ABSTRACT
Magra sheep reared around the Bikaner area of Rajasthan, India is famous for producing lustrous fleece as compared to other wool-producing native sheep breeds. Three clips of Magra wool, namely, February, July, and October months of the year are showing variations in luster and gross appearance. Present work has been conducted to unravel seasonal variations, in transcript expression patterns in major type I and type II keratin genes, that occur in wool follicles of lustrous wool-producing Magra sheep. Wool samples randomly collected were graded in high and low luster by Gloss 60° values. Wool follicles of the sheep with low (gloss 60° values; < 2.5) and high (gloss 60° values; > 2.5) luster were used for the analysis of type I and type II keratin transcripts. The results of Quantitative PCR had shown up-regulated expression of keratin genes in high-lustrous sheep as compared to low-lustrous sheep. K32, K40, and K82 transcript expression differed significantly (P ≤ .05) between high- and low-lustrous sheep. Nucleotide sequence analysis of the K40 and K82 genes revealed five and four non-synonymous mutations, respectively. From the present study, it can be speculated that the higher amount of keratin protein may play a crucial role in determining the luster property of the wool.
摘要
印度拉贾斯坦邦比坎纳地区饲养的马格拉羊,与其他产羊毛的本地绵羊品种相比,以产有光泽的羊毛而闻名. Magra羊毛的三个片段,即每年的二月、七月和十月,呈现出不同的光泽和外观. 目前的工作是为了揭示主要的I型和II型角蛋白基因转录表达模式的季节性变化,这些基因发生在有光泽的产毛马格拉羊的毛囊中. 随机收集的羊毛样品通过光泽度600值进行高光泽和低光泽分级。使用具有低光泽(光泽600值;<2.5)和高光泽(光泽度600值;>2.5)的绵羊毛囊分析I型和II型角蛋白转录物. 定量PCR结果显示,与低光泽绵羊相比,高光泽绵羊角蛋白基因表达上调. 高光泽羊和低光泽羊的K32、K40和K82转录表达差异显著(P ≤ .05). K40和K82基因的核苷酸序列分析分别显示了5个和4个非同义突变. 根据目前的研究,可以推测,较高量的角蛋白可能在决定羊毛的光泽特性方面起着关键作用.
Introduction
Wool is a commercial commodity derived from sheep and primarily made up of keratin and keratin- associated proteins. These structural phosphoproteins constitute intermediate filament (IF) as a major structural component. Keratin proteins are categorized as type I (acidic) and type II (neutral to basic) proteins (Yu et al. Citation2018). A total of 11 type I- and 7 type II-keratin proteins are reported in sheep (Plowman Citation2018). Type I keratins are further differentiated into three groups and those expressed within the cortex and cuticle of wool fibers are called trichocyte keratins. These trichocyte keratin genes encode proteins, which are K31, K32, K33A, K33B, K34, K35, K36, K37, K38, K39, and K40; however, K37 is not expressed in sheep (Yu et al. Citation2011). Type II keratin genes includes K81, K82, K83, K84, K85, K86, and K87. These type II keratin genes are tightly linked to each other in the sheep genome (Powell and Beltrame Citation1994). K83 is highly homologous with K81 and K86 and expression of K83, specifically detected in the wool cortex, is linked with decreasing fleece quality in Chinese Tan Sheep (Liu et al. Citation2017). K83 is mapped to ovine chromosome 3 (Hediger, Ansari, and Stranzinger Citation1991). QTL study has revealed loci on the ovine chromosome 3 are associated with wool brightness (McKenzie Citation2001). The K83 gene is tightly linked to the K87 gene. Extensive variations were observed in 5’ untranslated regions of the K87 keratin gene, previously known as the KRT 2.13 gene (McKenzie, Arora, and Hickford Citation2012). Type I and type II keratins are strictly interdependent for assembly into 10 nm IF and regulated in a pairwise fashion (Kim and Coulombe Citation2007). Non-helical N- and C- terminal segments of type I and type II wool keratins exhibit different patterns which indicate diverse functional role further added by many types of type I and II keratin proteins (Sparrow et al. Citation1992).
