Abstract
Induced pluripotent stem cell (iPSC)-based technologies offer an unprecedented opportunity to perform high-throughput screening of novel drugs for neurological and neurodegenerative diseases. Such screenings require a robust and scalable method for generating large numbers of mature, differentiated neuronal cells. Currently available methods based on differentiation of embryoid bodies (EBs) or directed differentiation of adherent culture systems are either expensive or are not scalable. We developed a protocol for large-scale generation of neuronal stem cells (NSCs)/early neural progenitor cells (eNPCs) and their differentiation into neurons. Our scalable protocol allows robust and cost-effective generation of NSCs/eNPCs from iPSCs. Following culture in neurobasal medium supplemented with B27 and BDNF, NSCs/eNPCs differentiate predominantly into vesicular glutamate transporter 1 (VGLUT1) positive neurons. Targeted mass spectrometry analysis demonstrates that iPSC-derived neurons express ligand-gated channels and other synaptic proteins and whole-cell patch-clamp experiments indicate that these channels are functional. The robust and cost-effective differentiation protocol described here for large-scale generation of NSCs/eNPCs and their differentiation into neurons paves the way for automated high-throughput screening of drugs for neurological and neurodegenerative diseases.
Introduction
Research in neuropsychiatry and neurodevelopmental disorders has been hampered by difficulty in obtaining neuronal cells from affected individuals that can be cultured and amplified in vitro. Human embryonic stem (hES) cells and human iPSCs Citation1 have revolutionized such research, as they can be differentiated into neurons in vitro from specific individualsCitation2Citation-4, potentially enabling personalized medicine by overriding the problems of allogenic recognition. Compelling evidence now indicates that iPSC-based models can be used to model selected aspects of neurological and neurodegenerative disorders.Citation5-7 Besides their potential to provide important molecular insights into pathogenic mechanisms, iPSC-based cellular platforms can also be used for drug discovery in specific differentiated cell types.Citation8 Such platforms require replicable, efficient, and cost effective protocols to generate uniform cultures of neurons in sufficient numbers to enable screening of potentially thousands of different compounds. For high-throughput screening, NSCs or NPCs from hES cells or iPSCs are currently transferred into multi-well culture plates for neuronal differentiation.Citation9,-Citation11 Different strategies are employed to derive the NSCs or NPCs, such as generation of EB followed by differentiation into neural rosettes.Citation12-14 EBs are 3-dimensional spherical aggregates that recapitulate several aspects of early embryogenesis. EBs generated from hES cells and human iPSCs efficiently differentiate into neural rosettes when cultured in specific selective culture media with growth factor supplements,Citation15 but EBs can have variable differentiation outcomes based on factors such as their initial size.Citation16 To enable more homogenous differentiation, microwell arrays have been specifically developed to allow the formation of EBs with uniform size (StemCell Technologies, Inc. Inc.).
The drawbacks related to EBs motivated differentiation protocols based on adherent culture systems that eliminate the EB generation step. Another advantage of adherent culture systems is that more uniform exposure to morphogens and growth/differentiation factors is achieved.Citation12 Neural rosettes (which represent a distinct class of NSCs) Citation17 generated in these adherent culture systems are isolated mechanically, then transferred and cultured into low attachment plates, where they form spherical cell aggregates called neurospheres that can be propagated as 3-dimensional structures,Citation18 or expanded as monolayer cultures of NSCs/NPCs Citation11 However, neurospheres are not ideal for large-scale production of neurons in multi-well plates for high-throughput screening because of technical difficulties in loading uniform numbers of spheres with uniform size into individual wells. Thus, monolayer cultures of NSCs/NPCs would be advantageous. Several protocols to derive NSCs/NPCs efficiently from hES cells or iPSCs currently utilize Noggin to induce neuronal differentiation,Citation19,20 but the need for Noggin significantly increases the cost of the differentiation process.
We have recently developed a method for efficient differentiation of human iPSCs into neurons Citation4 that does not involve generation of EBs. Using an adherent culture system that does not require Noggin, we generate NSCs/eNPCs in a scalable manner. This process takes about 4 weeks and approximately 4 additional weeks are required for differentiation mainly into VGLUT1 positive neurons. Here, we describe a detailed step-by-step protocol that allows efficient, robust, cost effective and large-scale generation of neurons. This differentiation protocol has been successfully applied to 6 different iPSC lines.
Results
Outline of differentiation protocol and nomenclature
The steps of large-scale generation of NSCs/eNPCs from iPSCs and their differentiation into neurons are schematically described in . Human iPSC cultures are exposed for 5 d to Neural Precursors (NP) Selection medium, and successively for 7 d in NP expansion medium. Following this phase, differentiating iPSC cultures are characterized by the presence of cellular aggregates that include emerging neural rosettes. These cells express NSCs markers, NSCs markers, such as NESTIN,Citation17 SOX1,Citation21 and MUSASHI Citation22 (). The expression of PAX6 Citation17 is limited to the areas forming neural rosettes (). This observation is in line with data by Suter et al. Citation23 showing that PAX6 expression follows the expression of SOX1. The cellular aggregates progressively develop into increasing numbers of neural rosettes.
Figure 1. Schematic flow diagram to depict the stages of differentiation into neurons from induced pluripotent stem cells (iPSCs). NSCs: neural stem cells; NPCs: neural progenitor cells; NLSs: neurosphere-like structures.
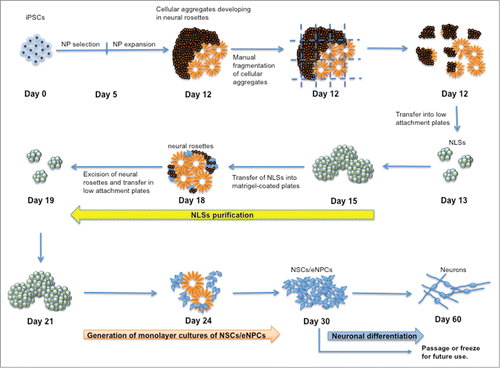
Figure 2. Expression of neural stem cell markers in cellular aggregates developing into neural rosettes observed in differentiating iPSCs cultured in NP expansion medium. Forming neuronal rosettes are more clearly distinguishable in Hoechst staining where nuclei of each neural rosette are arranged radially around a central cavity. Scale bar is 50 μm.
