ABSTRACT
Isolated primary hepatocytes from the liver are very similar to in vivo native liver hepatocytes, but they have the disadvantage of a limited lifespan in 2D culture. Although a sandwich culture and 3D organoids with mesenchymal stem cells (MSCs) as an attractive assistant cell source to extend lifespan can be used, it cannot fully reproduce the in vivo architecture. Moreover, long-term 3D culture leads to cell death because of hypoxic stress. Therefore, to overcome the drawback of 2D and 3D organoids, we try to use a 3D printing technique using alginate hydrogels with primary hepatocytes and MSCs. The viability of isolated hepatocytes was more than 90%, and the cells remained alive for 7 days without morphological changes in the 3D hepatic architecture with MSCs. Compared to a 2D system, the expression level of functional hepatic genes and proteins was higher for up to 7 days in the 3D hepatic architecture. These results suggest that both the 3D bio-printing technique and paracrine molecules secreted by MSCs supported long-term culture of hepatocytes without morphological changes. Thus, this technique allows for widespread expansion of cells while forming multicellular aggregates, may be applied to drug screening and could be an efficient method for developing an artificial liver.
1. INTRODUCTION
Artificial organ transplantation is an outstanding recent challenge in medicine as liver transplantation is deemed the best therapeutic method for severe liver diseases. However, most transplant patients die due to surgical complications, donor organ shortage and rejection risk.Citation1,Citation2 To resolve this, many scientists are attempting to develop artificial organs. In the case of an artificial liver, a major obstacle is the limited culture time for primary hepatocytes. After isolation for 3 days, the apoptotic pathway becomes activated in primary hepatocytes and they differentiate into fibrotic cells.Citation3 For this reason, increasing the culture time for primary hepatocytes is a major challenge that must be overcome in order to make artificial livers.
The functional support mesenchymal stem cells (MSCs) provide to primary hepatocytes has been demonstrated in many previous studies.Citation4-7 It is known that paracrine factors are secreted by MSCs including transforming growth factor β1 (TGF-β1), leukemia inhibitory factor (LIF) and hepatocyte growth factor (HGF). These molecules stabilize function and help with the maintenance of isolated hepatocytes both in vivo and in vitro.
Three-dimensional (3D) tissue engineering techniques have been shown to resolve issues inherent to 2D culture systems. In general, the formation of a 3D structure is the first step in the construction of specific target organs and tissues because the microenvironment and cell-cell contact in the organ are very important for function maintenance.Citation8-10 Thus, 3D bio-printing technology applied to tissue engineering has become one of the most efficient and useful tools for manufacturing artificial tissues and organs.Citation11-13
In this study, a 3D bio-printing technique was utilized to manufacture hepatic architectures. To accomplish this aim, we used primary hepatocytes with MSCs to support hepatocyte function and viability time in 3D structures. 3D bio-printing resolved loss of function and morphology issues in primary hepatocyte cultures.
2. MATERIALS AND METHODS
2.1. Animals
Both male and female Tg(CAG-DsRed*MST)1Nagy/J mice weighing 20–30 g were used for the isolation of primary hepatocytes. The mice were a kind gift from Won-Il Jeong, Graduate School of Medical Science & Engineering, KAIST, Dajeon, Korea, and were cared for based on guidelines established by Hanyang University, with water and food ad libitum while housed at the proper temperature, light intensity and humidity. This study was approved by the Ethics Committee of Hanyang University.
2.2. Isolation of mouse primary hepatocytes
Mouse primary hepatocytes were isolated using a two-stem collagenase perfusion method. Mouse primary hepatocyte were obtained from 6 to 8 weeks old Tg(CAG-DsRed*MST)1Nagy/J mice to easily trace the primary hepatocyte in 3D printed structures.Citation14 Briefly, the liver was perfused at 6 ml/min with solution A consisting of EDTA as a calcium chelating agent for 5 min at 37°C through the portal vein. After perfusion with solution A, the liver was perfused with solution B comprised of a trypsin inhibitor and collagenase for 8 min at 37°C. Subsequently, the extracted liver was finely cut in a dish using a surgical knife to separate the cells from the liver. Then, the liver pieces and the cells were filtered using 100 um cell strainers. The filtered mouse primary hepatocytes were washed 3 times with Williams' Medium E (Gibco, NY, USA) at 500 rpm and treated with 25% Percoll (GE Healthcare, Bucks, UK) to remove the dead cells. Primary hepatocytes thus obtained showed 80–90% viability.