Magra sheep reared around the Bikaner area of Rajasthan, India is famous for producing lustrous fleece as compared to other wool-producing native sheep breeds. Sheep in this region is usually undergone shearing thrice, namely, first in February, second in July, and third in October months of the year. Three clips of Magra wool are showing variations in luster and gross appearances (Kumar et al. Citation2019). Wool luster is a surface phenomenon and a measure of the specular reflection of wool fibers. It is measured as a ratio of reflectance at 45° (specular reflection) to reflectance at a 10° (or at an angle of 10 degree) keratin angle (Hunter Citation1975). Methods were described to measure wool luster, it includes subjective as well as objective procedures (Kumar et al. Citation2019, Citation2021). Studies were conducted to unravel molecular changes occurring in wool follicles of lustrous wool-producing sheep. Transcript expression analyses in felting Merino sheep has indicated that KAP 6.1, KAP 7.0, and KAP 8.0 gene were downregulated in lustrous wool follicles, while KAP 2.12 and KAP 4.2 genes were upregulated (Li et al. Citation2009). In another study, a type 1 hair cortex K33A (K1.2) transcript was upregulated in lustrous fleece-producing wool follicles of Magra sheep (Kumar et al. Citation2020). In a wool fiber bundle, the spatial arrangement of different keratin intermediate filaments and their molecular interaction with keratin-associated proteins are speculated to determine the physical attributes of wool and hair fiber (Powell and Rogers Citation1997). These macrofibril orientations and packing get affected by compositional variation in and its associated proteins. Wool traits are known to be controlled by multiple genes and not by individual keratins and keratin-associated proteins. In the present study, transcript expression differences in type I and type II keratin genes in wool follicles of Magra sheep around three seasons shearing has been analyzed.
Materials and methods
Collection of wool samples, wool follicles, and measurement of wool luster
Literature survey suggesting seasonal variation in wool luster and based on that three clips of wool samples were collected. Wool samples were randomly collected from Magra sheep and graded in high- and low-lustrous by Gloss 60° values as described elsewhere (Kumar et al. Citation2021). Briefly, scoured wools first, cleaned off to residual impurities, and combed to a stock of parallel fiber bundle. A Parallel fiber fringe of 35–40 mg weight has been prepared from the combed stock and used for luster estimation. To neutralize the variability (caused due to variation in fiber diameter from wool root to tip and scale direction of fiber surface), evaluation of fiber fringe was carried out at four different angles of 0°, 45°, 90°, and 135°. The measured Gloss 60° values for four different angles were averaged to represent the Gloss 60° values of an individual fiber fringe. Clip-wise luster evaluation study of the Magra sheep at Arid Regional Campus (ARC), Bikaner of CSWRI has been conducted. The lustrous animals were selected from the flock of Magra sheep and wool samples for all three clips have been collected. Gloss 60° values of the collected wool samples were evaluated and categorized into high (>2.5; Gloss 60° values) and low luster (<2.5; Gloss 60° values). Wool samples for all the three season clips, namely, February, July, and October have been evaluated for luster. Further, about 100 wool follicles from sheep with low (designated as control) and high (designated as treatment) values of luster (Gloss 60° degree) were collected in RNAlater (Sigma-Aldrich) storage reagent and preserved for further analysis.
