Abstract
Before the secretion of hard dental tissues, tooth germs undergo several distinctive stages of development (dental lamina, bud, cap and bell). Every stage is characterized by specific proliferation patterns, which is regulated by various morphogens, growth factors and homeodomain proteins. The role of MSX homeodomain proteins in odontogenesis is rather complex. Expression domains of genes encoding for murine Msx1/2 during development are observed in tissues containing highly proliferative progenitor cells. Arrest of tooth development in Msx knockout mice can be attributed to impaired proliferation of progenitor cells. In Msx1 knockout mice, these progenitor cells start to differentiate prematurely as they strongly express cyclin-dependent kinase inhibitor p19INK4d. p19INK4d induces terminal differentiation of cells by blocking the cell cycle in mitogen-responsive G1 phase. Direct suppression of p19INK4d by Msx1 protein is, therefore, important for maintaining proliferation of progenitor cells at levels required for the normal progression of tooth development. In this study, we examined the expression patterns of MSX1, MSX2 and p19INK4d in human incisor tooth germs during the bud, cap and early bell stages of development. The distribution of expression domains of p19INK4d throughout the investigated period indicates that p19INK4d plays active role during human tooth development. Furthermore, comparison of expression domains of p19INK4d with those of MSX1, MSX2 and proliferation markers Ki67, Cyclin A2 and pRb, indicates that MSX-mediated regulation of proliferation in human tooth germs might not be executed by the mechanism similar to one described in developing tooth germs of wild-type mouse.
Introduction
During the early odontogenic sequence, tooth germs undergo several histologically distinctive stages of development starting with appearance of thickenings of embryonic oral ectoderm (dental lamina), and progressing through bud, cap and bell stages before the secretion of hard dental tissues (i.e., enamel and dentin) takes place. Every stage of development is characterized by a specific spatial pattern of proliferative activity which is tightly regulated by the differential action of various morphogens, growth factors and homeodomain proteins.Citation1-5 The role of Muscle Segment Homeodomain proteins 1 and 2 (MSX1/MSX2) in odontogenesis is rather complex. Msx1 and Msx2 (murine homologues of human MSX1/2) are structurally and functionally related transcription factors which generally act as transcriptional suppressors.Citation6 This is executed either directly by DNA-binding via homeodomain to target gene's promoter region, or indirectly by protein-protein interactions with the target gene's activators (other transcription factors), which results with formation of inactive complexes between the individual Msxs and activators rendering the target gene transcription suppressed.Citation7 It seems that protein-protein interactions might be the main mechanism through which Msx1 and Msx2 exert transcriptional suppression in vivo, but the opposite effect of transcriptional activation is also possible depending on the context in which these protein-protein interactions occur.Citation8,9 The list of protein interactors with Msx1 and Msx2 includes not only homeodomain proteins such as Pax9, Dlx2, Dlx5 (which act as transcription activators), but also certain members of the four major signaling parthways (Shh, Wnt, Bmp, Fgf) involved in organogenesis, meaning that Msx1 and Msx2 can coordinate multiple signaling cues regulating development of various tissues and organs.Citation10-16
Analysis of expression domains of genes encoding for Msx1/2 during odontogenesis show that these transcription factors are predominantly present in areas of tooth germ containing highly proliferative mesenchymal and epithelial progenitor cells. Expression domains of Msx1/2 completely overlap only during the dental lamina stage when both genes are expressed in the mesenchyme underlying the dental lamina. Subsequently, the expression domain of Msx2 shifts to epithelial compartment of the tooth germ, whereas Msx1 remains to be expressed in the underlying dental mesenchyme.Citation2, 17-19 This occurs in period when, due to epithelial-to-mesenchymal interactions, instructive potential for odontogenesis switches from epithelial part of the tooth germ to dental mesenchyme, and is therefore reflected by diverse phenotypes observed in individual Msx gene knockouts. While tooth development is arrested early in the dental lamina stage in Msx1/2 double knockout mice, in Msx1 knockout mice tooth development arrests later during the bud-to-cap transition.Citation14, 20 Similarly, in humans, loss-of-function mutations of MSX1 gene are responsible for various forms of non-syndromic hereditary hypodontia affecting permanent dentition.Citation21, 22 On the other hand, Msx2 knockout mice display late tooth phenotype with severe hypoplastic defects of enamel and low resistance of all teeth to occlusal forces.Citation23
Various phenotypes seen in individual Msx gene knockout mice do have a common theme and that is impaired proliferation of progenitor cells in developing tissues where these genes (and the proteins they encode) are normally expressed. The arrest of tooth development in Msx1 knockout mice during the late bud stage is due to severe reduction of cell proliferation in dental mesenchyme. Thus, dental mesenchyme remains populated by insufficient number of progenitor mesenchymal cells and is, therefore, unable to properly condense around the tooth bud. Expression profiling of progenitor mesenchymal cells revealed that they specifically started to express differentiation markers for neuronal cell-lineage, along with cyclin-dependent kinase inhibitor p19INK4d.Citation24 p19INK4d belongs to a family of INK4 cyclin-dependent kinase inhibitors (CKI) whose function is to induce terminal differentiation of cells by blocking the progression of cell cycle during the G1 phase.Citation25 It does so by binding to cyclin-dependent kinases 4 and 6 (Cdk4/6) rendering them unable to form active complexes with D-type cyclins. Generally, every phase of the cell cycle is characterized by different set of complexes between cyclins and Cdks responsible for deactivation of Retinoblastoma protein (Rb) by phosphorylating it at specific residues in a step-wise manner, since hypophosphorylated form of Rb inhibits progression of the cell cycle and commitment to mitosis.Citation26-28 While p19INK4d and other members of INK4 CKIs (p15INK4b, p16INK4a and p18INK4c) are equally potent inhibitors of the cell cycle, only p18INK4c and p19INK4d are found to be ubiquitously expressed during organogenesis when they regulate physiological withdrawal from the cell cycle and terminal differentiation of multiple progenitor cell populations.Citation29, 30 In developing tooth germs of wild-type mouse, Msx1-mediated suppression of p19INK4d activity in dental mesenchyme seems to be the key mechanism by which proliferation of progenitor cells is maintained at levels required for the normal progression of odontogenic sequence. According to a recent report, Msx1 exerts that effect directly by binding to promoter region of p19INK4d gene.Citation31
In this study, we examined the expression patterns of MSX1, MSX2 and p19INK4d in human incisor tooth germs during the bud, cap and early bell stages of development. While MSX1/2 displayed similar expression domains to those of Msx1/2 in murine tooth germs, they also partially overlapped with expression domain of p19INK4d throughout the investigated period. This implies that MSX-mediated regulation of proliferation during the early stages of human odontogenesis might not be executed by the mechanism similar to one described in developing tooth germs of wild-type mouse.