To facilitate further differentiation into NSCs/eNPCs, cellular aggregates are manually fragmented and cultured in ultra-low attachment plates where they round up, forming spherical cellular aggregates ( and ). Microscopic and immunocytochemical analyses of the spherical cellular aggregates show features of neurospheres similar to those generated by culturing human sub-ventricular zone from postmortem brains Citation24 ( and Fig. S1). Because it is unclear whether these spherical cellular aggregates possess all the characteristics of neurospheres, they are termed neurosphere-like structures (NLSs) in this report. Note that most of the NLSs contain multiple neural rosettes. The NLSs are further purified by transferring them into matrigel-coated plates (). When the NLSs are thus flattened, it is possible to clearly observe the neural rosettes () that are subsequently dissected and transferred into new ultra-low attachment plates where they round up overnight to form new purified NLSs (). Purified NLSs can then be expanded as monolayer cultures of NSCs/eNPCs () and further differentiated into neurons ( and ).
Figure 3. Generation of NLSs and neural rosettes. (a) NLSs generated from cellular aggregates developing into neural rosettes. (b) NLSs generated from single cell suspension of cellular aggregates in Aggrewell plates. (c) Microphotograph of neural rosettes observed 12 hours after plating purified NLSs. Scale bar is 50 μm.
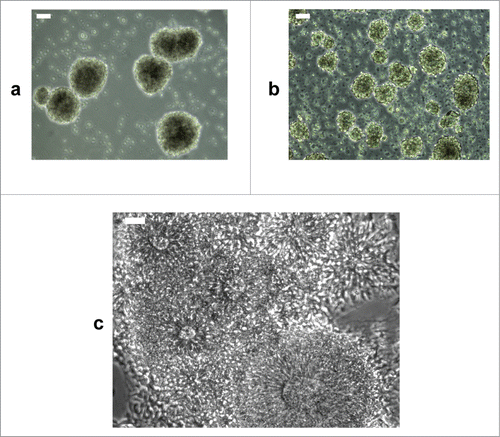
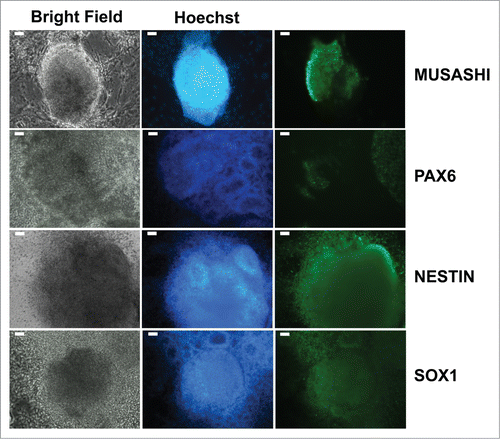
Figure 4. Characterization of iPSC-derived NSCs/eNPCs. (a) Left: Immunoblots showing the expression of markers specific for NSCs (MUSASHI, PAX6, NESTIN and SOX1), late NPCs (NEUROD1) and neurons (CALRETININ, TUJ1, and MAP2) in iPSC-derived NSCs/eNPCs. Right: Normalized expression of NSC markers. The data represent an average of 3 independent experiments. (b) Immunocytochemical analysis of iPSC-derived NSCs/eNPCs. Scale bar is 50 μm. (c) Quantification of PAX6 expression using High Content Analysis. RFU: relative fluorescence unit. A01-A03 denote biological replicates. The distribution of PAX6 labeling among NSCs/eNPCs is shown in the histograms. The histograms indicate RFU intensity in the vertical axis and number of cells at that intensity in the horizontal axis. The points outside the histograms indicate outliers. The tables below the histograms indicate the number of cells measured in the populations (Count), the average (Avg) and standard deviation (StdDev) of the overall PAX6 labeling in the population. Greater than 99% of the cells have PAX6 FITC levels above background. The average background level in the well is <400 gray levels. The minimum level in the cell is around 1,000 to 2,000. More than 99% of the cells show levels greater than 500.
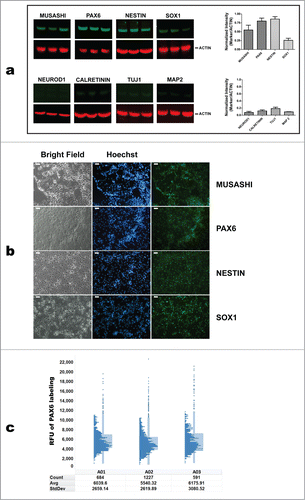
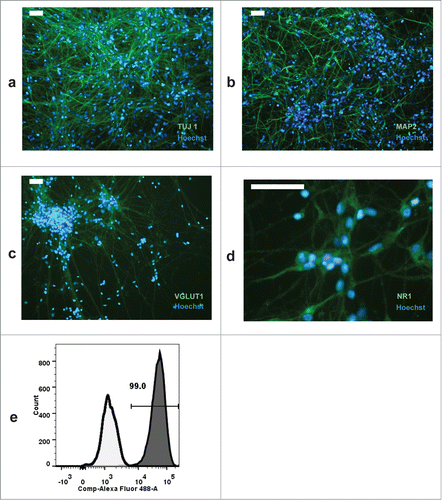
Figure 5. Immunostaining of iPSC-derived neurons with TUJ1 (a), MAP2 (b), VGLUT1 (c), and NR1 subunit of the NMDA receptor (d). The percentage of TUJ1 positive neurons was analyzed by flow cytometry; cells were permeabilized with Cytofix/Cytoperm (Becton Dickinson) and dead cells were excluded from the analysis. Filled histogram shows the fraction of TUJ1 stained cells compared to secondary antibody staining (e). Scale bar is 50 μm in a-c, and 75 μm in d.