RNA extraction and cDNA synthesis
A total of 36 wool follicle samples were processed for RNA extraction. Magra sheep (12) samples (6 from high lustrous and 6 from low lustrous) from each season/clip, namely, February, July, and October were used. Samples were centrifuged at 4000 rpm for 5 minutes at room temperature (RT). The supernatant was discarded and 1 ml TRI Reagent solution (Sigma) was added to each sample and vortexed for 15–20 minutes for complete lysis of wool follicle cells. Samples were incubated at RT for 5 minutes. Chloroform (200 µl) was added, mixed, and incubated at RT for 15 minutes. Samples were centrifuged at 12,000×g for 15 minutes at 4°C. The aqueous phase was transferred to a fresh RNase free 1.5 ml centrifuge tube and 0.5 ml of 2-propanol was added for RNA precipitation. The sample was allowed to stand for 10 minutes at RT. Again, the sample was centrifuged at 12,000×g for 10 minutes at 4°C. The supernatant was discarded and the RNA pellet was washed with 1 ml of 75% ethanol. The RNA pellet was reconstituted in RNase-free distilled water (20 µl). RNA (250 ng) was used for the synthesis of complementary DNA (cDNA) copies using high-capacity cDNA reverse transcription kit (Applied Biosystems) following the manufacturer’s instruction. The quality of cDNA was checked by amplification of the GAPDH gene and cDNAs were stored at −80°C (Eppendorf) till further use.
Quantitative PCR analysis
Quantitative PCR (qPCR) reaction mix containing 200 ng of cDNA, 0.2 µM each forward and reverse primers, 1.5 mM Magnesium chloride, 1-unit Taq DNA polymerase, 0.2 mM dNTPs, 1× PCR buffers adjusted to a total volume of 20 µl with PCR-grade water. Wool follicles from lustrous Magra sheep have been analyzed for transcript differences in major keratin type I and type II genes. The primers used for the study are given in .
Table 1. List of primers used for real-time PCR analysis and amplification of coding regions of type I and type II keratin genes.
PCR reaction was carried out in a real-time PCR system (CFX 96, Bio-Rad) with initial activation at 95°C for 4 minutes, followed by 45 cycles of 95°C for 0.20 sec, 55°C for 0.30 sec, and 72°C for 0.35 sec. Melt curve analysis was carried out in a continuous run at 65°C for 0.05 sec followed by 95°C for 0.50 sec. Quantitative PCR for each gene was run in duplicates in 10 µl reaction volume in 96 well Hard-Shell PCR plates (Bio-Rad; Cat#HSP9601). Each experiment was repeated twice. GAPDH primers referenced from elsewhere (Li et al. Citation2009) were used as internal reference gene. The qPCR results were normalized for the level of input RNA by assay of the GAPDH transcript and transcript copy numbers in high-lustrous Magra sheep (designated as Treatment; T) were expressed relative to low-lustrous Magra sheep (designated as control; C). Student t-tests were carried out to examine the significance (P ≤ .05) of the difference between mean transcript (▲▲Ct) levels in low- and high- lustrous Magra sheep.
Amplification, cloning, and sequence analysis of K40 and K82 CDS from lustrous Magra sheep
Since ovine mRNA sequence information for keratin 40 and 82 genes was lacking at the beginning of the study, primers for amplification of both mRNAs were designed using bovine mRNA sequences available in GenBank (NM_001105418.1 and NM_001082616.1; respectively). A total of 12 wool follicle samples for each gene; 6 from low-lustrous (gloss 60° value<2.5) and 6 from high-lustrous (gloss 60° value>2.5) have been used for sequence analysis. The PCR reaction was prepared with 1× PCR buffer, 1.5 mM MgCl2, 200 μM dNTPs, 0.2 μM of each forward and reverse primer, five units of Taq DNA Polymerase (Sigma) with 50–100 ng of template cDNA. The PCR reaction was run in 50 µl volume. The thermal cycler conditions were an initial denaturation at 94°C for 5 minutes followed by 35 cycles of 94°C for 0.45 sec, 59°C (K40), and 56°C (K82) for 0.45 sec and 72°C for 1 minute with a final extension at 72°C for 7 minutes. The PCR products were resolved on agarose (1.5%) gel and excised from the gel on UV illuminator. PCR products were gel purified using the gel purification kit (Thermo Scientific) following the manufacturer’s instructions. Purified PCR products were ligated with a pDrive cloning vector provided with the PCR cloning kit (Qiagen) following the manufacturer instructions and were transformed in Escherichia coli (DH5α). The bacterial colonies were cultured overnight in LB broth at 37°C with 150 RPM in an incubator shaker and recombinant plasmids were isolated using a plasmid purification kit (Thermo Scientific). The true clones were screened using colony PCR. Four plasmids from each sample were sequenced.