Results
Following the appearance of dental lamina by the end of the 5th gestational week, tooth germs undergo histologically distinctive stages of development (bud, cap and bell). In this study, the expression patterns of MSX1, MSX2, p19INK4d, and proliferation markers Ki67, Cyclin A2, and pRb were investigated in human incisor tooth germs in period between 7th and 14th gestational week by means of single and double immunofluorescence.
Expression patterns of p19INK4d, Ki67, Cyclin A2 and pRb in human incisor tooth germ during the bud stage and bud-to-cap transition
In the 7th gestational week, human incisor tooth germ is in the bud stage of development. Moderate cytoplasmic expression of p19INK4d can be observed in tooth bud and adjacent strip of embryonic oral epithelium, whereas very weak expression of p19INK4d can be observed in the region of jaw mesenchyme surrounding the tooth bud (dental mesenchyme) (, d1-e1, Subset 1). Ki67-positive cell nuclei can be seen throughout the tooth bud and in the dental mesenchyme (, b1-c1, Subset 1). Numerous Cyclin A2-positive and pRb-positive cell nuclei are distributed throughout the dental mesenchyme, and can also be seen in most of the tooth bud (, b2-e2, Subset 2).
Figure 1. Expression patterns of Ki67, p19INK4d, Cyclin A2, phosphorylated Rb, MSX1 and MSX2 in human incisor tooth germs during the bud stage and bud-to-cap stage transition. Expression patterns of p19INK4d, MSX1, MSX2 and proliferation markers Ki67, Cyclin A2 and pRb in human incisor tooth germ in the bud stage and during the bud-to-cap transition (Magnification: × 40, scale bar: 25 µm); (a1-6) DAPI staining of nuclei (inverted – red color); (b1-6, d1-6) expression patterns of investigated factors in epithelial and mesenchymal compartments of human incisor tooth germ; (c1-6, e1-6) merged image doublets of investigated factors' expression patterns with DAPI; approximation of expression domains for Ki67 and p19INK4d (Subsets 1, 4), Cyclin A2 and pRb (Subsets 2, 5), MSX1 and MSX2 (Subsets 3, 6), expression domains are displayed in magenta color (expression intensity range covered – mild to strong). Designations: oral epithelium (oe), dental lamina (dl), tooth bud (tb), jaw mesenchyme (m), outer enamel epithelium (oee), inner enamel epithelium (iee), stellate reticulum (sr), stratum intermedium (si), cervical loop (cl), dental papilla (dp).
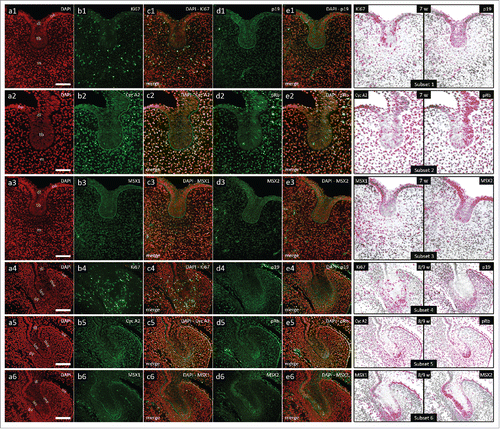
Bud-to-cap transition occurs between the 8th and 9th gestational week. During this period, central portion of the epithelial part of human incisor tooth germ is now enclosed by the inner and outer enamel epithelia (, a4-6). Dental mesenchyme starts to condense under the inner enamel epithelium designating formation of dental papilla. Moderate cytoplasmic expression of p19INK4d can be observed in the outer enamel epithelium, at the confluence of inner and outer enamel epithelium, and also in the condensing dental mesenchyme (, d4-e4, Subset 4). Ki67-positive cell nuclei can be seen in all tissues of the tooth germ (, b4-c4, Subset 4). Cyclin A2-positive and pRb-positive cell nuclei are distributed within the p19INK4d expression domain, as well as in the dental lamina and jaw mesenchyme (, b5-e5, Subset 5).
Expression patterns of p19INK4d, Ki67, Cyclin A2 and pRb in human incisor tooth germ during the cap stage
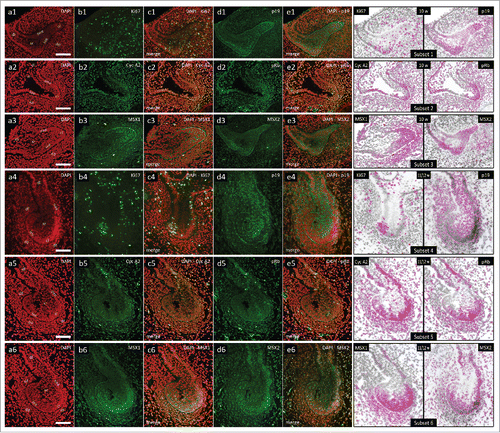
In the 10th gestational week, human incisor tooth germ is in the early cap stage of development. Central portion of the epithelial part of the tooth germ is comprised of cells which will form stellate reticulum, whereas the confluence regions of inner and outer enamel epithelia define prospective cervical loops. In the mesenchymal part of the tooth germ, dental papilla and dental follicle are now clearly visible (, a1-6). Moderate to strong cytoplasmic expression of p19INK4d can be observed in both prospective cervical loops, sections of outer enamel epithelium and in significant portion of dental papilla (, d1-e1, Subset 1). Ki67-positive cell nuclei are numerous in all tissues of the tooth germ (, b1-c1, Subset 1). Cyclin A2-positive and pRb-positive cell nuclei are still distributed within the p19INK4 expression domain, as well as in the dental follicle and jaw mesenchyme surrounding the tooth germ (, b2-e2, Subset 2). As the cap stage progresses (period of mid-to-late cap stage between the 11th and 12th gestational week), expression domain of p19INK4d displays dramatic shift in both epithelial and mesenchymal parts of the tooth germ. p19INK4d is now expressed mostly in the stellate reticulum and outer enamel epithelium, whereas only mild expression could be observed in the outer rim of dental papilla (, d4-e4, Subset 4). Furthermore, expression of Cyclin A2 now displays both nuclear and cytoplasmic. However, the distribution of Cyclin A2 expression domain (and pRb-positive cell nuclei) is similar to the one observed in the 10th gestational week, meaning it is complementary to the expression domain of p19INK4d (, b5-e5, Subset 5).
