ABSTRACT
The increasing demand for organs for transplantation necessitates the development of substitutes to meet the structural and physiological functions. Tissue decellularization and recellularization aids in retaining the three-dimensional integrity, biochemical composition, tissue ultra-structure, and mechanical behavior, which makes them functionally suitable for organ transplantation. Herein, we attempted to rebuild functional liver grafts in small animal model (Wistar rat) with a potential of translation. A soft approach was adopted using 0.1% SDS (Sodium Dodecyl Sulfate) for decellularization and primary hepatocytes were used as a potential cell source for recellularization. The decellularization process was evaluated and confirmed using histology, DNA content, ultra-structure analysis. The resultant scaffold was re-seeded with the rat hepatocytes and their biocompatibility was assessed by its metabolic functions and gene expression. The structural components of the Extracellular matrix (ECM) (Laminins, Collagen type I, Reticulins) were conserved and the liver cell-specific proteins like CK-18, alpha-fetoprotein, albumin were expressed in the recellularized scaffold. The functionality and metabolic activity of the repopulated scaffold were evident from the albumin and urea production. Expression of Cytokeratin-19 (CK-19), Glucose 6-Phosphatase (G6P), Albumin, Gamma Glutamyl Transferase (GGT) genes has distinctly confirmed the translational signals after the repopulation process. Our study clearly elucidates that the native extracellular matrix of rat liver can be utilized as a scaffold for effective recellularization for whole organ regeneration.
Introduction
Solid organ regeneration is an emerging area in tissue engineering, providing tissue constructs with functional remodeling, and potential to re-integrate into host systems.Citation1 Recent tissue engineering approaches provide biological scaffolds composed of extracellular matrix (ECM) controlling cell functions and promoting new tissue formation. Biomaterials were employed as scaffolds for the incorporation of required cell types which intrinsically modulate the tissue microenvironment. Various natural biopolymers have been used as scaffolds to mimic the extra cellular matrix of the tissues in vitro. Our previous study exhibited that these biopolymer-based scaffolds in a three-dimensional (3D) culture systems conserve the cell phenotype and support their proliferation.Citation2,Citation3 The biological signaling for cell adhesion and proliferation is constantly regulated by the scaffold proteins which are being modulated based on the metabolic and functional requirements of the healthy tissues and organ.Citation4 Intact three-dimensional extracellular matrices from different allogeneic and xenogeneic tissue sources are known to preserve the cellular phenotypes with the help of obligatory ligands and bioactive molecules during the process of tissue regeneration.Citation5,Citation6 Further, the natural ECM of the tissues is advantageous over conventional two-dimensional and three-dimensional culture systems, as they maintain and retain the complex microstructure and functional proteins.Citation7
The current focus is on the use of native biological scaffolds as a transplantable organ which can be employed for clinical use. In this context, numerous techniques for decellularization have been developed for various tissues/organs. This helps to remove cellular and nuclear content, along with the immunogenic antigens, while preserving the ECM architecture and structural components, such as collagen, elastin networks along with the biochemical constituents.Citation8
There are a variety of approaches such as chemical, enzymatic, and physical methods for effective decellularization of whole organs.Citation9 However, other bioactive ligands such as integrins, cadherins, are also important for cell–cell interaction, cell–ECM interaction, and new ECM formation.Citation10 The tissue ultra-structure, matrix density, and architecture vary among different types of organs and hence play a major role in the utilization of decellularization techniques. In case of a complex and dense organ such as liver, decellularization is usually achieved through the use of the hepatic vascular network and dynamic culture systems which can support the delicate vasculature of the organ during processing.Citation11 Completely decellularized organs or tissues can eradicate antagonistic immune response elicited by cell membrane epitopes, allogeneic or xenogeneic DNA, and damage-associated molecular pattern molecules.Citation12
The process of decellularization and recellularization for tissues and organs regeneration has emerged as a remarkably promising approach and several attempts are made to mimic the in vivo conditions, using perfusion techniques.Citation13 The progression of organ development technology involves appropriate tissue niche or microenvironment and well-integrated and vascularized extracellular matrices. The recellularization of the native ECM can be done by various cell types like stem cells, iPSCs, etc. However, freshly isolated primary hepatocytes (epithelial cells) were utilized to investigate the various aspects of liver recellularization and metabolism.Citation14
The hepatocytes comprise 80% of all the liver cells and they have a fundamental role in liver function. The recellularization strategies are being improved by finding clinically relevant cell type from a renewable tissue source and a competent cell seeding method.Citation15,Citation16 Thus, we optimized in vitro methods to reconstitute the acellular liver scaffold, with cell populations that distinctively re-integrates within the biomimetic ECM to generate functional hepatic tissue with a potential treatment approach for chronic liver diseases.
The density of the liver along with the fragility of hepatic ECM necessitates the use of mild detergents for quick decellularization for effective cell removal. We tried to optimize and simplify the technique of decellularization in order to make it more economical with minimal damage to liver ECM. The native biological liver scaffold facilitates cues for migration, adhesion, and differentiation of liver epithelial cells for their development and hepatic functioning governed by cell-ECM interactions. Thus, we utilized portal vein (PV) as a cell seeding route and primary rat hepatocytes as a cell source for recellularization. We also assessed the stability, integrity of ECM components and metabolic functionalities of the recellularized liver grafts.