Statistical analyses
One-way repeated measures ANOVA was used to compare gloss 60° luster values of the three seasons clips followed by Tukey HSD procedure for pairwise comparisons for measure of significance (P < .05). The qPCR data were analyzed for fold expression in wool follicles. Ct values obtained in quadruplets were used for analysis and the GAPDH gene was used for the normalization of the expression of transcripts. To find out significant (P ≤ .05) differences between mean values of normalized Ct values of transcript levels in two groups, namely, low- and high-lustrous Magra sheep, paired t-test was used. Multiple sequence analysis of the nucleotide sequences has been conducted using Clustal Omega web-based program.
Results and discussion
Magra sheep of India is well known for producing lustrous fleece among native wool-producing sheep. The genetic relationship with the lustrous phenotype in sheep is not yet fully unraveled. Previous studies had elaborated keratin and keratin-associated protein (KAPs) mRNA expression differences in wool follicles of lustrous and non-lustrous sheep (Kumar et al. Citation2020; Li et al. Citation2009); however, detailed analysis of gene expression studies in different seasons across the year is missing. In the present work, wool follicles were collected just before three seasons of shearing, their luster (gloss 60° values) evaluated, and transcript expression differences of major keratin genes in high (gloss 60° value; > 2.5) and low-lustrous (gloss 60° value; < 2.5) wool producing Magra sheep were determined. A total of 43 wool samples were evaluated for the different clips, out of which, 25 wool samples belonged to the same animal for all three clips. The clip-wise evaluated Gloss 60° values of 25 sheep are provided in .
Table 2. Clip wise Gloss 60° values of selected Magra animals (wool samples). This list includes only those animals available for three seasons sampling. The Gloss 60° value>2.5 was categorized as high-luster wool for February and July clip whereas October clip pooled average Gloss 60 value is≤2.5 and categorized as low-luster wool. One way ANOVA test for independent measures followed by Tukey HSD procedure to compare gloss 60° luster values of three seasons, namely, February (T1), July (T2), and October (T3) was done. One-way repeated measures ANOVA revealed that gloss 60° values between seasons (treatments) are significant (P < 0.00001) between sample means.
The objective luster value (Gloss 60°) of wool samples for three shearing seasons, namely, February, July, and October were found significantly (P < .05) different between seasons. The pooled (25 numbers) Gloss 60° values are observed in descending order of 2.75 > 2.54 > 2.40 for February, July, and October clips, respectively. Pairwise comparison revealed February and July clips as well as February and October clip was differed significantly (). The Gloss 60° value>2.5 was categorized as high-luster wool for February and July clips whereas; the October clip pooled average Gloss 60° value is≤2.5 and categorized as low-luster wool. The low gloss 60° values of the October clip are associated with its canary coloration which causes due to pigment in the suint (dried perspiration of sheep) enter in the wool fiber structure and staining it permanently under the conditions of high humidity and UV radiation after rainfall.
Gene expression from 36 RNA samples (12 from each season) has been analyzed for transcript differences in major keratin type I (K31, K32, K33A, K33B, K34, K35, K36, K38, K39, and K40) and keratin type II (K81, K82, K83, K84, K85, K86, and K87) genes using gene-specific primers. Primers producing single intact PCR products were used for real-time PCR analysis. Overall results of transcript expression had shown up-regulated expression of keratin genes in lustrous Magra sheep as compared to low-lustrous Magra sheep ().
Figure 1. Line charts showing normalized relative expression (X-axis) of different keratin genes (Y-axis) in high-lustrous (gloss 60° value>2.5) and low-lustrous (gloss 60° value<2.5) Magra sheep’s wool follicles collected in February, July and October months.