Figure 2. Expression patterns of Ki67, p19INK4d, Cyclin A2, phosphorylated Rb, MSX1 and MSX2 in human incisor tooth germs during the cap stage. Expression patterns of p19INK4d, MSX1, MSX2 and proliferation markers Ki67, Cyclin A2 and pRb in human incisor tooth germ during the cap stage (Magnification: × 20; scale bar: 40 µm); (a1-6) DAPI staining of nuclei (inverted – red color); (b1-6, d1-6) expression patterns of investigated factors in epithelial and mesenchymal compartments of human incisor tooth germ; (c1-6, e1-6) merged image doublets of investigated factors' expression patterns with DAPI; approximation of expression domains for Ki67 and p19INK4d (Subsets 1, 4), Cyclin A2 and pRb (Subsets 2, 5), MSX1 and MSX2 (Subsets 3, 6), expression domains are dispalyed in magenta color (expression intensity range covered – mild to strong). Designations: oral epithelium (oe), dental lamina (dl), tooth bud (tb), jaw mesenchyme (m), outer enamel epithelium (oee), inner enamel epithelium (iee), stellate reticulum (sr), stratum intermedium (si), cervical loop (cl), dental papilla (dp).
Expression patterns of p19INK4d, Ki67, Cyclin A2 and pRb in human incisor tooth germ during the early bell stage
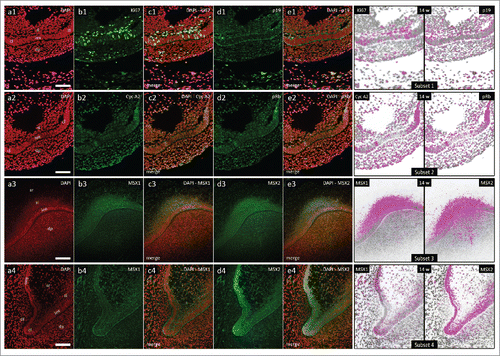
In the 14th gestational week, human incisor tooth germ is in the early bell stage of development. Epithelial part of the tooth germ is now defined as the enamel organ and is significantly enlarged (, a1-3). In the central portion of the enamel organ, stellate reticulum is populated by loosely distributed cells. Located between the stellate reticulum and the inner enamel epithelium there are several layers of densely packed spindle-shaped cells comprising stratum intermedium. Cervical loops begin to turn downwards as they continue to proliferate and enclose dental papilla. Mild to moderate cytoplasmic expression of p19INK4d can be observed in the inner enamel epithelium, both cervical loops, sections of the outer enamel epithelia adjacent to cervical loops and in the outer rim of dental papilla (, d1-e1, Subset 1; , d1-e1, Subset 1). Ki67-positive cell nuclei are present in all of these tissues, but they can also be seen in great numbers in the stratum intermedium (, b1-c1, Subset 1; , b1-c1, Subset 1). Cyclin A2-positive and pRb-positive cell nuclei are partially distributed within the p19INK4d expression domain (, b2-e2, Subset 2; , b2-e2, Subset 2). They can also be observed in the stratum intermedium and dental lamina. However, the distribution of Cyclin A2-positive cell nuclei displays somewhat asymmetric pattern in enamel organ when compared with the distribution of pRb-positive cell nuclei (, Subset 2).
Figure 3. Expression patterns of Ki67, p19INK4d, Cyclin A2, phosphorylated Rb, MSX1 and MSX2 in human incisor tooth germs during the early bell stage. Expression patterns of p19INK4d, MSX1, MSX2 and proliferation markers Ki67, Cyclin A2 and pRb in human incisor tooth germ in the early bell stage (Magnification: × 10; scale bar: 100 µm); (a1-3) DAPI staining of nuclei (inverted – red color); (b1-3, d1-3) expression patterns of investigated factors in epithelial and mesenchymal compartments of human incisor tooth germ; (c1-3, e1-3) merged image doublets of investigated factors' expression patterns with DAPI; approximation of expression domains for Ki67 and p19INK4d (Subset 1), Cyclin A2 and pRb (Subset 2), MSX1 and MSX2 (Subset 3), expression domains are displayed in magenta color (expression intensity range covered – moderate to strong). Designations: oral epithelium (oe), dental lamina (dl), tooth bud (tb), jaw mesenchyme (m), outer enamel epithelium (oee), inner enamel epithelium (iee), stellate reticulum (sr), stratum intermedium (si), cervical loop (cl), dental papilla (dp).
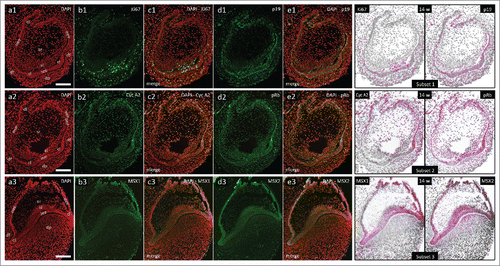
Figure 4. Expression patterns of Ki67, p19INK4d, Cyclin A2, phosphorylated Rb, MSX1 and MSX2 in human incisor tooth germs during the early bell stage (epithelial-mesenchymal interface and cervical loops). Expression patterns of p19INK4d, MSX1, MSX2 and proliferation markers at the inner enamel epithelium-dental papilla interface and in cervical loops of human incisor enamel organ in the early bell stage (Magnification: × 20; scale bar: 40 µm); (a1-a4) DAPI staining of nuclei (inverted – red color); (b1-4, d1-4) expression patterns of investigated factors in epithelial and mesenchymal compartments of human incisor tooth germ; (c1-4, e1-4) merged image doublets of investigated factors' expression patterns with DAPI; approximation of expression domains for Ki67 and p19INK4d (Subset 1), Cyclin A2 and pRb (Subset 2), MSX1 and MSX2 (Subsets 3, 4), expression domains are displayed in magenta color (expression intensity range covered – mild to strong). Designations: oral epithelium (oe), dental lamina (dl), tooth bud (tb), jaw mesenchyme (m), outer enamel epithelium (oee), inner enamel epithelium (iee), stellate reticulum (sr), stratum intermedium (si), cervical loop (cl), dental papilla (dp).