Characterization of iPSC-derived NSCs/eNPCs and neurons
Western blotting analysis of NSCs/eNPCs showed a relatively low expression level of NeuroD1, indicating the absence of, or the presence of only a minor fraction of late NPCs Citation25 (). Hence, NPCs were denoted as “early” (eNPCs). Expression of NCS markers MUSASHI, PAX6, NESTIN, and SOX1 is depicted in . Analysis of PAX6 expression in NSCs/eNPCs by high-content imaging in individual wells of a 96-well plate showed that greater than 99% of the cells have PAX6 FITC levels above background ().
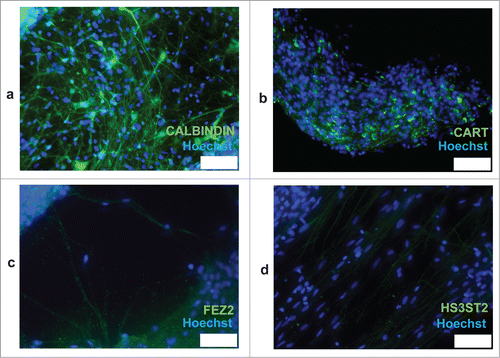
Following culture in neurobasal medium supplemented with B27 and BDNF for 4 weeks (see Reagents setup), more than 90% of differentiating cells were TUJ1 positive (). Immunocytochemical analysis showed that in our culture conditions, neuronal cells are also prevalently VGLUT1 positive (). Expression of the NR1 subunit of NMDA receptors was detected in a substantial fraction of neurons (). A small fraction of neurons were GAD1 positive.Citation4 No dopaminergic neurons were observed following immunocytochemistry. A subpopulation of neurons stained for CALBINDIN (), a cytosolic calcium-binding protein with increased expression in layer 3 of pyramidal cells in the dorsolateral prefrontal cortex (DLPFC) compared with layer 5 pyramidal cells in the same region.Citation26 In addition, expression of the neuropeptide CART, which is increased layer 3 versus layer 5 pyramidal cells (Dr David Lewis, unpublished), was observed only in discrete areas of the neuronal cultures (). Conversely, low-level staining was observed for the layer markers FEZ2 and HS3ST2, which have higher expression in layer 5 pyramidal cells vs. layer 3 cells (Dr David Lewis, unpublished) (). Taken together, these results indicate that neurons obtained with our differentiation procedure possess features of DLPFC layer 3 pyramidal neurons.
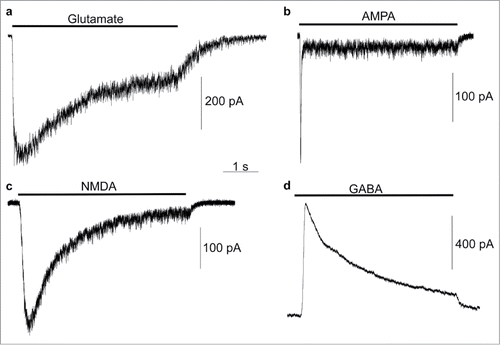
In whole-cell patch-clamp experiments, currents were recorded from ligand-gated channels after 2 months of neuronal differentiation of NSCs/eNPCs in neurobasal medium. Glutamate receptor-mediated currents were recorded at a holding potential of −85 mV in response to application of 100 μM glutamate + 100 μM glycine (6 of 10 cells; in responding cells peak current range was −88.5 to −771.1 pA, average −376.6 pA; ), 100 μM AMPA (8 of 13 cells; in responding cells peak current range was −15.5 to −481.8 pA, average −186.6 pA; ) and 100 μM N-methyl-d-aspartate (NMDA) + 100 μM glycine (4 of 7 cells; in responding cells peak current range was −80.0 to −320.0 pA, average −233.5 pA; ). GABA receptor-mediated currents were recorded at a holding potential of 15 mV in response to application of 1 mM GABA (11 of 12 cells; in responding cells peak current range was 75.5 to 3958.8 pA, average 736.9 pA; ).
Figure 7. Electrophysiological recordings of iPSC-derived neurons. Example traces of whole-cell voltage-clamp recordings from HFF1-S cells in response to application of the indicated agonists. Currents were recorded in a Mg+-free extracellular solution at a holding potential of −85 mV (a–c) or 15 mV (d) during approximately 5 s applications (indicated by horizontal bar above each trace) of 100 μM glutamate + 100 μM glycine (a, peak current −494.3 pA), 100 μM AMPA (b, peak current = −261.8 pA), 100 μM NMDA + 100 μM glycine (c, peak current = −324.4 pA), or 1 mM GABA (d, peak current = 851.8 pA).
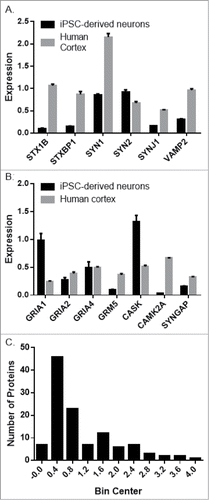
We observed robust expression of numerous neuronal proteins, both pre- and post-synaptic in iPSC-derived neurons (). In general, synaptic protein expression was higher in human cortex tissue compared to iPSC-derived neurons per μg total protein (). This is unsurprising, as the cellular composition of human cortex and neuronal cultures generated from iPSCs is very different. It does indicate, however, that the iPSC-derived neurons express many neuron specific genes.
Figure 8. Targeted proteomic comparison of iPSC-derived neurons and human cortex gray matter. Expression of representative presynaptic (A) and post-synaptic (B) proteins in iPSC-derived neurons (NSCs/eNPCs cultured in neurobasal medium for 6 weeks) and human cortex gray matter. (C). Distribution of the averaged difference between the 2 groups expressed as a ratio of iPSC-derived neurons: Cortex gray matter.