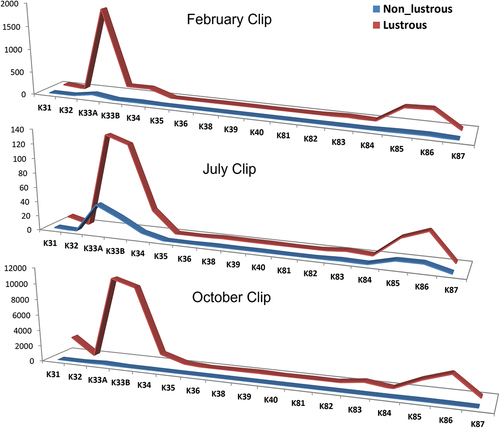
Expression of K32, K40, and K82 genes in July season’s wool follicles of lustrous Magra sheep was significantly (P ≤ .05) upregulated; however, only the K82 gene was significantly (P ≤ .05) upregulated in October wool follicles of lustrous Magra sheep as compared to low-lustrous wool follicles of Magra sheep ().
Table 3. Relative level of Keratin gene transcripts normalized (GAPDH) to level of input total RNA (Paired t-test); Cmeans = mean Ct values in low-lustrous (gloss 60° value<2.5) Magra sheep (control group); Tmeans = mean Ct values in high-lustrous (gloss 60° value>2.5) Magra sheep (treatment group).
The type I trichocyte keratin genes (K31 to K40) are expressed within the cortex and cuticle of wool fiber. K32 gene is grouped with K35 and K36 genes and exhibits more heterogeneity in head domains of IF. K37 is not expressed in sheep (Yu et al. Citation2018) which has been further confirmed in the present study. The type II keratins like K82 and K84 are abundantly expressed in the wool cuticles and in the present study K82 was significantly (P ≤ .05) upregulated in high-lustrous wool-producing sheep as compared with low-lustrous wool-producing sheep (). Previous work had shown that over-expression of some of the type II keratins to transgenic sheep enhances the overall gloss/luster of the wool produced from them (Bawden et al. Citation1998). Wool contains about 95% by weight of pure keratins (Cardamone et al. Citation2009). Both type I and type II keratin form the microfilament, also called IF in wool. It forms an obligate hetero-polymer where each type I keratin is combined with an associated type II molecule, which polymerize further to give 10 nm diameter IF. These type I (40 to 57 kDa; acidic; pI 4.5–6.0) and type II (50–70 kDa; neutral to basic; pI 6.5–8.5) keratin intermediate filament proteins are affecting structure and functions of wool and are regulated in a pair-wise fashion (Jacob et al. Citation2018). It is speculated that a wide range of keratin subtypes in the type I and type II families provides an opportunity for wide permutations and combinations of diverse functions of keratin proteins. Any alteration in the quantity of a particular keratin protein might affect the wool’s physical properties like luster.
From the present study it can be surmised that a higher amount of keratin proteins may play a crucial role in determining the luster property of the wool. The low expression of certain keratin-associated proteins (Kumar et al. Citation2020; Li et al. Citation2009) and high expression of keratin proteins in lustrous fleece are speculated to link with luster phenotypes. There are certain factors that trigger cellular pathways to upregulate the expression of the keratin family of genes. The variation in wool phenotype may be due to changes in ligand and receptor interactions. The results of the present work provide targets for the investigation of signaling pathway (s) involved and its expression differences in high- and low-lustrous wool phenotype. Transcripts of three genes, namely, K40, K82, and K84 were known to present only in wool fiber cuticles (Yu et al. Citation2011) and in the present study K40 and K82 transcripts expression patterns were found to differ significantly between high- and low-lustrous sheep () henceforth, K40 and K82 cDNAs were further sequence characterized. K40 (ENSOARG00020006496) gene is located in ovine chromosome 11 and has 7 exons. The cds (1296 bp) of keratin type I cytoskeletal 40 gene encode a 431 amino acid residues protein which had shown 98.84%, 96.52%, and 96.96% homology with goat-, buffalo- and taurine cattle-K40 protein, respectively. Deduced amino acid sequences were further aligned to find out missense mutations in amplified sequences. Five amino acid changes were identified (Supplementary file 1). Variant sequences of K40 mRNA were submitted to NCBI GenBank with accession numbers MZ173540-MZ173543 (4). K82 (ENSOARG00000017063) gene also called krt HB2 gene, is located in ovine chromosome 3. K82 transcript has 9 exons, which encode 1545 bp mRNA, which gives 514 amino acids (a. a.) long protein. In the present study, 1531 bp long mRNA was amplified, which is partial at the 3’ end and composed of 505 a. a. residues. Nucleotide sequence analysis revealed 9 single nucleotide polymorphisms in ORFs, out of which five are synonymous and four are non-synonymous (Supplementary file 1). Variant sequences of K82 mRNA were submitted to NCBI GenBank with accession numbers MN635610- MN635612 (3). In the present work, any association between observed polymorphisms and gloss 60° values could not be established, perhaps due to the low sample size. These hard alpha-keratins are low sulfur proteins. Keratins expressed specifically in the cuticle and cortical region of wool are of paramount importance in contributing to wool luster. Further studies in the protein profile of wool and wool follicles among low and high-luster wool-producing Magra sheep would be of great value to support the present finding.