2.3. Cell culture
Primary hepatocytes were cultured in Williams' Medium E supplemented with Cell Maintenance Supplements CM4000 (Life Technologies, CA, USA) at 37°C in a CO2 incubator. Mesenchymal stem cells derived from human umbilical cord blood (hUCB-MSCs) were cultured in Minimum Essential Media (GIBCO, NY, USA) supplemented with 10% Fetal Bovine Serum (GIBCO, NY, USA) and 1% penicillin/streptomycin. Mouse embryonic fibroblasts (MEFs) were isolated from an E13.5 fetal liver. MEFs were cultured in Dulbecco Modified Eagle Medium (GIBCO, NT, USA) supplemented with 10% Fetal Bovine Serum and 1% penicillin/streptomycin. The medium was changed every 2 days. MSCs were infected with a GFP expressing lentivirus to trace their behavior. Primary hepatocytes and MSCs printed in 3D architectures were cultured in Williams' Medium E supplemented with Cell Maintenance Supplements CM4000 at 37°C in a CO2 incubator.
2.4. Mesenchymal stem cells differentiation
Cells cultured for 5 passages were treated with adipogenic and osteogenic medium for 3–4 weeks with change of medium twice weekly. The adipogenic medium consisted of DMEM supplemented with 10% FBS, 2 mM insulin (Sigma), 500 mM 3-isobutyl-1-methylxanthine (Sigma), 1 mM dexamethasone (Sigma) and 200 mM indomethacin (Sigma). To evaluate differentiation, cells were fixed with 4% paraformaldehyde for 20 min at room temperature and stained with 0.5% Oil Red O in isopropanol for 30 min at room temperature. The osteogenic medium consisted of DMEM supplemented with 10% FBS, 0.1 mM dexamethasone, 10mM 3-glycerophosphate and 50 mg/ml ascorbic-2-phosphate (Sigma). Alizarin red staining was performed to confirm osteogenic differentiation.
2.5. Retrovirus generation and transduction
Retroviral particles were generated from gp-293 cells that stably expressed packaging plasmids.Citation15 In brief, cells were seeded at high confluence (>80%) into 10-cm dishes and transfected with 12 μg of each retroviral vector (GFP) plus 4-μg VSV-G plasmid using Lipofectamine 2000 (Invitrogen, California, USA). For virus infection, human mesenchymal stem cells were seeded in 6-well plates at 5 × 104 cells per well 1 day before transduction. The culture medium was changed into virus-containing supernatant, incubated for 6 hours with 8-μg polybrene, and then further cultured with fresh culture media.
2.6. FACS sorting
GFP-positive cells were re-suspended in PBS containing 10% FBS and 2 mM EDTA, and sorted on FACS AriaIII (BD Bioscience) equipped with Turbo Sort for high speed sorting. After sorting, more than 90% viability was determined for each cell population by trypan blue exclusion.
2.7. Manufacturing of 3D hepatic architectures
The 3D hepatic architectures were manufactured using a 3D bio-printing technique (Korea Institute of Machinery and Materials). The 3D printer was composed of a computer-aided system with a pressure controller, nozzle, syringe and 3-axes stage. 3% alginate and 1% calcium chloride were dissolved in Williams' Medium with the cells to fabricate cell-alginate architectures. Hydrogels consisting of cells and alginate were stacked up through the nozzle layer-by-layer. 4 × 107 primary hepatocytes and 4 × 106 hUCB-MSCs were printed in each layer. The cell-alginate hepatic architectures were 25 mm x 25 mm and composed of 5 layers. After printing, the architectures were treated with 5% calcium chloride for 1 min to induce cross-links between the alginate and calcium chloride and then washed with PBS.
2.8. qRT-PCR
Total RNA was extracted with TRIzol Reagent (Ambion, CA, USA) and chloroform. cDNAs were synthesized with 1 μg of RNA using a Transcriptor First Strand cDNA Synthesis Kit (Roche, Mannheim, Germany). Real-time PCR was conducted using 0.5 μl of each forward and reverse specific oligonucleotide primer, 10 μl of the qPCR PreMix (Dyne bio, Seongnam, Seoul), 8 μl of DW and 1 μl of cDNA. Reactions were analyzed in triplicate for each gene. Primer information is shown in . PCR consisted of 40 cycles at 95°C for 20 sec each and 60 cycles for 40 sec each.