Discussion
The relatively simple culture media described in this protocol allows a large-scale generation of NSCs/eNPCs. Variability in the yield of NCSs migrating away from neural rosettes has been noticed among iPSC lines from different individuals. Furthermore, fibroblast-like cells are observed during the expansion of NSCs/eNPCs, but these cells are manually eliminated. One 6-well plate of confluent NSCs/eNPCs is sufficient to generate approximately 15 384-well plates of highly enriched neuronal cultures (more than 90% of TUJ1 positive cells). High content analysis has shown a uniform distribution of NSCs/eNPCs over the different wells in 384-well plates. The relative proportions of astrocytes and oligodendrocytes have not been investigated because the goal of this study was to provide a generalizable method for large-scale generation of NSCs/eNPCs that can be further differentiate into neurons in microtiter well format for high throughput analyses. The cost to generate a neuronal culture in a 384-well plate (where NSCs/eNPCs have been cultured in neurobasal medium for 4 weeks) starting from iPSCs is approximately $190.
The protocol detailed here enables a scalable and cost-effective derivation of NSCs/eNPCs that can be further differentiated into VGLUT1 positive neurons displaying features of layer 3 pyramidal cells. Synaptic protein expression was, on average, less in iPSC-derived neurons compared to human cortex tissue. This finding is to be expected considering the numerous differences between cell culture and tissue. Further studies are needed to evaluate whether NSCs/eNPCs generated with this protocol can be differentiated into other neuronal cell types.
Materials and Methods
Human iPSC cell lines
Lines HFF1-S, 73-56010-02, 73-56010-01, 71-500-9000, and 71-500-9001 were generated at NIMH funded Rutgers University Cell and DNA Repository (RUCDR) from skin fibroblasts. ATR1, another line was derived from T lymphocytes. Cells were reprogrammed with Sendai viral vectors expressing the transcription factors Sox2, c-Myc, Klf4, and Oct4. Following QC measures described earlier,Citation4 iPSCs were maintained in 6-well plates with the supplemented mTeSRTM1 medium (Stem Cell Technologies) changed daily, allowing for manual cleaning of differentiated cells. The protocol was approved by the University of Pittsburgh Institutional Review Board and written informed consent was obtained from all participants.
Reagent setup
NP selection medium
Dulbecco's minimum essential medium/F12 medium supplemented with 0.5% N2 (GIBCO), 1 mM L-glutamine, 1% nonessential amino acids, 50 U/ml penicillin G, 50 mg/ml streptomycin, and 0.1 mM 2-mercaptoethanol.Citation18
NP expansion medium
Dulbecco's minimum essential medium/F12 medium supplemented with 1% N2, 20 ng/ml rhFGFβ (Invitrogen), 1 mM L-glutamine, 1% nonessential amino acids, 50 U/ml penicillin G, 50 mg/ml streptomycin and 0.1 mM 2-mercaptoethanol.Citation18
Neurobasal medium
Neurobasal medium supplemented with 2% B27 (GIBCO), 10 ng/ml BDNF (PeproTech), 50 U/ml penicillin G, and 50 mg/ml streptomycin.
Matrigel aliquot preparation: Thaw a 5 ml container of matrigel qualified for use with mTeSR™1 medium (BD) on ice. Alternatively, matrigel can be thawed by placing it on ice and leaving it at 4°C overnight. Prepare aliquots by adding 500 μl matrigel into cryovials on ice. Store aliquots at −20°C for use.
Differentiation procedure
Generation and passaging of feeder-free iPSC cultures
We recommend using iPSCs at low passage number in order to reduce the probability of karyotypic abnormalities.Citation27 We transfer one iPSC colony from feeder-based culture into a 6-well matrigel-coated plate as follows.
Matrigel Preparation. Thaw a matrigel aliquot on ice. Pipette 50 ml PBS into a disposable falcon 50 ml tube. Add 1 ml of the PBS into the cryotube containing 500 μl matrigel. Mix quickly and transfer back to the 50 ml falcon tube. Mix matrigel/PBS mixture by pipetting up and down with a 10 ml pipette and dispense into plates. To coat one well of a 6-well plate, use 1 ml matrigel/PBS mixture.
NOTE: Thawing the matrigel requires approximately 2 hours. Keep matrigel aliquot on ice at all times.
iPSC seeding. Examine iPSC culture under a microscope to select undifferentiated colonies. Using a marking pen, draw a circle around undifferentiated iPSC colonies on the bottom of the plate (differentiated iPSC are characterized by non-uniform cell morphology in any portion of the colonies). Replace the medium in which iPSCs are cultured with pre-warmed (37°C) mTeSR™1 medium. Cut selected colonies into pieces of uniform size using a bent 20 gauge needle; harvest the iPSC clumps with a 10 ml pipette, and transfer into new culture well coated with matrigel, prepared as described in step 1(i). Culture iPSCs in mTeSRTM1 medium for 5–7 d in an incubator under standard conditions (5% CO2, 37°C, and 100% humidity). Change culture medium every day. Eliminate unwanted differentiated iPSCs from each well. When the iPSC colonies become confluent (colony size approximately 1.5–2 mm), passage iPSC colonies 1:6 as described above.
Derivation of NSCs/eNPCs from iPSCs. DURATION: 12 d
Wash iPSC colonies with 2 ml 1X PBS at room temperature twice. Add 2 ml of pre-warmed NP selection medium and incubate under standard conditions. Change medium every other day. At day 5, replace NP selection medium with NP expansion medium and culture for 7 d After 3-5 d in NP expansion medium, 50–70% of colonies show the presence of cellular aggregates expressing NSCs markers NESTIN,Citation17 SOX1,Citation21 and MUSASHI Citation22 (). If there is paucity of neural structures after 7 d culture in NP expansion medium, continue to culture the cells for 5–7 more days.
Generation of NLSs. DURATION: 3–4 d
After 7 d in NP expansion medium, proceed to generate NLSs as follows.