Conclusion
Magra sheep of India is known for producing lustrous fleece among native wool-producing sheep. Type I and type II keratin form the microfilament also called IF in wool, regulated in a pair-wise fashion, and its spatio-temporal variation in expression along with keratin-associated proteins affects the structure and function of wool. The objective luster value (Gloss 60°) of wool sampled from three seasonal clips was found significantly (P < .05) different among the three seasons. The overall result of real-time PCR expression of mRNA has shown up-regulated expression of keratin genes in high-lustrous wool-producing sheep as compared to low-lustrous wool-producing sheep. Expression of K32, K40, and K82 genes in July season’s wool follicles of lustrous Magra sheep was significantly (P ≤ .05) upregulated; however, only the K82 gene was significantly (P ≤ .05) upregulated in October wool follicles of lustrous Magra sheep as compared to low-lustrous Magra sheep. It can be speculated that a higher amount of keratin proteins may play a crucial role in determining the luster property of the wool. Keratins expressed specifically in the cuticle and cortical region of wool is of paramount importance in contributing to wool luster. Sequence analysis of K40 and K82 genes revealed non-synonymous mutations affecting amino acids composition; however, no association was established with wool luster.
Highlights of the work
Magra sheep reared around Bikaner area of Rajasthan, India is famous for producing lustrous fleece as compared to other wool producing native sheep breeds.
Wool samples randomly collected were graded in high-lustrous and low-lustrous wool by Gloss 600 values.
Seasonal variations in transcript expression patterns in major type I and type II keratin genes occur in wool follicles of Magra sheep were analyzed.
Quantitative PCR results had shown up-regulated expression of keratin genes in high-lustrous Magra sheep as compared to low-lustrous Magra sheep. K40 and K82 transcript expression differed significantly between low-lustrous and high-lustrous sheep. Nucleotide sequence analysis of K40 and K82 gene revealed five and four non-synonymous mutations, respectively.
From present study, it can be speculated that higher amount of keratin proteins may play crucial role in enhancing the luster property of the wool.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bawden, C. S., B. C. Powell, S. K. Walker, and G. E. Rogers. 1998. Expression of wool intermediate filament keratin transgene in sheep fibre alters structure. Transgenic Research 7:273–10. doi:10.1023/A:1008830314386.
- Cardamone, J. M., A. Nuñez, R. A. Garcia, and M. Aldema-Ramos. 2009. Characterizing wool keratin. Advances in Materials Science and Engineering 2009:5. doi:10.1155/2009/147175.
- Hediger, R., H. A. Ansari, and G. F. Stranzinger. 1991. Chromosome banding and chromosome localization support extensive conservation of chromosome structure between cattle and sheep. Cytogenetics and Cell Genetics 57 (2–3):127–34. doi:10.1159/000133131.
- Hunter, R. S. 1975. The measurement of luster. London: Wiley Interscience.