Materials and method
SDS and Triton X-100 were purchased from Sigma-Aldrich, St. Louis, MO, USA. The required concentrations of SDS and Triton X-100 were made in autoclaved distilled water. Collagenase and dispase were procured from Gibco, Life technologies, USA. The required antibodies were procured from Abcam (Cambridge, United Kingdom) and Novus Biologicals (Colorado, United States).
Animals
Adult Wistar rats weighing 100–150 g were used for the development of decellularized liver scaffolds. Pathogen-free rats were maintained under controlled environmental conditions with free access to food and water.
Rat liver decellularization by detergent-based perfusion method
Wistar rats (n = 5) were anesthetized based on IACUC (Institutional Animal Care and Use Committee) guidelines for anesthesia using ketamine (100 mg/kg) and xylazine (10 mg/kg) per body weight using intra-peritoneal route, followed by topical skin disinfection. All the animals received necessary care based on criterions by Guide for the care and use of laboratory animals. A transverse incision was made in the abdomen, which exposed the portal vein, the bile duct, and the inferior vena cava. The portal vein was successfully cannulated with a 24-gauge (BD, Biosciences, USA) needle and connected to the infusion pump (Infusomat®P B Braun). The liver was initially perfused with 1X PBS for an hour then continuously perfused with 0.1% SDS in distilled water for 3 hr at a flow rate of 5 mL/m. Subsequently, the liver was washed with distilled water for 15 min, followed with 0.1% Triton X-100 for 30 min to remove other bound nucleic acids. The completely decellularized liver was washed extensively and preserved in PBS containing penicillin (100 IU/mL), streptomycin (100 IU/mL), gentamycin (50 IU/mL), amphotericin B (2.5 μg/mL), at 4°C until further use.
Quantification of DNA and collagen of decellularized liver matrix
Decellularized liver matrices and fresh rat liver tissues were used for the quantification of total DNA. Briefly, total DNA was isolated from equal volumes of decellularized and fresh liver tissues using a commercially available kit (DNeasy Blood & Tissue Kit, QIAGEN). The DNA concentration was estimated at 260 nm using a NanoDrop spectrophotometer (ND-2000 c; Thermo, USA). (n = 5)
The collagen content in the native tissue and decellularized tissue was quantified using Sirius red colorimetric assay. Briefly, tissues were homogenized using a tissue homogenizer and the homogenate was centrifuged at 12,000 x g for 30 min at 4°C. The pellet was dried using a vacuum concentrator and dissolved in 1X PBS and plated for further estimation. The collagen standards ranging from 15−90 µg were made as a serial dilution in PBS and added to each well in triplicates. 65 µg Sirius red stain (Direct Red 80, Sigma) in saturated picric acid was added and incubated for 30 min at 37°C on a rocker. The plates were spun at 10,000x g for 30 min at 4°C. The pellet was dissolved in 0.1 N NaOH for 30 min and read on a spectrophotometer at 530 nm absorbance. (n = 5)
Isolation of primary rat hepatocytes
Primary adult hepatocytes were isolated from Wistar rats weighing 100–150 gm, using a modified collagenase and dispase perfusion technique. Rats were anesthetized using ketamine (100 mg/kg) and xylazine (10 mg/kg) per body weight intraperitoneally, followed by cannulation. The liver was initially perfused with 1X PBS solution, followed by the perfusion with pre-warmed collagenase type IV (0.05%) and dispase (0.3%) combined solution at a rate of 300 ml/hr. After the liver turns mushy it was excised and minced gently in cold William’s E media (Gibco, USA). The suspension was filtered through 70 μm cell strainer (BD Biosciences, USA) and centrifuged at 50 × g for 5 min. The cell pellet was re-suspended in complete William’s E media and viability was assessed using trypan blue.
Reseeding of the decellularized liver scaffold
The decellularized liver matrix was washed with PBS and antibiotics and perfused using complete William’s E media supplemented with 10% FBS and 0.0036 μg/mL insulin (sigma, USA), 10 ng/mL epidermal growth factor (Sigma-aldrich, USA), penicillin (100 IU/mL), streptomycin (100 IU/mL), gentamycin (50 IU/mL) through the portal vein prior to recellularization. A total of 60 million cells was resuspended in 20 mL of culture medium and perfused into the scaffold slowly at a flow rate of 1 mL/min using an infusion pump (Infusomat®P B Braun). The injected cells were retained in the liver scaffold as a static culture and then complete media was perfused into the scaffold continuously using the perfusion pump at a flow rate of 0.5 mL/min. The entire perfusion system was placed in 5% CO2 incubator at 37°C. The perfusate was collected and replaced with the fresh media every day for 14 days.
Characterization of the decellularized and recellularized liver matrix
Histological staining
Normal fresh liver, decellularized liver matrix, and recellularized liver scaffolds were fixed in 10% neutral buffered formalin at room temperature for 24 h (n = 5). The paraffin fixed tissue samples were sectioned into 5 μm sections followed by staining with Hematoxylin and Eosin (H&E) and Masson’s trichrome (MT) as per standard staining protocols.