Aspirate the medium, wash the cells twice with 1 ml DMEM/F12 and add 2 ml of pre-warmed NP expansion medium. With the plate under the microscope (use the 4X objective), use a bent 20-gauge needle to dissect cellular aggregates into pieces of approximately 40–50 μm diameter.
Collect the NP expansion medium containing the dissected cell aggregates and transfer into an ultra-low attachment 6-well plate. Culture in standard incubator conditions overnight.
On the following day, cell aggregates should round up into floating spherical cellular aggregates denoted NLSs ().
Culture NLSs for 2–3 d in NP expansion medium. Change half the culture medium every day as follows. Collect NLSs in suspension from the plate and transfer into a 15 ml conical tube. Allow NLSs to settle to the bottom of the conical tube for 10 min. Replace half of the culture medium with fresh medium. Transfer the NLSs back to the 6-well ultra-low attachment plate. To distribute NLSs uniformly on low attachment plates, re-suspend NLSs by pipetting up and down 3–4 times with a 5 ml pipette. As the NLSs tend to sink to the bottom rapidly, transfer into 2 wells at a time, resuspending each time. Continue culturing until the NLSs reach a diameter of approximately 300 μm.
Alternative method for size-controlled formation of NLSs. DURATION: 4–5 d
Derivation of size-controlled NLSs is more expensive than the method described above. However, the percentage of NLSs differentiating into neural cells is higher with this method. To generate size-controlled NLSs, dissect cellular aggregates and transfer them into a 15 ml conical tube. Pellet the cells by spinning down for 30 sec at 1,200 rpm. Discard the supernatant and add 2 ml of StemPro Accutase (Life Technologies). Incubate at 37°C for 10 min (dissociate cells thoroughly by pipetting with a P1000 Pipetman every 2–3 min). Add 8 ml of DMEM/F12 and mix by inverting the tube 2–3 times. Leave the conical tube for 10 minutes to enable the undissociated clumps to settle to the bottom of the tube. Transfer the single cell suspension into a new 15 ml conical tube and centrifuge for 5 min at 1,200 rpm. Discard the supernatant and re-suspend the cells in 5 ml of NP expansion medium. Count viable cells using the trypan blue method. Adjust the volume of cell suspension to a density of 2 × 106 cells/ml. Add 2 ml of NP expansion medium to each well of an Aggrewell plate (Stem cell Technologies, Vancouver BC). Centrifuge the plate for 5 min at 1,200 rpm and discard the medium. Transfer 1 ml of cell suspension into each well to be used and centrifuge the plate as described above. Incubate plate overnight under standard culture conditions. Following overnight incubation, the cells should form spherical cell aggregates of approximately 40–50 μm (). After 24 hours, collect the NLSs by using a transfer pipette and transfer to an ultra-low attachment 6-well plate. Culture the NLSs as described above until they reach the desired size (approximately 200–250 μm). To efficiently dislodge NLSs from single micro-wells of the Aggrewell plate, apply mild positive pressure on different regions at the bottom of the well, using a transfer pipette bulb. After harvesting NLSs, ensure that all the NLSs have been collected by reviewing the donor plate under the microscope. Repeat the previous steps if necessary.
Purification of NLSs. DURATION: 6–7 d
Transfer the NLSs floating in the NP expansion medium into a 15 ml falcon tube and allow them to settle for 10 min. Aspirate the supernatant and re-suspend the NLSs into 12 ml of NP expansion medium. Distribute the NLS suspension (2 ml/well) in a matrigel-coated 6-well plate. Incubate in standard conditions. Following overnight incubation, NLSs tend to adhere to the bottom of the culture plate and flatten. At this stage, neural rosettes can be observed (). Place the culture plate under an inverted microscope. Dissect and collect the neural rosettes following the same procedure described in steps 3(i) and 3(ii). Incubate in standard conditions. On the following day, neural rosettes should round up into NLS. Culture NLSs for 2–3 d until the NLSs reach a size of approximately 300 μm. Change half the culture medium every other day as described in step 3(iv).
Generation of monolayer cultures of NSCs/eNPCs. DURATION: 9 d
Collect the NLSs into NP expansion medium and transfer into a matrigel-coated 6-well plate. Incubate in standard conditions. After a few hours, NLSs adhere to the surface of the culture plate and NSCs migrate away. At this stage, heterogeneous cultures will be generated, consisting of NSCs and eNPCs. Fibroblast-like cells may still be observed and should be removed under the inverted microscope by scraping them with a pipette tip.
Passaging NSCs/eNPCs
Aspirate NP expansion medium and rinse with DMEM/F12. For 6-well cultures, add 1 ml of StemPro Accutase per well and incubate for 3 min at 37°C. Add 2 ml of DMEM/F12 medium and gently detach cells by pipetting up and down with a p1000 tip. Transfer the cell suspension into a 15 ml conical tube. Centrifuge 5 min at 1,200 rpm. Discard the supernatant, re-suspend the cells with 12 ml of NP expansion medium, and culture in standard conditions in the appropriate plates coated with matrigel. On the next day, replace NP expansion medium with mTeSRTM1 medium and culture until the cells are confluent (). When confluent, the cultures in 6-well plates should contain approximately 3 × 106 cells/well. From this stage onwards, NSCs/eNPCs are cultured and passaged in mTeSRTM1 medium. The NSCs/eNPCs can be stored at −135°C or in liquid nitrogen. We do not recommend expanding NSCs/NPCs for more than 12 passages to reduce the risk of chromosomal aberration.Citation28 Freezing and thawing procedures of NSCs/eNPCs are described in Supplementary Information.
Generating neuronal cultures in 96- and 384-well plates. DURATION: 4 weeks.
