., Lee H. W., Chen Y., Louis C. F., Yang J. J. Identification of the calmodulin binding domain of connexin 43. J Biol Chem 2007; 282: 35005–35017