SEM analysis
Scanning electron microscopy (SEM) was used to observe the microstructure of the ECM in the decellularized liver and recellularized as well as the fresh rat livers (n = 5). Samples were fixed in Karnowsky’s fixative (pH 7.2) for 24 h at 4°C. Following washing with 0.1 M phosphate buffer, samples were dehydrated in a graded ethanol-water series up to 100% ethanol. Specimens were dried at a critical point using CO2 and images were recorded with a scanning electron microscope (S-3400N, HITACHI).
Immunostaining
The ECM components in the decellularized matrix were determined by fluorescent staining for the presence of collagen type I, laminin and reticulins. Recellularized scaffolds were characterized for the presence of hepatocyte-specific proteins like CK-18, Alpha-fetoprotein (AFP), Glucose 6 phosphate dehydrogenase (G6PD) and Albumin. Briefly, the liver scaffolds were fixed in 4% formaldehyde solution and embedded in paraffin. The paraffin blocks were sectioned at 5 μm thickness. Slides were rehydrated, washed with 1X PBS and blocked using Bovine Serum Albumin (BSA) for 30 min at 37°C. Decellularized tissue sections and sections from fresh rat liver tissues were incubated with primary antibodies such as collagen type I (dilution 1:200), laminin (dilution 1:200), and reticulins (dilution 1:100) overnight at 4°C.
Recellularized scaffold sections were incubated with primary antibodies G6PD (2 μg/mL concentration), albumin (dilution 1:500), AFP (20 μg/mL concentration). After washing in PBS, slides were incubated with secondary goat anti-rabbit Alexa 568 and Alexa 488 for 1 hr at 37°C. Cytokeratin-18 antibody (CK-18) (dilution 1:200) was already tagged with Alexa 488 fluorochrome and thus, recellularized liver sections were incubated for 1 h at 37°C. Later, all the tissue sections were mounted using mounting medium (Vectashield containing DAPI) (4’, 6’-diamidino-2-phenylindole dihydrochloride) (Abcam, Cambridge, UK) and incubated for 2–3 min at 37°C in dark. The imaging was performed using a confocal laser microscope (Leica TCS, CLSM) and the images were processed using Leica software. (n = 5)
Assessment of recellularized liver function
The culture medium of the continuously perfused recellularized engrafted liver (3D) culture system and the 2D culture system were collected periodically. Albumin and urea content in the culture media were measured using the Agilent bioanalyzer (Agilent Technologies, USA) according to the manufacturer’s instructions.
Quantitative reverse transcription PCR
Total RNA was extracted using a TRIZOL method and 100–200 ng of total RNA was reverse transcribed into cDNA using ProtoScript First strand cDNA conversion kit (New England Biolabs, France) as per manufacturers’ instructions. Primers for the amplification of albumin, G6P, CK19, GGT, and actin-beta (Act B) were generated as shown in . Quantitative RT-PCR assay was performed using SYBR-green PCR Master Mix (Applied Biosystems, USA) on the StepOneTM Real-Time PCR system. Each experiment was performed in triplicate, and the gene expression levels were normalized to the levels of Act B.
Table 1. List of primers.
Statistical analysis
All data were analyzed using SPSS statistical software and using Student’s t-test. All values and graphs represented as mean ± standard deviation (SD). The error bars represent the standard error of the mean. Statistical significance was defined as p ≤ 0.05.
Results
Decellularized liver scaffolds
The cannulated liver gradually turned light brown color to white and translucent with the removal of cellular material upon perfusion with 0.1% SDS, an anionic detergent that lyses cells and solubilizes cytoplasmic components and membrane lipid. 0.1% Triton X-100, a nonionic surfactant for the recovery of membrane components under mild non-denaturing conditions assisted the removal of bound nucleic acids as manifested from the visible vasculature of slender veins. The microvascular networks and blood vessels were preserved in the decellularized matrix as evident from .
Figure 1. Decellularized whole rat liver. (a) Schematic representation of the decellularization process the native rat liver turns light brown after 1X PBS perfusion and gradually turns white and then translucent with 0.1% SDS after 4 h. The complete decellularization was evident from the appearance of visible vascular networks and clear veins. (b) Hematoxylin and Eosin staining of native rat liver and decellularized rat liver showing presence and absence of nucleic material (blue), respectively. Masson’s trichrome staining of native rat liver showing the presence of collagen (blue), pink cytoplasm while the decellularized rat liver shows the presence of abundant collagen but absence of cytoplasm and nuclei, respectively. The arrows indicating the presence of blood vessels. SEM images of native rat liver tissue show the presence of populated hepatocytes throughout the liver matrix. Decellularized rat liver matrix without the cellular content along with networks of ECM fibers with lamellar structures was observed.
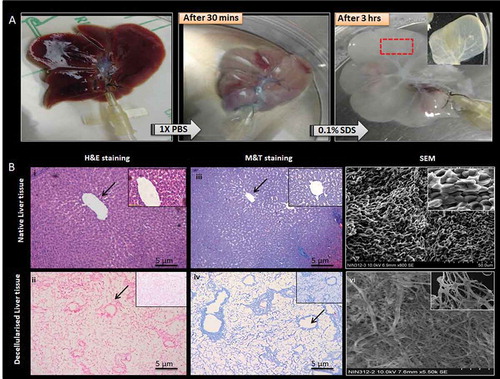
The H&E staining of the resultant liver scaffold showed no residual cells. Whereas the Masson’s trichrome staining showed the presence of collagen fiber maintained as tubular structures in the liver ECM. However, no cellular staining was seen in the liver matrix. The normal fresh rat liver was used as a control and the cellular components were stained by H&E and MT staining along with the blue color which stained conserved collagen matrix. The decellularization was also confirmed by SEM where no cells were observed within the decellularized liver matrix. The extracellular matrix showed networks of collagen fibers with lamellar structures with intact large and small vessels. The matrix had arrangements of connective tissue fibers clearly defining parenchymal-free spaces. The rat liver samples without treatment retained their extracellular matrix with fibers arranged in honeycomb-like pattern with proper distribution of the cellular components and hepatocytes ().