Expression patterns of MSX1 and MSX2 in human incisor tooth germ during the investigated period
During the investigated period, expression domains of MSX1 and MSX2 displayed significant overlapping. In the bud stage, MSX1 and MSX2 are moderately expressed in both the tooth bud and the underlying dental mesenchyme with mixed nuclear and cytoplasmic expression pattern. Nevertheless, MSX1 exhibits predominance in the dental mesenchyme, as opposed to MSX2 which is more expressed in the epithelial part of the tooth germ (, b3-e3, Subset 3). During the bud-to-cap transition, the expression domain of MSX1 extends over both sides of the epithelial-mesenchymal interface, whereas the expression domain of MSX2 shifts to the epithelial part of the tooth germ (, b6-e6, Subset 6). Similar distribution of their expression domains is maintained during the succeeding cap stage (, b6-e6, Subset 6). It is intriguing that mid-to-late cap stage is the only period when expression domains of both MSX1 and MSX2 do not significantly overlap with expression domain of p19INK4d (, Subset 4, Subset 6). Namely, MSX1 and MSX2 are expressed in the inner and outer enamel epithelia where the expression of p19INK4d seems to be down-regulated. However, MSX2 (and not MSX1) displays nuclear expression pattern in both enamel organ epithelia at that period (, b6, d6; ). In the early bell stage, MSX1 and MSX2 are expressed throughout the inner and outer enamel epithelia and stratum intermedium, but only MSX2 is expressed from both sides of the interface between the inner enamel and dental papilla at the site of future cusp tip (, b3-e3, Subset 3; , b3-e3, Subset 3). Furthermore, the expression domain of MSX1 in enamel organ does not extend as much as the expression domain of MSX2. The intensity of expression of MSX1 seems to differ in the opposing cervical loops in contrast to symmetric expression pattern displayed by MSX2 (, Subset 3; , b4-c4, Subset 4). It should be noted that the expression patterns MSX1 and MSX2 proteins described here correspond well with the expression patterns of MSX1 and MSX2 genes in developing human tooth germs previously reported in other studies.Citation32, 33
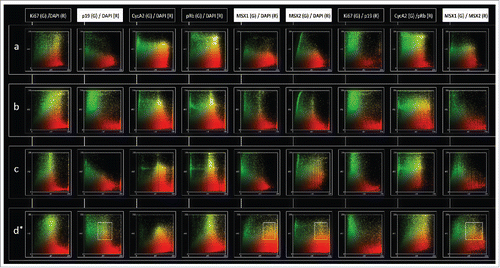
Figure 5. Intensity correlation analysis of MSX1, MSX2, p19INK4d and proliferation markers in human incisor tooth germs during the investigated period. Intensity correlation analysis of MSX1, MSX2, p19INK4d and proliferation markers in human incisor tooth germ during bud (row a), bud-to-cap (row b), cap (row c) and early bell stage (row d) of development; Expression patterns of investigated factors displayed on color scatterplots are split into red (R) (x-axis) and green (G) channels (y-axis). Yellow color on scatterplots designates positive correlation between the two channels, meaning that both factors are not only expressed in the same tissue (overlapping expression domains/co-occurrence), but also in the same cellular compartment (nuclear or cytoplasmic expression pattern/co-localization). Scatterplots on the left show the type of expression pattern specifically for each factor (DAPI columns). Intensity correlation between the expression patterns of pairs of investigated factors is also shown on (columns Ki67/p19; Cyc A2/pRb; MSX1/MSX2). Note that expression pattern of Ki67, Cyclin A2 and pRb is predominantly nuclear (rows a-d; columns Ki67/DAPI, Cyc A2/DAPI and pRb/DAPI), whereas p19INK4d and MSX1 display cytoplasmic expression pattern (rows a-d; columns p19/DAPI and MSX1/DAPI). Similar correlations can be made for MSX2, except for a shift from cytoplasmic to nuclear expression pattern observed in the cap stage (row c; column MSX2/DAPI). Positive intensity correlation between the expression patterns of MSX1 and MSX2 can only be seen during the bud (row a; column MSX1/MSX2) and early bell stage (row d, column MSX1/MSX2). Framed areas (row d; columns p19/DAPI, MSX1/DAPI, MSX2/DAPI) show correlation bias since p19INK4d, MSX1 and MSX2 display perinuclear and/or cytoplasmic expression patterns during the early bell stage. Correlation bias is due to low magnification of merged image doublets (× 10) on which the intensity correlation analysis has been performed. However, positive correlation between the expression patterns of MSX1 and MSX2 in the early bell stage is valid since both factors have cytoplasmic expression pattern and their expression domains significantly overlap (row d; column MSX1/MSX2, framed area).
Discussion
Despite the fact that Msx proteins are structurally and functionally similar transcriptional suppressors, much of their biological function (as well as the effects caused by the loss of that function) depends upon the molecular context in which they operate. During murine odontogenesis, the expression of Msx1/2 genes is regulated by reciprocal cues from key signaling pathways (Wnt, Shh, Fgfs, Bmps) whose activity and effects are, in turn, modulated by the action of Msx proteins. In most cases, Msx proteins achieve this through protein-protein interactions with particular members of those signaling pathways and other homeodomain proteins, whereas the direct transcriptional repression of Msx1/2 target genes by DNA-binding is rarely described in vivo.Citation6, 34 In developing tissues, Msx1/2 protein expression is found specifically in regions populated by mitotically active progenitor cells.Citation12, 35 Interestingly, a significant fraction of these cells are kept in prolonged G1 phase of the cell cycle, which explains why Msx1/2 by themselves exert no mitogenic effect, but rather function to keep the progenitor cells in mitogen-responsive state until the microenvironment conditions are favorable for completion of cell division.