TABLE 1. Oligonucleotide primers for real-time PCR.
2.9.Albumin secretion and Urea synthesis
Amount of Mouse Albumin secretion into the media over a 24 hr was measured with mouse albumin ELSIA kit (Bethyl Laboratory, Texas, USA). Urea synthesis produced by cultured hepatocytes was measured using Urea Assay kit (Abcam, Cambridge, USA). The experimental methods were followed according to the protocol of the companies.
2.10. Hematoxylin & eosin staining
For paraffin blocks, 3D bio-printed architectures fixed in 10% formalin were dehydrated with a series of 1-hour washes in 70%, 80%, 90%, 95% and 100% ethanol. The 3D architectures were cleared with xylene twice for one hour each. Next, the 3D alginate architectures were infiltrated with paraffin for 4 hours, and embedded. For staining, multiple 4 μm sections were cut and transferred to slides. The 3D sections were immersed in xylene for 15 minutes to deparaffinize and then rehydrated with 100% ethanol three times for 7 min each. After hydration, the sections were stained with hematoxylin for 3 min. Next, they were counterstained with eosin for 1 min and washed with running water for 5 min. Sections were subsequently immersed in 50%, 70%, 90% and 100% ethanol for 3 min each. Lastly, the samples were soaked with xylene and fastened with a mounting solution (Southern Biotech, Birmingham, UK).
2.11. Cell viability assay
Cell viability was confirmed using Live/Dead Viability/Cytotoxicity kit (Molecular Probes, Oregon, USA). The cells cultured 3D structure were washed three times with a PBS and incubated with solution consisted of 2 μm calcein AM and 4 μm EthD-1 for 15 min at 37°C. Dead cells expressed red fluorescence and live cells stained green under a fluorescence microscope.
2.12. Confocal laser scanning microscopic analysis
Cell-alginate structures were first sectioned as above, and samples were deparaffinized with xylene for 15 min. After deparaffinization, all sections were rehydrated with graded ethanol. Antigen retrieval was performed by incubating samples in 10 mM sodium citrate buffer (pH 6.0) in an autoclave. The slides were then incubated in distilled water for 5 min. Afterwards, the samples were triple stained by incubating with the following primary antibodies: anti-chicken green fluorescent protein antibody (1:200 dilution; Abcam), anti-rabbit red fluorescent protein antibody (1:200 dilution; Abcam) and anti-goat serum albumin antibody (1:200 dilution; Abcam) at 4°C overnight. After an overnight incubation, the samples were rinsed 3 times with PBS and then incubated with Hoechst 33342 (1:500 dilution; Life Technologies, Eugene, OR, USA) and the appropriate secondary antibodies labeled with Alexa Fluor 488, 546 and 633 for 1 hour at room temperature. Samples were mounted with mounting solution and examined under a confocal laser scanning microscope (TCS SP5, Leica, Heidelberg, Germany).
2.13. Statistical analysis
Quantitative data were obtained from at least three independent donors each of triplicate and presented as means ± standard error and inferential statistics (p-values). Statistical significance was evaluated using paired two-tailed Student's t-tests and is indicated by *P < 0.05, **P < 0.01, ***P < 0.001.
3. RESULTS
3.1. Fabrication of 3D printed alginate architecture consisted of mouse primary hepatocytes and human UCB-MSCs
To generate the 3D hepatic architecture, primary hepatocytes and MSCs were isolated and cultured, respectively. In a standard 2D monolayer culture system, primary hepatocytes and MSCs each have unique morphological characteristics (, ) and fluorescence (, ). Isolated hepatocytes confirmed the expression of hepatic specific genes such as Albumin, AFP, CK18 and CYP1A2 by immunostaining (, ). Also, to validate the MSCs characteristics, MSCs were differentiated into adipogenic and osteogenic linages. (, ). Therefore, these results suggest that isolated primary hepatocytes and MSCs represented their own characteristics well.