Evaluation of ECM protein components of decellularized liver
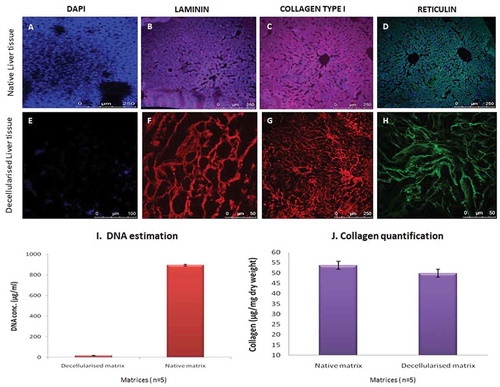
These matrix proteins were preserved as indicated by structural and basement membrane components of the ECM in the decellularized liver scaffolds. More specifically, laminin was predominantly found in the basement membrane of larger blood vessels and the liver matrix whereas, collagen type I was preserved in the sinusoids and the portal ducts. The reticulin staining demonstrated the presence of reticular fibers in the parenchymal matrix of the decellularized tissues without any nuclear component (no DAPI staining). The native fresh rat liver also demonstrated similar expression of extracellular matrix proteins along with the preserved nucleus as shown by DAPI staining ().
Figure 2. Determination of ECM and cellular components. Specific ECM proteins and the cellular components of the acellular liver bio-scaffold comparison with fresh rat liver tissue were studied. (a-d) Immunostaining results of the fresh liver showed collagen type I, laminins and reticulins mostly around larger vessels and also throughout the parenchymal spaces consistent with their localization of nuclear staining with DAPI. (e-h) similarly, the ECM proteins (Collagen type I and laminins) were observed around vascular basal membrane and parenchymal areas of the acellular liver bioscaffold. Reticulin staining demonstrated the presence of reticular fibers in the parenchymal matrix of the decellularized tissues. However, the nuclear component was not visible in the decellularized liver matrix upon staining with DAPI. (i) The residual DNA content decreased significantly in the decellularized rat livers (11 ± 4 µg/ml) as compared to normal rat livers (980 ± 18 µg/ml) measured per mg wet tissue weight (n = 5). (j) Collagen content in the native liver tissue was 53 ± 7 μg/mg wet tissue whereas, the collagen content of the decellularized matrix decreased to 49 ± 6 μg/mg wet tissue yet indicating that the fibrillary collagen of the native liver was retained after decellularization process (n = 5).
DNA and ECM collagen content
The residual DNA content was significantly decreased in the decellularized matrix as compared to normal untreated rat liver tissues, 11 ± 4 µg/mL and 980 ± 18 µg/mL, respectively. DNA quantification also confirmed the decellularization process indicating the absence of the nuclear material in the scaffold as compared to the fresh liver tissue ().
The collagen content was estimated in the liver matrix after decellularization. The standards were fit to a 4-parameter curve, R2 = 0.99 (n = 5). However, collagen content in the decellularized matrix slightly decreased when compared to the native tissue, i.e.,, 49 ± 6 μg/mg wet tissue and 53 ± 7 μg/mg wet tissues, respectively, as shown in . This indicates that the enzymatic decellularization processes maintained the original native tissue structure and components to a major extent without altering the extracellular matrix component.
Re-seeding liver matrix with adult hepatocytes
Primary rat hepatocytes were successfully isolated by one step collagenase and dispase perfusion method. An average yield of 60–70 × 106 cells/mL was obtained and >85% cell viability was confirmed by trypan blue. The adult freshly isolated hepatocytes were infused through the portal vein and the cells were aggregated in the liver graft gradually in order to prevent the shear stress and minimize the mechanical damage to engrafted cells. The decellularized liver scaffold was initially primed with culture medium aiding homogenous distribution and proliferation of the hepatocytes in the liver scaffold ().
Figure 3. Re-population and characterization of rat liver scaffold. (a) The well-preserved liver scaffold was initially infused with the hepatocyte culture medium. Freshly isolated primary rat hepatocytes were infused into the scaffold through the PV of the decellularized liver. The decellularized liver was fully populated with hepatocytes and continuously perfused with the culture medium for the proliferation and functioning of the engrafted cells. (b) The H&E and MT staining of the reseeded liver graft showed uniform distribution of the hepatocytes throughout the liver section. The hepatocytes were repopulated around the vessels and the surrounding parenchymal spaces throughout ECM as evident from SEM analysis. The arrow indicating the group of hepatocytes in the scaffold. (c)The liver-specific proteins like albumin, CK 18, G6P, and AFP expression were observed (green fluorescence) on the cell surfaces and the nucleus was stained with DAPI (blue) of repopulated liver section. (d) Staining of the native rat liver as positive control and staining of the nucleus with DAPI as negative control without primary antibody.