Based on a previous report about roles of Msx1 in regulation of proliferation in mouse tooth germ, it seems that this is done by Msx1-mediated transcriptional repression of p19INK4d gene which encodes p19INK4d, an important CKI whose expression induces the exit from the cell cycle at G1 phase in cells destined to terminally differentiate.Citation24 According to our findings, the regulation of proliferation in human tooth germ by MSX1 (human homologue of murine Msx1) might not be conducted through direct transcriptional repression of p19INK4d gene. Namely, the expression domains of MSX1, MSX2 and p19INK4d partially overlapped in epithelial and mesenchymal compartment of human tooth germ throughout the investigated period. On a cellular level, MSX1 displayed predominantly cytoplasmic expression which implies that, in developing human tooth germ, MSX1 probably mediates its effects most through protein-protein interactions. Since the promoter region of human p19INK4d gene has also been shown to contain MSX1-binding sites, and since the modes of action of MSX proteins are highly context-dependent, we cannot exclude that MSX1-mediated transcriptional repression of p19INK4d does not occur in other organs during human organogenesis as the actual mechanism of regulation of proliferation. For example, we have observed a clear demarcation of expression domains of both MSX1 and MSX2 with expression domain of p19INK4d in developing eye lens, where MSX1 and MSX2 were expressed in highly proliferative marginal region, while p19INK4d was expressed in central portion which contains already differentiating cells (article in preparation). Recently, MSX1 has been implicated in tumorigenesis of anterior lobe of human pituitary gland during which expression of MSX1 makes dramatic shift from cytoplasmic to predominantly nuclear expression pattern.Citation36 Although we are not aware of any studies about expression patterns of p19INK4d in human pituitaries, it is intriguing that p19INK4d knockout mice exhibit hyperplasia and tumors of the anterior lobe of pituitary, which is exactly where both p19INK4d and Msx1 are normally expressed in wild-type animals.Citation37
The observation that expression domain of p19INK4d in human tooth germ partially overlapped with expression domains of proliferation markers Ki67, cyclin A2 and pRb throughout the investigated period can be interpreted from several different aspects. Firstly, normal progression of the cell cycle in proliferating cells depends on oscillating levels of the main cell cycle regulators, i.e., cyclins and CKIs.Citation38,39 Namely, every phase of the cell cycle is characterized by the expression of different subsets of cyclins, where, for example, D-type cyclins are strongly expressed in mitogen-responsive G1 phase, as opposed to A-type cyclins which accumulate during the S phase. Interestingly, p19INK4d is the only member of INK4 CKI which displays dynamic expression pattern during the cell cycle, and is, therefore, present at incrementally higher levels as the cell cycle progresses, especially during the S and G2 phases.Citation25 Secondly, findings from cell culture studies indicate that p19INK4d protein has quite a rapid turnover and, in contrast to p18INK4c which is more prone to post-transcriptional regulation, changes in p19INK4d protein levels almost faithfully reflect the activity of its encoding gene.Citation40 Thirdly, although Cdk4/6 are the only downstream targets of p19INK4d identified thus far, there is circumstantial evidence that during the very early stages of embryogenesis p19INK4d might operate through cyclin D-Cdk4/6-Rb independent mechanism.Citation41 Namely, withdrawal from the cell cycle and terminal differentiation of multiple progenitor cells in developing and adult tissues are the main effects mediated by p19INK4d due to its ability to bind and inhibit Cdk4/6, which, in turn, prevents formation of active cyclin D-Cdk4/6 complexes required for sequential phosphorylation of growth suppressive Rb and progression of the cell cycle in proliferating cells. However, continuous expression of both p19INK4d protein and p19INK4d mRNA in Xenopus laevis embryos in period prior to mid-blastula transition (MBT), shows that active transcription of p19INK4d gene can even occur in cells which, by default, contain very high levels of inactive, hyperphosphorylated form of Rb (pRb), and whose cell cycle consists of only S and M phases.Citation42 Unfortunately, there are no data about molecular mechanisms regulating expression of p19INK4d or possible p19INK4d downstream targets unrelated to cyclin D-Cdk4/6-Rb cascade during the very early stages of development.
With regard to this, the described overlapping of expression domain of p19INK4d with expression domains of proliferation markers Ki67, cyclin A2 and pRb in developing human tooth germ does not seem surprising. It is intriguing, though, why these expression domains overlap only partially. We could assume that evaluation of expression patterns of p18INK4c (another member of INK4 family of CKIs, which is also expressed during organogenesis), might provide more insight into this matter since functionally related factors frequently display complementary expression patterns during odontogenesis.Citation43,44 In contrast to what has been reported about involvement of p19INK4d in regulation of proliferation during murine odontogenesis, the results presented in this study indicate that p19INK4d plays more active role in regulation of proliferation during human odontogenesis. Tooth germs of wild-type mouse do not express p19INK4d in bud and cap stage, whereas p19INK4d is clearly expressed in specific areas of both mesenchymal and epithelial parts of human tooth germs during those stages. Furthermore, the distribution of expression domains and patterns of MSX1, MSX2, p19INK4d with those of proliferation markers Ki67, cyclin A2 and pRb, indicates that MSX-mediated regulation of proliferation in human tooth germs might not be executed by the same mechanisms described in tooth germs of wild-type mice. Apart from inter-species differences, the fact that certain loss-of-function mutations of MSX1 gene in humans cause hypodontia in permanent dentition only, points out that these mechanisms might not even be the same in development of primary and secondary teeth.
Materials and Methods
Tissue procurement and processing
For the purpose of this study, we used tissue samples from ten human fetuses aged 7, 8/9, 10, 11/12 and 14 weeks of gestation, which were obtained after spontaneous abortions and tubal pregnancies from the Department of Pathology, University Hospital in Split, Croatia. The samples were stored in the form of histological sections and kept at −24°C as a part of human tissue archival collection of the Department of Anatomy, Histology and Embryology, School of Medicine, University of Split. Tissue processing approval was given by the Ethical and Drug Committee of University Hospital in Split (Class: 033–081/11-03/0005, No: 2181-198-03-04/10-11-0024, 2011) in accordance with Helsinki Declaration.Citation45 External measurements were used for the assessment of gestational age of the human fetuses.Citation46 Immunohistochemical analysis was performed on fetal tissues from head area and/or parts of jaws which contained tooth germs. Paraffin-embedded fetal tissues were cut in transversal or frontal planes (serial 7 µm sections) and tissue sections were mounted on glass slides for microscopic examination using Olympus BX51 light microscope (Olympus, Tokyo, Japan). Tissue preservation and presence of structures of interest was confirmed by the examination of control sections stained with haematoxylin and eosin. After that, sections were processed for single and double immunofluorescence staining.