FIGURE 1. Characterization of hepatocytes and hUCB-MSCs and Fabrication of 3D printed alginate structure. (A). (a) Confirmation of hepatocyte morphology, (b) fluorescence expression in CAG-dsRed primary hepatocytes, (c) Alb (green), AFP(red), (d) CK18 (green), and CYP1A2 (red). (e) Morphology of hUCB-MSCs, (f) GFP-tagged, and (g and f) confirmation of differentiation ability into adipogenic and osteogenic linages of hMSCs. Bars, 50 μm (c-d), 100 μm (a-b, e-f), and 200 μm (g-h). (B). A schematic diagram of a 3D printed architecture composed of mouse primary hepatocytes and human MSCs. The 3D hepatic architecture manufactured to be 25 mm x 25 mm in size and composed of 5 layers. (C). Distribution of CAG-dsRed primary hepatocytes and GFP-tagged hMSCs in the 3D hepatic architecture under fluorescence microscopy at day 1. Bars, 100 μm.
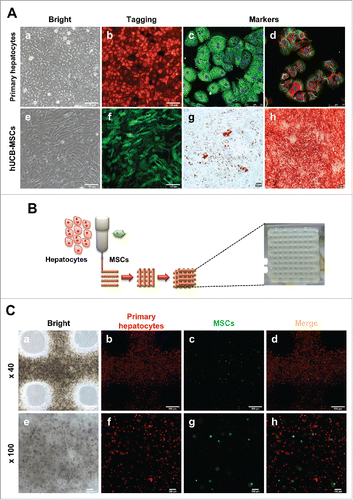
To fabricate the 3D structure, these cells were mixed with a 3% alginate solution, and printed onto a culture dish at a ratio of 10:1 primary hepatocytes:MSCs at a size of 25 mm x 25 mm for 5 layers (). These 3D hepatic architectures were incubated and analyzed for 7 days. To evaluate the behavior of the primary hepatocytes and MSCs in the 3D hepatic architecture, the fluorescence of each cell type was detected. Under a bright field, hepatocytes and MSCs showed a homogenous distribution in the alginate architecture (, ). Also, hepatocytes (red fluorescence, , ) and MSCs (green fluorescence, , ) were visualized very well under a fluorescent microscope, and confirmed that these two cell types were evenly distributed (, ). These results suggest that alginate hydrogels are adequate for generating 3D hepatic architecture, and primary hepatocytes and hUCB-MSCs are evenly distributed and survived in alginate architectures.
3.2. Maintenance of morphology in 3D alginate structures
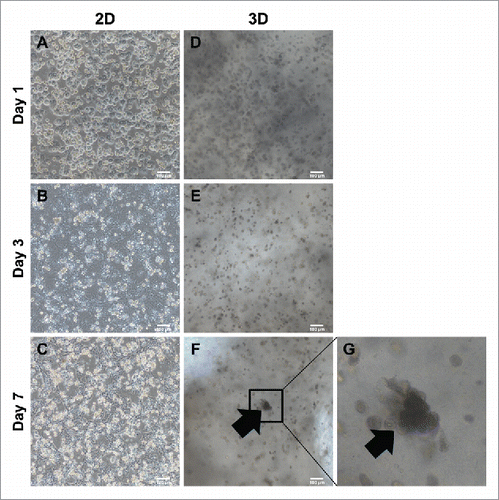
Generally, primary hepatocytes lose their function and cuboidal morphology in 2–3 days after isolation. To compare the behavior of primary hepatocytes in 2D and 3D culture systems, we cultured hepatocytes and MSC onto either a culture dish or a 3D hepatic architecture. After 3 days of isolation on a 2D culture dish, primary hepatocytes changed from an epithelial morphology () to a fibrotic morphology () and completely lost their hepatic shape after 7 days (). In contrast, primary hepatocytes were found as 13 cells within the alginate hepatic structure after 1–3 days in culture (, ). After culturing for 7 days, primary hepatocytes aggregated and expanded over time into hepatic-like organoids (, enlarged in G). These assemblages were found at various locations in the 3D hepatic architecture. This suggested that primary hepatocytes cultured in 3D architectures were able to mimic hepatic form and could survive for more than 1 week.
FIGURE 2. Comparison of morphology between 2D culture dish and 3D alginate architecture. (A). ∼ (C) Morphological changes onto 2D culture dish. Cuboidal hepatic morphology is forfeited as time goes on until day 7. (D)∼(F) Morphological changes compared between 2D. Cuboidal hepatic morphology is sustained until day 7 in 3D alginate architecture. (G) Magnification of (F). Black arrows indicate areas mimicking hepatic structure in 3D architectures. Bars, 100 μm.