- Jacob, J. T., P. A. Coulombe, R. Kwan, and M. B. Omary. 2018. Type I and type II keratin intermediate filaments. Cold Spring Harbor Perspectives in Biology 10 (4):a018275. doi:10.1101/cshperspect.a018275.
- Kim, S., and P. A. Coulombe. 2007. Intermediate filament scaffolds fulfil mechanical, organizational, and signaling functions in the cytoplasm. Genes & Development 21 (13):1581–97. doi:10.1101/gad.1552107.
- Kumar, R., A. S. Meena, A. Chopra, and A. Kumar. 2020. Keratin gene expression differences in wool follicles and sequence diversity of high-glycine tyrosine keratin-associated proteins (Kaps) in Magra sheep of India. Journal of Natural Fibers 17 (9):1257–63. doi:10.1080/15440478.2018.1558157.
- Kumar, A., R. K. Sawal, H. K. Narula, S. Kumar, and R. Kumar. 2019. Subjective and objective/machine evaluation of wool luster in Magra sheep vis-a-vis wool grading and animal selection. Journal of Natural Fibers 16:644–51. doi:10.1080/15440478.2018.1431996.
- Kumar, A., D. B. Shakyawar, R. Kumar, A. S. Meena, N. L. Meena, and A. Chopra. 2021. Objective evaluation of luster (Gloss 60°) of different Indian wool. Journal of Natural Fibers 19 (15):10490–98. doi:10.1080/15440478.2021.1994091.
- Li, S. W., H. S. Ouyang, G. E. Rogers, and C. S. Bawden. 2009. Characterization of the structural and molecular defects in fibres and follicles of the merino felting luster mutant. Experimental dermatology 18 (2):134–42. doi:10.1111/j.1600-0625.2008.00774.x.
- Liu, Y., X. Kang, W. Yang, M. Xie, J. Zhang, and M. Fang. 2017. Differential expression of KRT83 regulated by the transcription factor CAP1 in Chinese Tan Sheep. Gene 614:15–20. doi:10.1016/j.gene.2017.03.007.
- McKenzie, G. W. 2001. A search for quantitative trait loci affecting wool colour. Proceeding of the New Zealand Society of Animal Production 61:104–08.
- McKenzie, G. W., R. Arora, and J. G. H. Hickford. 2012. Genetic variation in the 5′UTR of the KRT2.13 gene of sheep. Animal Science Journal 83 (3):194–98. doi:10.1111/j.1740-0929.2011.00933.x.
- Plowman, J. E. 2018. Diversity of trichocyte keratins and keratin associated proteins. In The hair fibre: Proteins, structure and development. Advances in experimental medicine and biology, ed. J. Plowman, D. Harland, and S. Deb-Choudhury, Vol. 1054, 21–32. Singapore: Springer.
- Powell, B. C., and J. S. Beltrame. 1994. Characterization of a hair (wool) keratin intermediate filament gene domain. The Journal of Investigative Dermatology 102:171–77. doi:10.1111/1523-1747.ep12371758.
- Powell, B. C., and G. E. Rogers. 1997. Formation and structure of human hair, 59–148. Basel, Switzerland: Birkhauser Verlag.
- Sparrow, L. G., C. P. Robinson, J. Caine, D. T. W. Mcmohan, and P. M. Strike. 1992. Type II intermediate-filament proteins from wool. Biochemistry Journal 282 (1):291–97. doi:10.1042/bj2820291.
- Yu, Z., J. E. Plowman, P. Maclean, J. E. Wildermoth, R. Brauning, J. C. McEwan, and N. J. Maqbool. 2018. Ovine keratome: Identification, localization and genomic organisation of keratin and keratin-associated proteins. Animal Genetics 49 (5):361–70. doi:10.1111/age.12694.
- Yu, Z., J. E. Wildermoth, O. A. M. Wallace, S. W. Gordon, N. J. Maqbool, P. H. Maclean, A. J. Nixon, and A. J. Pearson. 2011. Annotation of sheep keratin intermediate filament genes and their patterns of expression. Experimental dermatology 20 (7):582–88. doi:10.1111/j.1600-0625.2011.01274.x.