Dissociate NSCs/eNPCs with StemPro Accutase as described above. Count the cells and re-suspend them in an appropriate volume of mTeSRTM1 medium. Distribute cells in 96-well plates at 104 cells/well or in 384-well plate at 2.5 × 103 cells/well. On the next day, change mTeSRTM1 to neurobasal medium and culture for 4 weeks. Replace half of the culture medium with fresh medium every other day by using a multi-channel pipette. Under these conditions, highly enriched cultures of neurons are generated ().
Immunocytochemistry and Microscopy
Immunocytochemistry was performed as previously described.Citation29 Primary antibodies used were mouse anti-human NESTIN monoclonal antibody (R&D Systems, 1:200 dilution), rabbit polyclonal anti-SOX1 antibody (Abcam, 1:200 dilution), rabbit polyclonal anti-MUSASHI antibody (Abcam, 1:200 dilution), rabbit polyclonal anti-PAX6 antibody (Abcam, 1:200 dilution), mouse anti-β TUBULIN III monoclonal antibody (clone Tuj-1, R&D System, 1:50 dilution), mouse monoclonal anti-MAP2 (Millipore, 1:200 dilution), rabbit polyclonal anti-VGLUT1 antibody (Synaptic Systems, 1:200 dilution), rabbit anti-NMDAR1 monoclonal antibody (Abcam, 1:400 dilution), rabbit polyclonal to CALBINDIN antibody (Abcam, 1:200 dilution), rabbit polyclonal to FEZ2 antibody (Abcam, 1:200 dilution), rabbit polyclonal to HS3ST2 antibody (Novus Biologicals, 1:50 dilution), mouse monoclonal to CART antibody (Abcam, 1:100 dilution). Alexa Fluor 488 goat anti-rabbit secondary antibody (Life Technologies, 1:200 dilution) and Alexa Fluor 488 goat anti-mouse secondary antibody (Life Technologies, 1:200 dilution) were used for detection. Counterstaining was done with Hoechst 33342 ((Life Technologies, 1:1000 dilution). Images were acquired using a Leica IL MD LED inverted fluorescence microscope.
Western blots
Cell lysates were prepared with 10 volumes of RIPA buffer (25 mM Tris–HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, and 0.1% SDS), denatured by heating at 95°C and then separated by electrophoresis on 4–12% gradient SDS-poly-acrylamide gels, transferred onto Immobilon membranes (Millipore). Immunoblots were probed with monoclonal antibodies similar to those employed in immunocytochemistry analysis; other antibodies used were rabbit anti-CALRETININ monoclonal antibody (Abcam, 1:800 dilution), rabbit anti-NEUROD1 polyclonal antibody (Abcam, 1:200 dilution). The blots were probed with fluorescently labeled secondary antibodies (Li-Cor antibodies) to detect immunoreactivity. Fluorescent signal intensities were then quantified using an Odyssey Imaging scanner (Li-Cor).
Quantification of PAX6 expression using high content analysis
NSC/eNPCs were fixed with 4% paraformaldehyde and labeled with rabbit polyclonal anti- PAX6 antibody followed by Alexa Fluor 488 goat anti-rabbit secondary antibody, and with Hoechst 33342 to identify individual cellular nuclei. Cells were imaged on the ImageXpress Ultra (IXU) High Content imager with 20x objective using the 405 nm excitation/447 nm emission to visualize the Hoechst nuclear stain, and 488 nm excitation/514 nm emission filter to visualize the PAX6 Alexa Fluor 488 stain. Images were analyzed using the Multiwavelength Cell Scoring application within the MetaXpress software that runs the IXU, and the average intracellular intensity of the PAX6 stain was determined for each cell.
Electrophysiological recordings
Plate iPSC-derived NSCs/eNPCs onto matrigel-coated 18 mm coverslips and culture for 8 weeks in neurobasal medium for neuronal differentiation. Visualize neuronal cells on 18 mm coverslips with a Zeiss inverted microscope. The external recording solution contains 140 mM NaCl, 2.8 mM KCl, 1 mM CaCl2, 0.01 mM EDTA (to eliminate contaminating Zn2+, which inhibits some NMDA receptor subtypes), 10 mM HEPES, pH 7.2 ± 0.05 adjusted with NaOH, osmolality 290 ± 10 mmol/kg adjusted with sucrose, and agonists added at indicated concentrations. The pipette solution contains 130 mM K-gluconate, 10 mM NaCl, 1 mM CaCl2, 1.1 mM EGTA, 10 mM HEPES, 2 mM ATP-Na2, 2 mM MgCl2, pH 7.2 ± 0.05 adjusted with KOH, osmolality 275 ± 10 mmol/kg.
Whole-cell recordings from iPSC-derived neurons are performed using pipettes pulled from borosilicate glass and fire-polished (5–8 MΩ). Membrane currents are recorded with an Axopatch 200B amplifier (Molecular Devices) in voltage-clamp mode, low-pass filtered at 5 kHz, and digitized at 10 kHz with a Digidata 1322a (Molecular Devices). Series resistance is compensated at 75–85%. For data presented here, recordings were rejected if holding current at −85 mV exceeded −600 pA (Molecular Devices). Rapid solution exchange is achieved using an in-house-fabricated fast perfusion system Citation30 connected to gravity-fed reservoirs. All experiments are performed at room temperature. Peak currents are measured relative to baseline current prior to agonist application using Clampfit 10.3 (Molecular Devices).