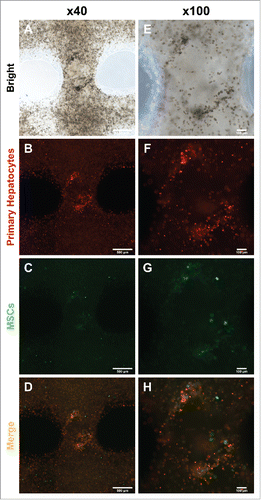
To evaluate hepatocyte assembly with MSCs, we analyzed cell types under fluorescence microscopy. After 7 days in culture, cells assembled with each other (, ). Surprisingly, hepatocyte assembly was focused in areas where MSCs were present (, ). This result showed that MSCs had the ability to support primary hepatocyte aggregation into a hepatic-like structure.
FIGURE 3. Migration and colonization of mouse primary hepatocytes with hMSCs. (A) and (E) Bright microscope image of 3D printed mouse primary hepatocytes and hMSCs. (B) ∼ (D) and (F) ∼ (H) Fluorescence image of 3D printed mouse primary hepatocytes and hMSCs. Bars, (A) ∼ (D) is 500 μm and (E) ∼ (F) is 100 μm.
3.4. Expression level of hepatocyte specific genes and proteins in the 3D architecture
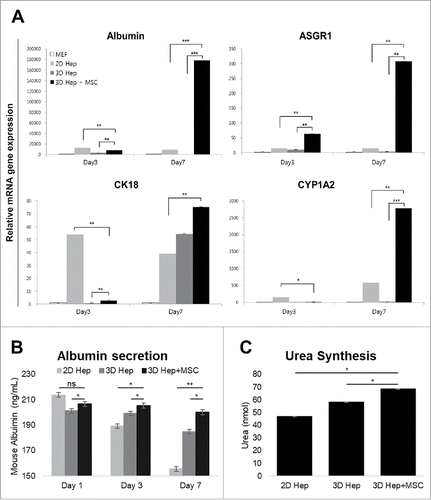
A major inquiry regarding primary hepatocytes cultured in 3D architectures was whether hepatic gene expression would be maintained. To confirm hepatic gene expression levels, we analyzed four major hepatic genes Albumin, Asialoglycoprotein receptor 1 (ASGR1), Cytokeratin 18 (CK18) and Cytochrome P450 1A2 (CYP1A2) using qRT-PCR. Expression of these genes in primary hepatocytes gradually decreased over time in 2D culture systems, as well as in 3D printed architectures without MSC co-culture. However, hepatic gene expression drastically increased on day 7 in 3D-printed architectures with MSC co-culture (). Even though mouse albumin secretion levels of 2D hepatocytes also dramatically reduced after day 7, 3D hepatocytes printed with MSCs preserved albumin secretion well (). Urea production level also higher on the 7 days in 3D hepatocyte with MSCs group than 2D hepatocyte group (). These data indicated that MSCs and the 3D culturing environment supported hepatic function in primary hepatocytes.
FIGURE 4. Comparison of hepatic functions between the 2D culture dish and 3D alginate architecture. (A). The expression of Albumin, ASGR1, CK18 and CYP1A2 genes gradually increased in primary hepatocytes cultured with hMSCs in a 3D hepatic structure. Gene expression continued for at least 7 days. MEF, mouse embryonic fibroblasts. *P < 0.05, **P < 0.01, ***P < 0.001. Data are mean ± SD. (B). Mouse Albumin secretion was measured in the cultured medium at day 1, 3, and 7. *P < 0.05, **P < 0.01. Data are mean ± SD. (C). Urea synthesis level at day 7 in hepatocytes cultured in 2D, 3D and 3D with MSCs. **P < 0.01, ***P < 0.001. Data are mean ± SD.
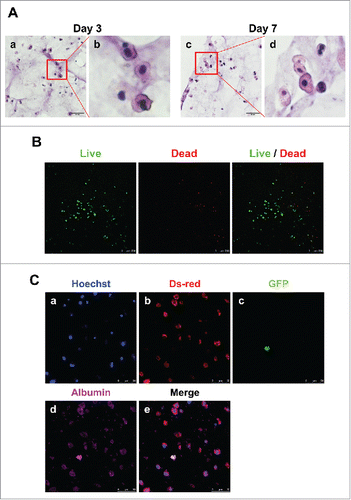
We also performed hematoxylin and eosin (H&E) staining, live/dead assay and immunofluorescence analysis to confirm the expression of hepatic proteins. Primary hepatocytes maintained their original morphology and characteristics such as one or two large euchromatic nuclei with abundant cytoplasm in the 3D architecture with MSC co-culture for 7 days (). The viability of 3D hepatic structure was confirmed to be alive even after 7 days (). Finally, we visualized their fluorescence to determine if albumin was still expressed in hepatocytes or MSCs. Albumin expression was detected in both primary hepatocytes and MSCs (). These results suggest that MSCs supported primary hepatocyte function, and 3D bio-printing allowed primary hepatocytes to retain their function and hepatic structure.