Immunofluorescence staining protocol
Tissue sections were deparaffinized by standard protocol in xylene and descending ethanol solutions, rehydrated in distilled water and left to incubate for 30 min in 0.1% H2O2 in order to suppress endogenous peroxidase activity. Sections were shortly washed in PBS before they were placed in sodium citrate buffer and heated at 95°C in microwave oven for 15 min. Before the 24 h incubation in dark chamber with primary antibodies, sections were left to cool down to room temperature. Primary antibodies used for immunofluorescence staining were as follows: mouse monoclonal anti-human Ki67 antibody (proliferation marker) (1:50; DAKO, Glostrup, Denmark), rabbit monoclonal anti-human p19INK4d antibody (1:100; ab102842, Abcam, UK), goat monoclonal anti-human MSX1 antibody (1:300; ab93287, Abcam, UK), rabbit monoclonal anti-human MSX2 antibody (1:300; ab190070, Abcam, UK), mouse monoclonal anti-human Cyclin A2 antibody (proliferation marker; somatic cells in the S/G2 phase of the cell cycle) (1:200; ab38, Abcam, UK), and rabbit monoclonal anti-human pRb (proliferation marker; somatic cells in the S/G2 phase of the cell cycle) (1:50; ab173289, Abcam, UK) for hyperphosphorylated form of Rb (phosphorylated at Serine780 residue). Single immunofluorescence staining was performed for each primary antibody, and following pairs were selected for double immunofluorescence staining: (1) anti-Ki67/anti-p19INK4d; (2) anti-MSX1/anti-MSX2; (3) anti-Cyclin A2/anti-pRb. After the incubation with primary antibodies, sections were prepared for 1 h incubation in dark chamber with secondary antibodies as follows: anti-goat Alexa Fluor 488 (GREEN; 1:400, ab150129, Abcam, UK), anti-rabbit anti-mouse Alexa Fluor 488 (GREEN; 1:400; ab150105, Abcam, UK), anti-rabbit Alexa Fluor 594 (RED; 1:300, ab, Abcam, UK), anti-mouse Alexa Fluor 594 (RED; 1:300, ab150108, Abcam, UK), and anti-rabbit Alexa Fluor 488 (GREEN; 1:300, ab150073, Abcam, UK). When the incubation in secondary antibodies was done, sections were washed in PBS and counterstained with 4′6′-diamidino-2-phenylindole (DAPI) for 1–2 min to stain nuclei. Sections were shortly rinsed in distilled water, air-dried, mounted and cover-slipped (Immuno-Mount; Shandon, Pittsburgh, PA). Negative control staining for antibody specificity was performed by omission of primary antibodies from the staining procedure. Secondary antibody signal bleeding was controlled by comparison of photo-micrographs from single and double immunofluorescence staining for each primary antibody used.
Data processing and photo-micrograph analysis
Immunofluorescence images were taken by SPOT Insight camera (Diagnostic Instruments, USA), mounted on Olympus BX61 fluorescence microscope (Olympus, Tokyo, Japan). Raw images were acquired using CellA® software. Raw images were further processed in Adobe Photoshop® CS6 as follows: blue-to-red inversion for blue immunofluorescence (DAPI nuclear staining), and red-to-green inversion for red immunofluorescence (p19INK4d, MSX2 and Cyclin A2 staining). Color inversion was not performed for green immunofluorescence (Ki67, MSX1, pRb staining). Merged image doublets (inverted primary antibody 1/inverted DAPI; primary antibody 2/inverted DAPI; primary antibody 1/primary antibody 2) were then assembled for expression domain approximation and intensity correlation analysis by using ImageJ software (NIH, Public domain) according to recommendations for analysis of immunofluorescence in biological microscopy.Citation47,48 Pixel intensity misalignment ratio was set at default value (50%) for cytoplasmic expression patterns. That was done in order to include minimal overlapping of the background halo around DAPI stained nuclei with cytoplasmic signals into approximation of expression domain. Intensity correlation analysis was performed with custom specification of region of interest (ROI) on merged image doublets in order to exclude tissues surrounding the tooth germs. In order to eliminate co-localization/co-occurrence bias between different signals, intensity correlation analysis was performed on merged image doublets at magnifications × 20 and × 4049.
TABLE 1. Semiquantification analysis of expression of MSX1, MSX2 and p19INK4d in epithelial and mesenchymal parts of human incisor tooth germ between 7 and 14 weeks of development.
ABBREVIATION
| MSX1 | = | Muscle Segment Homeodomain Protein 1 |
| MSX2 | = | Muscle Segment Homeodomain Protein 2 |
| p19INK4d | = | Ink4 Family Cyclin-dependent Kinase Inhibitor p19 |
| phospho Rb | = | phosphorylated Retinoblastoma Protein |
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
No potential conflicts of interest were disclosed.
AUTHOR CONTRIBUTIONS
Darko Kero designed the study, performed selection of primary antibodies and immunofluorescent staining of tissue sections (MSX1/MSX2/p19INK4d), acquired, interpreted and processed data, assessed the literature review reports and wrote the manuscript. Katarina Vukojevic performed double immunofluorescent staining (Cyclin A2/phospho Rb), acquired and interpreted data, reviewed part of the literature (cell cycle regulation) and wrote Materials and Methods and Results sections of the manuscript. Petra Stazic performed double immunofluorescent staining (Ki67/p19INK4d), acquired and interpreted data, reviewed part of the literature (p19INK4d and INK4 CDK cell cycle inhibitors/ functional studies) and wrote part of Results section of the manuscript. Danijela Sundov performed single immunofluorescent staining (MSX1/MSX2), reviewed part of the literature (MSX1 and MSX2 in organogenesis and odontogenesis), and wrote Result section of the manuscript. Snjezana Mardesic Brakus performed double immunofluorescent staining (Ki67/p19INK4d), and reviewed part of the literature (regulation of proliferation in organogenesis and odontogenesis). Mirna Saraga Babic designed the study, provided inputs on staining procedures and data interpretation, revised and co-edited the manuscript with Darko Kero.