Targeted Proteomics
Cell lysates were prepared as described above and total protein quantified by micro BCA (Pierce). Human postmortem cortex gray matter homogenates previously prepared from 3 normal subjects were obtained.Citation31 20 μg of iPSC-derived neurons proteins or human cortex gray matter tissue homogenate were mixed with 20 μg of a stable isotope labeled neuronal proteome standard, separated by electrophoresis on a 4–12% gradient SDS-poly-acrylamide gel, and subject to on-gel digestion as previously described.Citation32 Liquid Chromatograph – Selected Reaction Monitoring analyses were conducted on a TSQ Quantum triple stage quadrupole mass spectrometer (ThermoFisher Scientific) with an Ultimate 3000 HPLC (Dionex) and a PicoChip Column (New Objective). Two μl (∼1 μg protein) sample was loaded onto the column at 1 μl/min for 12 min, and eluted at 300 nl/min over a 25 min gradient from 3–35% mobile phase B (Acetonitrile containing 0.1% formic acid). SRM transitions were timed using 1 min retention windows, depending on the number of SRMs to be assayed. Transitions were monitored, allowing for a cycle time of 1 sec, resulting in a dynamic dwell time never falling below 10 msec. The MS instrument parameters were as follows: capillary temperature 275°C, spray voltage 1100 V, and a collision gas of 1.4 mTorr (argon). The resolving power of the instrument was set to 0.7 Da (Full Width Half Maximum) for the first and third quadrupole. Data were acquired using a Chrom Filter peak width of 4.0 sec. Peak areas and area ratios were calculated within Skyline.Citation33 Raw files were loaded into Skyline files containing target proteins/peptides/transitions. All individual SRM transitions and integration areas were manually inspected. Transitions for which the signal-to-noise ratio was below 3 were excluded from analysis. The ratios of the integrated areas for “light” endogenous peptides and “heavy” Citation13C6STD peptides were calculated to obtain peptide measures using multiple transitions per peptide. 165 peptides from 116 proteins were measured.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Figure_S1.pdf
Download PDF (2.1 MB)Supplementary_Information.docx
Download MS Word (14.2 KB)Acknowledgments
We thank our study participants and research staff. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Mental Health, the National Institutes of Health, the Department of Veterans Affairs, or the United States Government.
Funding
This project was funded from NIH grants MH63480 and MH045817, and MH071533, by Stanley Medical Research Foundation 07R-1712, and by the Ri.MED. We also thank China Scholarship Council for providing a fellowship to YZ and thank Indo-US Science & Technology Forum (IUSSTF) for providing a fellowship to DKD. The Biomedical Mass Spectrometry Center and UPCI Cancer Biomarker Facility are supported in part by award P30CA047904.
Supplemental Material
Supplemental data for this article can be accessed on thepublisher's website.
References
- Takahashi K, Yamanaka S. Induced pluripotent stem cells in medicine and biology. Development 2013; 140:2457-61; PMID:23715538; http://dx.doi.org/10.1242/dev.092551
- Beevers JE, Caffrey TM, Wade-Martins R. Induced pluripotent stem cell (iPSC)-derived dopaminergic models of Parkinson's disease. Biochem Soc Trans 2013; 41:1503-8; PMID:24256244
- Eigentler A, Boesch S, Schneider R, Dechant G, Nat R. Induced pluripotent stem cells from friedreich ataxia patients fail to upregulate frataxin during in vitro differentiation to peripheral sensory neurons. Stem Cells Dev 2013; 22:3271-82; PMID:23879205; http://dx.doi.org/10.1089/scd.2013.0126
- D'Aiuto L, Di Maio R, Heath B, Raimondi G, Milosevic J, Watson AM, Bamne M, Parks WT, Yang L, Lin B, et al. Human induced pluripotent stem cell-derived models to investigate human cytomegalovirus infection in neural cells. PLoS One 2012; 7:e49700; PMID:23209593
- Liu GH, Qu J, Suzuki K, Nivet E, Li M, Montserrat N, Yi F, Xu X, Ruiz S, Zhang W, et al. Progressive degeneration of human neural stem cells caused by pathogenic LRRK2. Nature 2012; 491:603-7; PMID:23075850; http://dx.doi.org/10.1038/nature11557
- Kaye JA, Finkbeiner S. Modeling Huntington's disease with induced pluripotent stem cells. Molecular and cellular neurosciences 2013; 56:50-64; PMID:23459227; http://dx.doi.org/10.1016/j.mcn.2013.02.005
- Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, Li Y, Mu Y, Chen G, Yu D, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature 2011; 473:221-5; PMID:21490598; http://dx.doi.org/10.1038/nature09915
- Boissart C, Poulet A, Georges P, Darville H, Julita E, Delorme R, Bourgeron T, Peschanski M, Benchoua A. Differentiation from human pluripotent stem cells of cortical neurons of the superficial layers amenable to psychiatric disease modeling and high-throughput drug screening. Transl Psychiatry 2013; 3:e294; PMID:23962924
- Charbord J, Poydenot P, Bonnefond C, Feyeux M, Casagrande F, Brinon B, Francelle L, Aurégan G, Guillermier M, Cailleret M, et al. High throughput screening for inhibitors of REST in neural derivatives of human embryonic stem cells reveals a chemical compound that promotes expression of neuronal genes. Stem Cells 2013; 31:1816-28; PMID:23712629; http://dx.doi.org/10.1002/stem.1430
- Efthymiou A, Shaltouki A, Steiner JP, Jha B, Heman-Ackah SM, Swistowski A, Zeng X, Rao MS, Malik N. Functional screening assays with neurons generated from pluripotent stem cell-derived neural stem cells. J Biomol Screen 2014; 19:32-43; PMID:24019252; http://dx.doi.org/10.1177/1087057113501869