FIGURE 5. Detection of hepatocytes and MSCs in 3D architecture by H&E staining, live/dead assay and immunostaining. (A). H&E staining at day 3 (a, enlarged in b) and 7 (c, enlarged in d). Bars, 60 μm. (B). Live/Dead Viability/Cytotoxicity Assay. Live cells (Green) and Dead cells (Red). Bars, 50 μm. (C). Immunofluorescent staining for DsRed (b), GFP (c) and albumin (d) at day 7. Hoechst (blue; a) labels all nuclei. Bars, 50 μm.
4. DISCUSSION
Studies on primary hepatocytes have been actively pursued to elucidate details on the metabolism and uptake specific to these cells.Citation16 The hepatic roles of primary hepatocytes including urea synthesis, albumin secretion and expression of cytochrome P450 have been utilized primarily in drug screening studies.Citation17 However, when primary hepatocytes are detached from the liver, they rapidly lose their hepatic functions.Citation18,Citation19 Many studies have shown that culture environment conditions, media supplements, collagen gel sandwich methods and an extracellular matrix can have an effect on primary hepatocyte function when culturing.Citation20
MSCs retain the ability to differentiate into hepatocytes and to regulate inflammation via a suite of mechanisms including cell-to-cell contact. Therefore, MSCs can instantiate and support regenerative microenvironments within injured organs and tissues.Citation21-23 Umbilical cord-derived MSCs have been found to provide advantageous factors for diverse cells in co-culture including primary hepatocytes.Citation24 The typical 2D culture method used for culturing primary hepatocytes and MSCs together has a few limitations with respect to retaining hepatocyte-like cell structure and function. Primary hepatocytes have been shown to lose their function and MSCs readily lose their ability to differentiate when grown together in 2D culture.Citation25 Furthermore, there are few studies on primary hepatocytes and MSCs in 3D co-cultures.
Recently, many reports have demonstrated that 3D architecture cultures can improve culture environments compared to 2D plate cultures. The state of cells cultured in 2D systems is poor due to inadequate microenvironmental effects on their function compared to natural 3D tissues.Citation12,Citation26 To overcome the disadvantages of a 2D co-culture system, we used 3D bio-printing technology to produce a 3D culture that closely mimicked a portion of an actual liver and enabled the support of greater numbers of cells compared to a 2D culture.Citation27 Also, 3D bio-printing confers the ability to print desired cell architectures and control cell type distributions. Thus, 3D bio-printing allows for the manufacture of a realistic 3D environment that can mimic organ microenvironments, including the liver.Citation28-30
When primary hepatocytes are cultured in a tissue system, they tend to assemble with each other.Citation31 Up to 3 days after culturing, primary hepatocytes were in a state of single cells in 3D structures. However, after 7 days, primary hepatocytes progressively congregated in areas where there were functional MSCs. According to this study's genetic data, expression of hepatic genes gradually increased and primary hepatocyte cell morphology was stable. These findings indicated that 3D culture environments and MSCs allowed primary hepatocytes to assemble into a liver-like form and retain stable hepatic function.
Until now, the collagen sandwich system has been considered the best technique for maintaining primary hepatocytes. This method is superior to 3D bio-printing for the investigation of morphologic alterations; however, the maintenance of hepatic function is similar in both of these culture methods.
In conclusion, we produced a liver-mimicking architecture with 3D bio-printing using primary hepatocytes, and demonstrated the benefits of 3D bio-printing with co-cultured primary hepatocytes and MSCs. This technology has immense potential for tissue engineering and will contribute significantly to toxicity and drug screening studies.
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
No potential conflicts of interest were disclosed.
Additional information
Funding
REFERENCES
- Yu SJ. A concise review of updated guidelines regarding the management of hepatocellular carcinoma around the world: 2010–2016. Clin Mol Hepatol. 2016;22:7–17. doi:10.3350/cmh.2016.22.1.7. PMID:27044761.