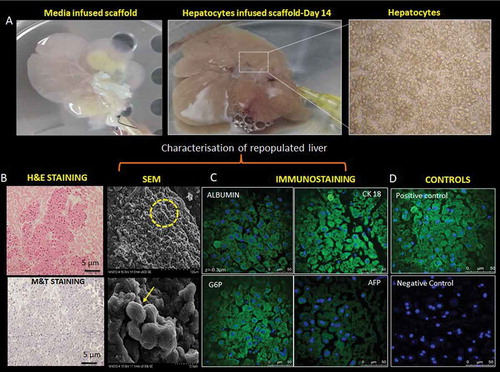
The histological evaluation by H&E and MT staining revealed that the majority of cells were distributed throughout the scaffold. Furthermore, engrafted cells were distributed around central veins, which indicated that hepatocytes entered the ECM near the portal area of the parenchyma and beyond. Subsequently, these cells reached the peri-central area across the lobular honey-combed ECM as evident from the SEM analysis. SEM images of the reseeded liver graft after 14 days of perfusion culture displayed that hepatocytes were engrafted around the vessels and repopulated the surrounding parenchymal area ().
Immunohistochemical staining of recellularized liver sections confirmed the localization of the hepatocytes, throughout the liver ECM. The liver epithelial cells, i.e., hepatocytes seeded through the PV were well engrafted within the liver scaffold which was well evident with the expression of liver-specific proteins like albumin, CK-18, GG6P, and AFP along with the nucleus stained with DAPI (). The positive and negative controls for the experiment are depicted in .
Biocompatible recellularized 3D Liver graft
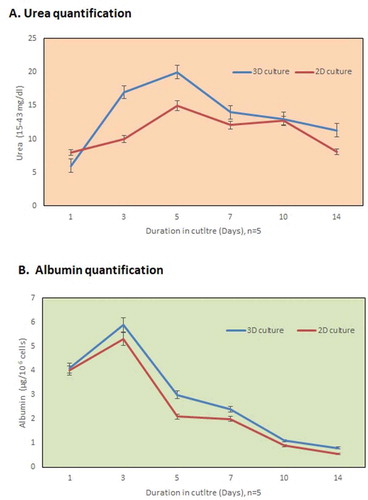
The functionality of the infused hepatic cells was analyzed by quantification of albumin and urea. The cumulative urea production increased remarkably after 5 days of culture, i.e.,, 20 ± 0.45 mg/106 cells and albumin synthesis improved to 6 ± 0.23 μg/106 cells after 2 days as shown in , respectively. As compared to the 2D culture system, the increase can be attributed to the proliferation of the hepatocytes within the engrafted liver matrix (3D culture system). (n = 5)
Figure 4. In vitro functional evaluation. (a) The urea synthesis by the recellularized liver graft increased continuously in the culture medium from day 3 to day 7 as compared to the 2D culture system. (b) The metabolism of the hepatocytes was well evident from the higher albumin production after 2 days (n = 5) in the 3D-engrafted culture system.
Gene expression of the recellularized liver
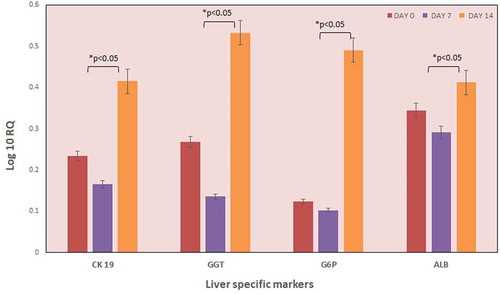
To characterize the kinetics of the changes in gene expression occurring as a consequence of hepatocyte infusion in the decellularized liver scaffold, we performed quantitative RT-PCR of well-known liver-specific genes, namely, CK-19, GG6P, Albumin, GGT, and Act B as house-keeping gene at day 0, 7 and 14. Gene expression has distinctly confirmed the translational signals after the repopulation process. Expression of CK-19, GGT, and G6P increased approximately 2-4-fold after 14 days of culture. Progressive increase of hepatocyte markers like albumin and CK19 during the culture period indicated appropriate repopulation of decellularized liver graft as demonstrated in . All the liver-specific genes have shown an increase in their expression heading toward the development of a recellularized liver graft. (n = 5)
Figure 5. Expression of liver-specific genes. The expression of liver-specific genes of the recellularized liver graft was evaluated by qPCR analysis (n = 5). The graph indicates the expression levels of various liver-specific genes on different days. The data revealed increased levels of ALB, CK 19, GGT, and G6P genes by day 10. However, GGT and G6P gene expression were significantly higher after 10 days culture as compared to the preliminary expression levels. Data is expressed as mean ± SD with significance at p < .05.
Discussion
Whole organ decellularization techniques have emerged as a new therapeutic strategy for organ regeneration by generating natural tissue matrices. The tissue architecture and composition along with the ECM regulates the maintenance of cell phenotypes and differentiation into a specific tissue. Nevertheless, the composition of ECM can get altered following various decellularization methods, which results in the amendment of cell proliferation and differentiation. Among the various decellularization processes being studied widely, continuous vascular perfusion is a technique that is intended to preserve the three-dimensional architecture of an organ while eliminating parenchymal cell population.Citation17 Hence, we present the perfusion through portal vein as an efficient method in the delivery of decellularizing agents into the tissue owing to the close proximity of the vascular network. The mild concentration of SDS (0.1%) has been standardized for continuous perfusion, considering the complex liver density with abundant cellular enzymes and delicate hepatic ECM. The decellularization process was aimed for a lesser duration with intent of maintaining the integrity and preserving the niche for subsequent recellularization process.