Acknowledgements
The authors wish to thank Mrs. Marica Maretic, mag.chem.ing., for expert technical assistance.
FUNDING
This work was supported by the Ministry of Science, Education and Sports of the Republic of Croatia (grant no. 021-2160528-0507, principal investigator prof. Mirna Saraga-Babic, MD, PhD).
REFERENCES
- Kero D, Novakovic J, Vukojevic K, Petricevic J, Kalibovic Govorko D, Biocina-Lukenda D, Saraga-Babic M. Expression of Ki-67, Oct-4, gamma-tubulin and alpha-tubulin in human tooth development. Arch Oral Biol. 2014; 59:1119-29. doi:10.1016/j.archoralbio.2014.05.025.
- Sharpe PT. Neural crest and tooth morphogenesis. Adv Dent Res. 2001; 15:4-7. doi:10.1177/08959374010150011001.
- Thesleff I. Epithelial-mesenchymal signalling regulating tooth morphogenesis. J Cell Sci. 2003; 116:1647-8. doi:10.1242/jcs.00410.
- Thesleff I, Mikkola M. The role of growth factors in tooth development. Int Rev Cytology. 2002; 217:93-135. doi:10.1016/S0074-7696(02)17013-6.
- Zhang Q, Davenport JR, Croyle MJ, Haycraft CJ, Yoder BK. Disruption of IFT results in both exocrine and endocrine abnormalities in the pancreas of Tg737(orpk) mutant mice. Lab Invest; A journal of technical methods and pathology. 2005; 85:45-64. doi:10.1038/labinvest.3700207.
- Hu G, Lee H, Price SM, Shen MM, Abate-Shen C. Msx homeobox genes inhibit differentiation through upregulation of cyclin D1. Development. 2001; 128:2373-84.
- Bendall AJ, Abate-Shen C. Roles for Msx and Dlx homeoproteins in vertebrate development. Gene. 2000; 247:17-31. doi:10.1016/S0378-1119(00)00081-0.
- Menezes ME, Mitra A, Shevde LA, Samant RS. DNAJB6 governs a novel regulatory loop determining Wnt/beta-catenin signalling activity. Biochem J. 2012; 444:573-80. doi:10.1042/BJ20120205.
- Thomas T, Kurihara H, Yamagishi H, Kurihara Y, Yazaki Y, Olson EN, Srivastava D. A signaling cascade involving endothelin-1, dHAND and msx1 regulates development of neural-crest-derived branchial arch mesenchyme. Development. 1998; 125:3005-14.
- Bei M. Molecular genetics of tooth development. Current Opinion Gene Dev. 2009; 19:504-10. doi:10.1016/j.gde.2009.09.002.
- Lallemand Y, Bensoussan V, Cloment CS, Robert B. Msx genes are important apoptosis effectors downstream of the Shh/Gli3 pathway in the limb. Dev Biol. 2009; 331:189-98. doi:10.1016/j.ydbio.2009.04.038.
- Lallemand Y, Nicola MA, Ramos C, Bach A, Cloment CS, Robert B. Analysis of Msx1; Msx2 double mutants reveals multiple roles for Msx genes in limb development. Development. 2005; 132:3003-14. doi:10.1242/dev.01877.
- Jernvall J, Thesleff I. Reiterative signaling and patterning during mammalian tooth morphogenesis. Mech Dev. 2000; 92:19-29. doi:10.1016/S0925-4773(99)00322-6.
- Maas R, Bei M. The genetic control of early tooth development. Critical reviews in oral biology and medicine: an official publication of the American Association of Oral Biologists. 1997; 8:4-39. doi:10.1177/10454411970080010101.
- Ohazama A, Haycraft CJ, Seppala M, Blackburn J, Ghafoor S, Cobourne M, Martinelli DC, Fan CM, Peterkova R, Lesot H, et al. Primary cilia regulate Shh activity in the control of molar tooth number. Development. 2009; 136:897-903. doi:10.1242/dev.027979.
- Thesleff I, Tummers M. Tooth organogenesis and regeneration. StemBook[Internet]. Cambridge (MA): Harvard Stem Cell Institute; 2008-2009 Jan 31. doi:10.3824/stembook.1.37.1.
- Kero D, Saraga-Babic M. Odontogenesis – A Masterful Orchestration of Functional Redundancy or What Makes Tooth Bioengineering an Intrinsically Difficult Concept. J Stem Cell Res Therapeutics. 2016; 1:7. doi:10.15406/jsrt.2016.01.00022.
- Thesleff I, Sharpe P. Signalling networks regulating dental development. Mech Dev. 1997; 67:111-23. doi:10.1016/S0925-4773(97)00115-9.
- Yamashiro T, Tummers M, Thesleff I. Expression of bone morphogenetic proteins and Msx genes during root formation. J Dent Res. 2003; 82:172-6. doi:10.1177/154405910308200305.
- Cobourne MT, Sharpe PT. Making up the numbers: The molecular control of mammalian dental formula. Semin Cell Dev Biol. 2010; 21:314-24. doi:10.1016/j.semcdb.2010.01.007.
- Abid MF, Simpson MA, Petridis C, Cobourne MT, Sharpe PT. Non-syndromic severe hypodontia caused by a novel frameshift insertion mutation in the homeobox of the MSX1 gene. Arch Oral Biol. 2017; 75:8-13. doi:10.1016/j.archoralbio.2016.11.018.
- Lidral AC, Reising BC. The role of MSX1 in human tooth agenesis. J Dent Res. 2002; 81:274-8. doi:10.1177/154405910208100410.
- Satokata I, Ma L, Ohshima H, Bei M, Woo I, Nishizawa K, Maeda T, Takano Y, Uchiyama M, Heaney S, et al. Msx2 deficiency in mice causes pleiotropic defects in bone growth and ectodermal organ formation. Nat Genet. 2000; 24:391-5. doi:10.1038/74231.