- Keeffe EB. Liver transplantation: current status and novel approaches to liver replacement. Gastroenterology. 2001;120:749–62. doi:10.1053/gast.2001.22583. PMID:11179248.
- Vinken M, Maes M, Oliveira AG, Cogliati B, Marques PE, Menezes GB, Dagli ML, Vanhaecke T, Rogiers V. Primary hepatocytes and their cultures in liver apoptosis research. Arch Toxicol. 2014;88:199–212. doi:10.1007/s00204-013-1123-4. PMID:24013573.
- Sgodda M, Aurich H, Kleist S, Aurich I, Konig S, Dollinger MM, Fleig WE, Christ B. Hepatocyte differentiation of mesenchymal stem cells from rat peritoneal adipose tissue in vitro and in vivo. Exp Cell Res. 2007;313:2875–86. doi:10.1016/j.yexcr.2007.05.020. PMID:17574236.
- Medicetty S, Bledsoe AR, Fahrenholtz CB, Troyer D, Weiss ML. Transplantation of pig stem cells into rat brain: proliferation during the first 8 weeks. Exp Neurol. 2004;190:32–41. doi:10.1016/j.expneurol.2004.06.023. PMID:15473978.
- Wu XB, Tao R. Hepatocyte differentiation of mesenchymal stem cells. Hepatobiliary Pancreat Dis Int. 2012;11:360–71. doi:10.1016/S1499-3872(12)60193-3. PMID:22893462.
- Liu M, Yang J, Hu W, Zhang S, Wang Y. Superior performance of co-cultured mesenchymal stem cells and hepatocytes in poly(lactic acid-glycolic acid) scaffolds for the treatment of acute liver failure. Biomed Mater. 2016;11:015008. doi:10.1088/1748-6041/11/1/015008. PMID:26836957.
- Symes MD, Kitson PJ, Yan J, Richmond CJ, Cooper GJ, Bowman RW, Vilbrandt T, Cronin L. Integrated 3D-printed reactionware for chemical synthesis and analysis. Nat Chem. 2012;4:349–54. doi:10.1038/nchem.1313. PMID:22522253.
- Shimizu TS, Le Novere N, Levin MD, Beavil AJ, Sutton BJ, Bray D. Molecular model of a lattice of signalling proteins involved in bacterial chemotaxis. Nat Cell Biol. 2000;2:792–6. doi:10.1038/35041030. PMID:11056533.
- Bailey MJ, Schulten K, Johnson JE. The use of solid physical models for the study of macromolecular assembly. Curr Opin Struct Biol. 1998;8:202–8. doi:10.1016/S0959-440X(98)80039-0. PMID:9631294.
- Jones N. Science in three dimensions: the print revolution. Nature. 2012;487:22–3. doi:10.1038/487022a. PMID:22763531.
- Murphy SV, Atala A. 3D bioprinting of tissues and organs. Nat Biotechnol. 2014;32:773–85. doi:10.1038/nbt.2958. PMID:25093879.
- Chia HN, Wu BM. Recent advances in 3D printing of biomaterials. J Biol Eng. 2015;9:4. doi:10.1186/s13036-015-0001-4. PMID:25866560.
- Klaunig JE, Goldblatt PJ, Hinton DE, Lipsky MM, Chacko J, Trump BF. Mouse liver cell culture. I. Hepatocyte isolation. In Vitro. 1981;017:913–25. doi:10.1007/BF02618288.
- Kim HJ, Jeong J, Park S, Jin YW, Lee SS, Lee SB, Choi D. Establishment of hepatocellular cancer induced pluripotent stem cells using a reprogramming technique. Gut Liver. 2017;11:261–9. doi:10.5009/gnl15389. PMID:27728962.
- Swift B, Brouwer KL. Influence of seeding density and extracellular matrix on bile Acid transport and mrp4 expression in sandwich-cultured mouse hepatocytes. Mol Pharm. 2010;7:491–500. doi:10.1021/mp900227a. PMID:19968322.
- Mathijs K, Kienhuis AS, Brauers KJ, Jennen DG, Lahoz A, Kleinjans JC, van Delft JH. Assessing the metabolic competence of sandwich-cultured mouse primary hepatocytes. Drug Metab Dispos. 2009;37:1305–11. doi:10.1124/dmd.108.025775. PMID:19251822.