Our results confirms the study published by Pan et al.Citation18 where the rat liver was decellularized using 0.1% SDS through the portal vein and hepatic artery. SEM images also confirmed complete decellularization of the liver tissue with compact three-dimensional architecture, lamellar structures, and intact large and small vessels as reported by Xu et al.Citation19 The ECM components, including collagens, are highly conserved between species and do not cause obvious immune responses and these needs to be retained in the process of whole organ decellularization in order to prevent the collapse of the internal porous structure and flexible deformation.Citation20,Citation21 The mild treatment with SDS preserved the collagen of the liver matrix which is important for the maintenance of native physiological and structural tissue compliance. Thus, equilibrium was attained between the removal of the cellular components and the vascular integrity of the resulted 3D liver scaffold.
The experimental data reported preserved matrix components (laminin, collagen type I, reticulins) of the decellularized liver which are required for the maintenance of tissue microenvironment. A previous study by Yeung et al.Citation22 was found similar to our results on the effect of SDS on extracellular matrix components of tissues.
The preserved structural components of liver ECM can support the remodeling of functional hepatic tissue with appropriate cell types. The hepatocytes are well known to perform most of the liver functions in conjugation with several other cell types for body metabolism and survival. Hepatocytes tend to lose their phenotype in 2D cultures but retain their characteristics in mechanically and chemically conserved 3D microenvironment.Citation23 3D culture systems provide an appropriate niche for cell-cell and cell-extracellular environment interactions as compared to 2D systems. The cells tend to change their morphology and mode of divisions, leading to loss of diverse phenotype and polarity in 2D cultures following our previous study on the encapsulation of chondrocytes within alginate hydrogel.Citation2 Thus, we studied an established therapeutic approach toward 3D recellularization process to achieve a functional biological liver graft.
Further, we introduced adult hepatocytes through portal vein perfusion into acellular liver matrix to assess their successful migration and re-population within liver parenchyma. The perfusion via portal vein allows access to the hepatic vessels enabling improved repopulation of vasculature throughout the scaffold. Our study suggests that rat liver scaffold provided the appropriate spatial distribution of cells for homing in the presence of oxygen and nutrient gradients. The continuous medium perfusion provides large interaction of proliferating hepatocytes with the ECM aiding in effective removal of metabolic waste.Citation24 The cells injected into the acellular rat liver scaffold expressed typical hepatic, endothelial, and biliary epithelial markers, comparable to the results by Griffith et al.Citation25 and previous studies on the differentiation of human liver stem cells into matured hepatocytes in acellular scaffolds.Citation26 We also observed the presence of hepatocytes in the parenchymal spaces outside the microvasculature as reported by Basak et al.Citation27 which was attributed to loss of sinusoidal integrity in the absence of the liver sinusoidal endothelium and other non-parenchymal cells.
The decellularized liver matrix was infused with approximately 60 million cells and was seeded efficiently via portal vein this was earlier reported adequate to restore liver function in animal models.Citation28 The retained ECM constituents like collagen, fibronectin, and laminin, orchestrated successful migration, adhesion of re-populated rat hepatocytes. The preserved structural and biochemical ECM components aided in ameliorating the biological function of the hepatocytes leading to significant albumin and urea production in the culture medium. Large numbers of hepatocytes are able to proliferate, partially differentiate and are functional based on the urea and albumin secretion by the hepatoblasts and prostacyclin secretion. Thus, allowing liver reconstruction by authentic cell-cell and cell-matrix interactions, which are essential for cell differentiationand maintenance of specialized function.Citation29 In another study by Butter et al.Citation30 it was suggested, to utilize the repopulated grafts soon after cell engraftment and graft re-organization to avoid loss of functional hepatocytes.
The gene expression of liver-specific cell types in the recellularized rat liver matrix also confirms the integration of the parenchymal cells. The 2-3-fold increase in the expression of albumin, CK-19 genes confirms the presence of required liver tissue microenvironment and vasculature for organ regeneration. The hepatocytes survival within the liver scaffold may be attributed to the vital cell-ECM interactions and structural stimuli directing their growth and differentiation. Balestrini et al.Citation31 also demonstrated that there is divergent matrix conservation with respect to species it is likely that species-dependent native cues inherent to liver ECM may have profound effects on the suitability of the rat hepatocytes. The major limitation of this indigenous perfusion device was the requirement of a large number of cells in contrast to the limited availability of primary hepatocytes. To confirm the development and maintenance of hepatic system and functions in the in vitro 3D scaffold systems, a long-term additional data and the direct comparison to the monolayer cultures would be required.