- Han J, Ito Y, Yeo JY, Sucov HM, Maas R, Chai Y. Cranial neural crest-derived mesenchymal proliferation is regulated by Msx1-mediated p19(INK4d) expression during odontogenesis. Dev Biol. 2003; 261:183-96. doi:10.1016/S0012-1606(03)00300-2.
- Canepa ET, Scassa ME, Ceruti JM, Marazita MC, Carcagno AL, Sirkin PF, Ogara MF. INK4 proteins, a family of mammalian CDK inhibitors with novel biological functions. IUBMB life 2007; 59:419-26. doi:10.1080/15216540701488358.
- Guo J, Sheng G, Warner BW. Epidermal growth factor-induced rapid retinoblastoma phosphorylation at Ser780 and Ser795 is mediated by ERK1/2 in small intestine epithelial cells. J Biol Chem. 2005; 280:35992-8. doi:10.1074/jbc.M504583200.
- Harbour JW, Luo RX, Dei Santi A, Postigo AA, Dean DC. Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell. 1999; 98:859-69. doi:10.1016/S0092-8674(00)81519-6.
- Ortega S, Malumbres M, Barbacid M. Cyclin D-dependent kinases, INK4 inhibitors and cancer. Biochimica et biophysica acta. 2002; 1602:73-87.
- Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol. 2006; 7:667-77. doi:10.1038/nrm1987.
- Zindy F, Quelle DE, Roussel MF, Sherr CJ. Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene. 1997; 15:203-11. doi:10.1038/sj.onc.1201178.
- Zhao M, Gupta V, Raj L, Roussel M, Bei M. A network of transcription factors operates during early tooth morphogenesis. Mol Cell Biol. 2013; 33:3099-112. doi:10.1128/MCB.00524-13.
- avideau JL, Demri P, Hotton D, Gu TT, MacDougall M, Sharpe P, Forest N, Berdal A. Comparative study of MSX-2, DLX-5, and DLX-7 gene expression during early human tooth development. Pediatr Res. 1999; 46:650-6. doi:10.1203/00006450-199912000-00015.
- Lin D, Huang Y, He F, Gu S, Zhang G, Chen Y, Zhang Y. Expression survey of genes critical for tooth development in the human embryonic tooth germ. Developmental dynamics: An official publication of the American Association of Anatomists. 2007; 236:1307-12. doi:10.1002/dvdy.21127.
- Newberry EP, Latifi T, Battaile JT, Towler DA. Structure-function analysis of Msx2-mediated transcriptional suppression. Biochemistry. 1997; 36:10451-62. doi:10.1021/bi971008x.
- Miletich I, Sharpe PT. Normal and abnormal dental development. Hum Mol Genet. 2003; 12(Spec No 1):R69-73. doi:10.1093/hmg/ddg085.
- Mizokami Y, Egashira N, Takekoshi S, Itoh J, Itoh Y, Osamura RY, Matsumae M. Expression of MSX1 in human normal pituitaries and pituitary adenomas. Endocr Pathol. 2008; 19:54-61. doi:10.1007/s12022-008-9021-7.
- Bai F, Chan HL, Smith MD, Kiyokawa H, Pei XH. p19Ink4d is a tumor suppressor and controls pituitary anterior lobe cell proliferation. Mol Cell Biol. 2014; 34:2121-34. doi:10.1128/MCB.01363-13.
- Murray AW. Recycling the cell cycle: cyclins revisited. Cell 2004; 116:221-34. doi:10.1016/S0092-8674(03)01080-8.
- Zindy F, Nilsson LM, Nguyen L, Meunier C, Smeyne RJ, Rehg JE, Eberhart C, Sherr CJ, Roussel MF. Hemangiosarcomas, medulloblastomas, and other tumors in Ink4c/p53-null mice. Cancer Res. 2003; 63:5420-7.
- Forget A, Ayrault O, den Besten W, Kuo ML, Sherr CJ, Roussel MF. Differential post-transcriptional regulation of two Ink4 proteins, p18 Ink4c and p19 Ink4d. Cell Cycle. 2008; 7:3737-46. doi:10.4161/cc.7.23.7187.
- Savatier P, Huang S, Szekely L, Wiman KG, Samarut J. Contrasting patterns of retinoblastoma protein expression in mouse embryonic stem cells and embryonic fibroblasts. Oncogene. 1994; 9:809-18.
- Cosgrove RA, Philpott A. Cell cycling and differentiation do not require the retinoblastoma protein during early Xenopus development. Developmental Biol. 2007; 303:311-24. doi:10.1016/j.ydbio.2006.11.015.
- Kero D, Cigic L, Medvedec Mikic I, Galic T, Cubela M, Vukojevic K, Saraga-Babic M. Involvement of IGF-2, IGF-1R, IGF-2R and PTEN in development of human tooth germ – an immunohistochemical study. Organogenesis. 2016; 12:152-67. doi:10.1080/15476278.2016.1197460.
- Kero D, Kalibovic Govorko D, Medvedec Mikic I, Vukojevic K, Cigic L, Saraga-Babic M. Analysis of expression patterns of IGF-1, caspase-3 and HSP-70 in developing human tooth germs. Arch Oral Biol. 2015; 60:1533-44. doi:10.1016/j.archoralbio.2015.07.004.
- Williams JR. The Declaration of Helsinki and public health. Bull World Health Organ 2008; 86:650-2. doi:10.2471/BLT.08.050955.
- O'Rahilly R. Guide to the staging of human embryos. Anat Anz. 1972; 130:556-9.
- Dunn KW, Kamocka MM, McDonald JH. A practical guide to evaluating colocalization in biological microscopy. Am J Physiol Cell Physiol 2011; 300:C723-42. doi:10.1152/ajpcell.00462.2010.
- Zinchuk V, Zinchuk O. Quantitative colocalization analysis of confocal fluorescence microscopy images. Curr Protoc Cell Biol. 2008; Chapter 4:Unit 4 19. doi:10.1002/0471143030.cb0419s39.
- Adler J, Parmryd I. Colocalization analysis in fluorescence microscopy. Methods Mol Biol. 2013; 931:97-109. doi:10.1007/978-1-62703-056-4_5.