- Boess F, Kamber M, Romer S, Gasser R, Muller D, Albertini S, Suter L. Gene expression in two hepatic cell lines, cultured primary hepatocytes, and liver slices compared to the in vivo liver gene expression in rats: possible implications for toxicogenomics use of in vitro systems. Toxicol Sci. 2003;73:386–402. doi:10.1093/toxsci/kfg064. PMID:12657743.
- Rodriguez-Antona C, Donato MT, Boobis A, Edwards RJ, Watts PS, Castell JV, Gomez-Lechon MJ. Cytochrome P450 expression in human hepatocytes and hepatoma cell lines: molecular mechanisms that determine lower expression in cultured cells. Xenobiotica. 2002;32:505–20. doi:10.1080/00498250210128675. PMID:12160483.
- Choi HJ, Choi D. Successful mouse hepatocyte culture with sandwich collagen gel formation. J Korean Surg Soc. 2013;84:202–8. doi:10.4174/jkss.2013.84.4.202. PMID:23577314.
- Jiang Y, Jahagirdar BN, Reinhardt RL, Schwartz RE, Keene CD, Ortiz-Gonzalez XR, Reyes M, Lenvik T, Lund T, Blackstad M, et al. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature. 2002;418:41–9. doi:10.1038/nature00870. PMID:12077603.
- Yagi H, Soto-Gutierrez A, Navarro-Alvarez N, Nahmias Y, Goldwasser Y, Kitaga`wa Y, Tilles AW, Tompkins RG, Parekkadan B, Yarmush ML. Reactive bone marrow stromal cells attenuate systemic inflammation via sTNFR1. Mol Ther. 2010;18:1857–64. doi:10.1038/mt.2010.155. PMID:20664529.
- Yagi H, Soto-Gutierrez A, Parekkadan B, Kitagawa Y, Tompkins RG, Kobayashi N, Yarmush ML. Mesenchymal stem cells: Mechanisms of immunomodulation and homing. Cell Transplant. 2010;19:667–79. doi:10.3727/096368910X508762. PMID:20525442.
- Arutyunyan I, Elchaninov A, Makarov A, Fatkhudinov T. Umbilical Cord as prospective source for mesenchymal stem cell-based therapy. Stem Cells Int. 2016;2016:6901286. doi:10.1155/2016/6901286. PMID:27651799.
- Cai J, Zhao Y, Liu Y, Ye F, Song Z, Qin H, Meng S, Chen Y, Zhou R, Song X, et al. Directed differentiation of human embryonic stem cells into functional hepatic cells. Hepatology. 2007;45:1229–39. doi:10.1002/hep.21582. PMID:17464996.
- Boland T, Mironov V, Gutowska A, Roth EA, Markwald RR. Cell and organ printing 2: fusion of cell aggregates in three-dimensional gels. Anat Rec A Discov Mol Cell Evol Biol. 2003;272:497–502. doi:10.1002/ar.a.10059. PMID:12740943.
- Ji R, Zhang N, You N, Li Q, Liu W, Jiang N, Liu J, Zhang H, Wang D, Tao K, et al. The differentiation of MSCs into functional hepatocyte-like cells in a liver biomatrix scaffold and their transplantation into liver-fibrotic mice. Biomaterials. 2012;33:8995–9008. doi:10.1016/j.biomaterials.2012.08.058. PMID:22985996.
- Jakab K, Norotte C, Marga F, Murphy K, Vunjak-Novakovic G, Forgacs G. Tissue engineering by self-assembly and bio-printing of living cells. Biofabrication. 2010;2:022001. doi:10.1088/1758-5082/2/2/022001. PMID:20811127.
- Marga F, Jakab K, Khatiwala C, Shepherd B, Dorfman S, Hubbard B, Colbert S, Gabor F. Toward engineering functional organ modules by additive manufacturing. Biofabrication. 2012;4:022001. doi:10.1088/1758-5082/4/2/022001. PMID:22406433.
- Kim Y, Kang K, Jeong J, Paik SS, Kim JS, Park SA, Kim WD, Park J, Choi D. Three-dimensional (3D) printing of mouse primary hepatocytes to generate 3D hepatic structure. Annals Surg Treatment Res. 2017;92:67–72. doi:10.4174/astr.2017.92.2.67.
- Arterburn LM, Zurlo J, Yager JD, Overton RM, Heifetz AH. A morphological study of differentiated hepatocytes in vitro. Hepatology. 1995;22:175–87. PMID:7601410.