The repopulated cells are known to essentially transform native ECM to reinstate the functional properties of regenerated organs by initialing the integration of native scaffold protein components.Citation32 Production of bioengineered livers for transplantation in patients with chronic liver failure would have an enormous impact to overcome the problem of donor organ shortage. Advancement in the identification of readily expandable cell type for repopulation is required to develop an appropriate functional tissue with minimal risk on transplantation.Citation33 Recently iPSCs are fabricated to develop human liver tissue for analyzing biological pathways and molecular interaction in liver diseases.Citation34 In conclusion, successfully recellularized liver scaffolds can be further aimed in designing natural 3D tissue constructs toward the fabrication of functional tissue and organ. We demonstrated a well-recognized and convincible protocol utilized for organ recellularization with its current research trajectory. The study had few limitations, including the lack of blood vessels which may affect the in vivo recellularization process. Additional investigation is required to justify the immunogenic effects of decellularized xenogeneic scaffolds and to determine the capability of the recellularized scaffolds as models for drug metabolism and in vivo whole organ regeneration.
Declaration of Potential Conflict of Interest
The authors declare that there is no conflict of interest. All the relevant data in the manuscript is available.
Statement of Ethics
All animal procedures were conducted with prior approval from Jeeva Life Sciences Pvt. Ltd, Hyderabad institutional animal ethical committee (IAEC) ref no. CPCSEA/IAEC/JLS/006/01/17/004 and all the protocols were followed as per the approved guidelines.
Acknowledgments
We acknowledge the Department of Science and Technology (DST), SERB (Science and Engineering Research Board), Govt. of India for providing research grant under National Post-doctoral Fellow Scheme (N-PDF), Grant no. PDF/2016/003465. We also thank Dr. K. Ravindranath, Dr. Suman Kapur, Pavan Mujawdiya, Dr. Meenakshi P, and Bala Kishan for their kind support.
Additional information
Funding
References
- Gilbert TW. Strategies for tissue and organ decellularization. J Cell Biochem. 2012;113(7):2217–22. doi:10.1002/jcb.v113.7.
- Debnath T, Shalini U, Kona LK, Vidya Sagar JVS, Kamaraju SR, Gaddam S, Chelluri LK. Development of 3D alginate encapsulation for better chondrogenic differentiation potential than the 2D pellet system. J Stem Cell Res Ther. 2015;5:276.
- Debnath T, Ghosh S, US P, Kona L, SR K, Sarkar S, Gaddam S, KC L. Proliferation and differentiation potential of human adipose-derived stem cells grown on chitosan hydrogel. PLoS One. 2015;10(3):e0120803. doi:10.1371/journal.pone.0120803.
- Hoshiba T, Lu H, Kawazoe N, Chen G. Decellularized matrices for tissue engineering. Expert Opin Biol Ther. 2010;10(12):1717–28. doi:10.1517/14712598.2010.534079.
- Ott HC, Matthiese TS, Goh SK, Black LD, Kren SM, Netoff TI, Taylor DA. Perfusion-decellularized matrix: using nature’s platform to engineer a bioartificial heart. Nat Med. 2008;14(2):213–21. doi:10.1038/nm1684.
- Petersen TH, Calle EA, Zhao L, Lee EJ, Gui L, Raredon MB, Gavrilov K, Yi T, Zhuang ZW, Breuer C, et al. Tissue-engineered lungs for in vivo implantation. Science. 2010;329(5991):538–41. doi:10.1126/science.1189345.
- Pei M, Li JT, Shoukry M, Zhang Y. A review of decellularized stem cell matrix: A novel cell expansion system for cartilage tissue engineering. Eur Cell Mater. 2011;22:333–43. doi:10.22203/eCM.v022a25.
- Patnaik SS, Wang B, Weed B, Wertheim JA, Liao J. Decellularized scaffolds: concepts, methodologies, and applications in cardiac tissue engineering and whole-organ regeneration. In Chapter 3. Tissue regeneration: where nanostructure meets biology. World Scientific Company; 2014;2.
- Fung YC. Biomechanics: mechanical Properties of Living Tissues. Second Edition. New York: Springer-Verlag; 1981.
- Chelluri LK. Stem cells and extracellular matrices. Morgan & Claypool Publishers: Science; 2012.
- Miyauchi Y, Yasuchika K, Fukumitsu K, Ishii T, Ogiso S, Minami T, Kojima H, Yamaoka R, Katayama H, Kawai T, et al. A novel three-dimensional culture system maintaining the physiological extracellular matrix of fibrotic model livers accelerates progression of hepatocellular carcinoma cells. Sci Rep. 2017;7(1):9827. doi:10.1038/s41598-017-09391-y.
- Lotze MT, Deisseroth A, Rubartelli A, Gilbert TW. Damage-associated molecular pattern molecules. Clin Immunol. 2007;124(1):1–4. doi:10.1016/j.clim.2007.02.006.
- Badylak SF, Taylor D, Uygun K. Whole-organ tissue engineering: decellularization and recellularization of three-dimensional matrix scaffolds. Annu Rev Biomed Eng. 2011;13(27–53):20. doi:10.1146/annurev-bioeng-071910-124743.
- Faulk DM, Wildemann JD, Badylak SF. Decellularization and cell seeding of whole liver biologic scaffolds composed of extracellular matrix. Journal of Clinical and Experimental Hepatology. 2014;5(1):69–80. doi:10.1016/j.jceh.2014.03.043.