- Zhao WN, Cheng C, Theriault KM, Sheridan SD, Tsai LH, Haggarty SJ. A high-throughput screen for Wnt/β-catenin signaling pathway modulators in human iPSC-derived neural progenitors. J Biomol Screen 2012; 17:1252-63; PMID:22923789; http://dx.doi.org/10.1177/1087057112456876
- Dhara SK, Stice SL. Neural differentiation of human embryonic stem cells. J Cell Biochem 2008; 105:633-40; PMID:18759328; http://dx.doi.org/10.1002/jcb.21891
- Muratore CR, Srikanth P, Callahan DG, Young-Pearse TL. Comparison and optimization of hiPSC forebrain cortical differentiation protocols. PLoS One 2014; 9:e105807; PMID:25165848
- Velasco I, Salazar P, Giorgetti A, Ramos-Mejía V, Castaño J, Romero-Moya D, Menendez P. Concise review: generation of neurons from somatic cells of healthy individuals and neurological patients through induced pluripotency or direct conversion. Stem Cells 2014; 32:2811-7; PMID:24989459; http://dx.doi.org/10.1002/stem.1782
- Kurosawa H. Methods for inducing embryoid body formation: in vitro differentiation system of embryonic stem cells. J Biosci Bioeng 2007; 103:389-98; PMID:17609152; http://dx.doi.org/10.1263/jbb.103.389
- Bauwens CL, Peerani R, Niebruegge S, Woodhouse KA, Kumacheva E, Husain M, Zandstra PW. Control of human embryonic stem cell colony and aggregate size heterogeneity influences differentiation trajectories. Stem Cells 2008; 26:2300-10; PMID:18583540; http://dx.doi.org/10.1634/stemcells.2008-0183
- Elkabetz Y, Studer L. Human ESC-derived neural rosettes and neural stem cell progression. Cold Spring Harb Symp Quant Biol 2008; 73:377-87; PMID:19204067; http://dx.doi.org/10.1101/sqb.2008.73.052
- Cho MS, Hwang DY, Kim DW. Efficient derivation of functional dopaminergic neurons from human embryonic stem cells on a large scale. Nat Protoc 2008; 3:1888-94; PMID:19008875; http://dx.doi.org/10.1038/nprot.2008.188
- Baharvand H, Mehrjardi NZ, Hatami M, Kiani S, Rao M, Haghighi MM. Neural differentiation from human embryonic stem cells in a defined adherent culture condition. Int J Dev Biol 2007; 51:371-8; PMID:17616926; http://dx.doi.org/10.1387/ijdb.072280hb
- Kirkeby A, Nelander J, Parmar M. Generating regionalized neuronal cells from pluripotency, a step-by-step protocol. Front Cell Neurosci 2012; 6:64; PMID:23316134
- Venere M, Han YG, Bell R, Song JS, Alvarez-Buylla A, Blelloch R. Sox1 marks an activated neural stem/progenitor cell in the hippocampus. Development 2012; 139:3938-49; PMID:22992951; http://dx.doi.org/10.1242/dev.081133
- Kim DS, Lee DR, Kim HS, Yoo JE, Jung SJ, Lim BY, Jang J, Kang HC, You S, Hwang DY, et al. Highly pure and expandable PSA-NCAM-positive neural precursors from human ESC and iPSC-derived neural rosettes. PLoS One 2012; 7:e39715; PMID:22911689; http://dx.doi.org/10.1371/journal.pone.0039715
- Suter DM, Tirefort D, Julien S, Krause KH. A Sox1 to Pax6 switch drives neuroectoderm to radial glia progression during differentiation of mouse embryonic stem cells. Stem Cells 2009; 27:49-58; PMID:18832594; http://dx.doi.org/10.1634/stemcells.2008-0319
- Gonzalez-Perez O. Neural stem cells in the adult human brain. Biol Biomed Rep 2012; 2:59-69; PMID:23181200
- Gao Z, Ure K, Ables JL, Lagace DC, Nave KA, Goebbels S, Eisch AJ, Hsieh J. Neurod1 is essential for the survival and maturation of adult-born neurons. Nat Neurosci 2009; 12:1090-2; PMID:19701197; http://dx.doi.org/10.1038/nn.2385
- Hayes TL, Lewis DA. Nonphosphorylated neurofilament protein and calbindin immunoreactivity in layer III pyramidal neurons of human neocortex. Cereb Cortex 1992; 2:56-67; PMID:1633408; http://dx.doi.org/10.1093/cercor/2.1.56
- Taapken SM, Nisler BS, Newton MA, Sampsell-Barron TL, Leonhard KA, McIntire EM, Montgomery KD. Karotypic abnormalities in human induced pluripotent stem cells and embryonic stem cells. Nat Biotechnol 2011; 29:313-4; PMID:21478842; http://dx.doi.org/10.1038/nbt.1835
- Nguyen TD, Widera D, Greiner J, Müller J, Martin I, Slotta C, Hauser S, Kaltschmidt C, Kaltschmidt B. Prolonged cultivation of hippocampal neural precursor cells shifts their differentiation potential and selects for aneuploid cells. Biol Chem 2013; 394:1623-36; PMID:24084358
- D'Aiuto L, Robison CS, Gigante M, Nwanegbo E, Shaffer B, Sukhwani M, et al. Human IL-12 p40 as a reporter gene for high-throughput screening of engineered mouse embryonic stem cells. BMC Biotechnol 2008; 8:52; PMID:18522747; http://dx.doi.org/10.1186/1472-6750-8-52
- Qian A, Antonov SM, Johnson JW. Modulation by permeant ions of Mg(2+) inhibition of NMDA-activated whole-cell currents in rat cortical neurons. J Physiol 2002; 538:65-77; PMID:11773317; http://dx.doi.org/10.1113/jphysiol.2001.012685
- Deo AJ, Goldszer IM, Li S, DiBitetto JV, Henteleff R, Sampson A, et al. PAK1 protein expression in the auditory cortex of schizophrenia subjects. PLoS One 2013; 8:e59458; PMID:23613712; http://dx.doi.org/10.1371/journal.pone.0059458
- MacDonald ML, Ciccimaro E, Prakash A, Banerjee A, Seeholzer SH, Blair IA, Hahn CG. Biochemical fractionation and stable isotope dilution liquid chromatography-mass spectrometry for targeted and microdomain-specific protein quantification in human postmortem brain tissue. Mol Cell Proteomics 2012; 11:1670-81; PMID:22942359; http://dx.doi.org/10.1074/mcp.M112.021766
- Lista A, Arbilla S, Langer SZ. Modulation of the electrically evoked release of 5-[3H]hydroxytryptamine from rat cerebral cortex: effects of alpidem, CL 218872, and diazepam. J Neurochem 1988; 51:1414-21; PMID:2844992