- Severgnini M, Sherman J, Sehgal A, Jayaprakash NK, Aubin J, Wang G, Zhang L, Peng CG, Yucius K, Butler J, et al. A rapid two-step method for isolation of functional primary mouse hepatocytes: cell characterization and asialoglycoprotein receptor-based assay development. Cytotechnology. 2012;64(2):187–95. doi:10.1007/s10616-011-9407-0.
- Kmiec Z. Cooperation of liver cells in health and disease. Adv Anat Embryol Cell Biol. 2001;161:1–151.
- Hussein KH, Park KM, Kang KS, Woo HM. Biocompatibility evaluation of tissue engineered decellularized scaffolds for biomedical application. Materials Science and Engineering: C. 2016;67:766–78. doi:10.1016/j.msec.2016.05.068.
- Pan MX, Hu PY, Cheng Y, Cai LQ, Rao XH, Wang Y, Gao Y. An efficient method for decellularization of the rat liver. J Formosan Med Assoc. 2014;113(10):680–87. doi:10.1016/j.jfma.2013.05.003.
- Xu B, Håkansson J, Kuna VK, Elebring E, Holgersson S. Assessing rat liver-derived biomatrix for hepatic tissue engineering with human fetal liver stem cells. Adv Tissue Eng Regen Med Open Access. 2016;1:00012.
- Baldwin HS. Early embryonic vascular development. Cardiovasc Res. 1996;31:e34e45.
- Wang X, Lin P, Yao Q, Chen C. Development of small-diameter vascular grafts. World J Surg. 2007;31(4):682–89. doi:10.1007/s00268-006-0731-z.
- Yeung BKS, Chong PYC, Petillo PA. Glycochemistry. In: Wang PG, Bertozzi CR, editors. Principles, synthesis, and applications. New York: Marcel Dekker; 2001. p. 425.
- Bale S, Geerts S, Jindal R, Yarmush ML. Isolation and co-culture of rat parenchymal and non-parenchymal liver cells to evaluate cellular interactions and response. Sci Rep. 2016;6(1):25329. doi:10.1038/srep25329.
- Gao R, Wu W, Xiang J, Lv Y, Zheng X, Chen Q, Wang H, Wang B, Liu Z, Ma F. Hepatocyte culture in autologous decellularized spleen matrix. Organogenesis. 2015;11(1):16–29. doi:10.1080/15476278.2015.1011908.
- Griffith CK, Miller C, Sainson RC, Calvert JW, Jeon NL, Hughes CC, George S. Diffusion limits of an in vitro thick pre vascularized tissue. Tissue Eng. 2005;11(1–2):257–66. doi:10.1089/ten.2005.11.257.
- Navarro-Tableros V, Herrera Sanchez MB, Figliolini F, Romagnoli R, Tetta C, Camussi G. Recellularization of rat liver scaffolds by human liver stem cells. Tissue Eng part A. 2015;21(11–12):1929–39.
- Basak EU, Alejandro SG, Hiroshi Y, Maria-Louisa I, Maria AG, Carley S, Jack MM, Naoya K, Arno WT, François B, et al. Organ reengineering through development of a transplantable recellularized liver graft using decellularized liver matrix. Nat Med. 2010;16(7):814–20. doi:10.1038/nm.2170.
- Fox IJ, Chowdhury JR, Kaufman SS, Goertzen TC, Chowdhury NR, Warkentin PI, Dorko K, Sauter BV, Strom SC. Treatment of the Crigler-Najjar syndrome type I with hepatocyte transplantation. N Engl J Med. 1998;338(20):1422–26. doi:10.1056/NEJM199805143382004.
- Baptista PM, Siddiqui MM, Lozier G, Rodriguez SR, Atala A, Soker S. The use of whole organ decellularization for the generation of a vascularized liver organoid. Hepatology. 2011;53(2):604–17. doi:10.1002/hep.24067.
- Butter A, Aliyev K, Hillebrandt KH, Raschzok N, Kluge M, Seiffert N, Tang P, Napierala H, Muhamma AI, Reutzel-Selke A, et al. Evolution of graft morphology and function after recellularization of decellularized rat livers. J Tissue Eng Regen Med. 2018;12(2):e807–e816. doi:10.1002/term.2383.
- Balestrini JL, Gard AL, Gerhold KA, Wilcox EC, Liu A, Schwan J, Le AV, Baevova P, Dimitrievska S, Zhao L, et al. Comparative biology of decellularized lung matrix: implications of species mismatch in regenerative medicine. Biomaterials. 2016;102:220–30. doi:10.1016/j.biomaterials.2016.06.025.
- He Y, Lu F. Development of synthetic and natural materials for tissue engineering applications using adipose stem cells. Stem Cells Int. 2016;2016:5786257. doi:10.1155/2016/5786257.
- Yesmin S, Paget MB, Murray HE, Downing R. Bio-scaffolds in organ-regeneration: clinical potential and current challenges. Curr Res Transl Med. 2017;65(3):103–13. doi:10.1016/j.retram.2017.08.002.
- Collin de l’Hortet A, Takeishi K, Guzman-Lepe J, Morita K, Achreja, Popovic B, Wang Y, Handa K, Mittal A, Meurs N, et al. Generation of human fatty livers using custom-engineered induced pluripotent stem cells with modifiable SIRT1 metabolism. Cell Metab. 2019;30(2):385–401. doi:10.1016/j.cmet.2019.06.017.
